Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast
Abstract
1. Introduction
2. Material and Methods
2.1. Strains and Culture Conditions
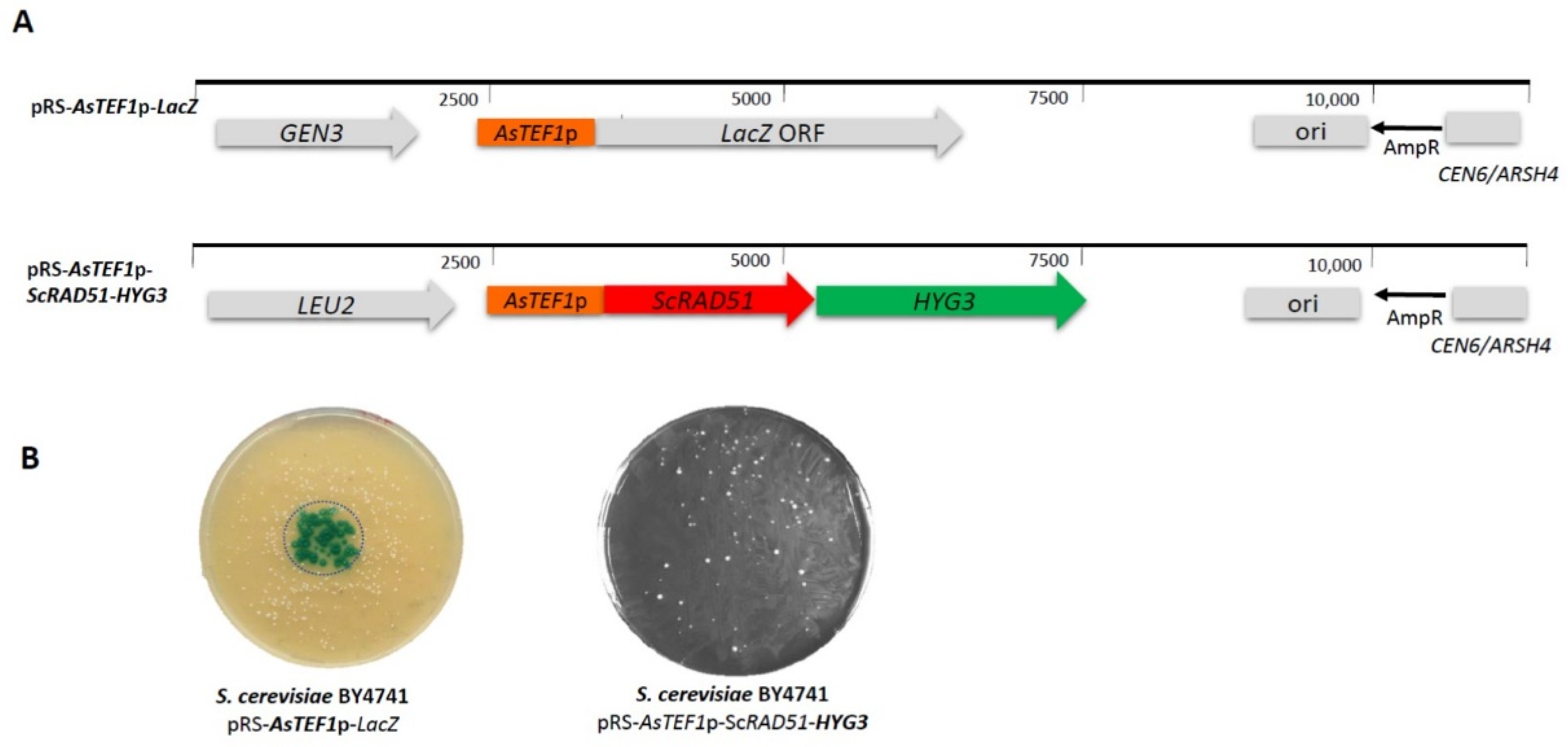
2.2. Plasmid Design and Constructions
2.3. Yeast Transformation
2.4. Verification of Lager Yeast Transformants
3. Results
3.1. Design and Testing of Synthetic Marker and Reporter Genes
3.2. Transformation of Lager Yeast
3.3. PCR-Based Gene Targeting is Enhanced by RAD51 Overexpression
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Hansen, E.C. Grundlagen zur Systematik der Saccharomyceten. Zentralbl. Bakteriol. II Natur. 1904, 12, 529–538. [Google Scholar]
- Walther, A.; Hesselbart, A.; Wendland, J. Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 Genes Genomes Genet. 2014, 4, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Wendland, J. Lager yeast comes of age. Eukaryot. Cell 2014, 13, 1256–1265. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.; Wang, B.; Bellora, N.; Peris, D.; Hulfachor, A.B.; Koshalek, J.A.; Adams, M.; Libkind, D.; Hittinger, C.T. The Genome Sequence of Saccharomyces eubayanus and the Domestication of Lager-Brewing Yeasts. Mol. Biol. Evol. 2015, 32, 2818–2831. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.; Liti, G. Saccharomyces pastorianus: Genomic insights inspiring innovation for industry. Yeast 2015, 32, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Okuno, M.; Kajitani, R.; Ryusui, R.; Morimoto, H.; Kodama, Y.; Itoh, T. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 2016, 23, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Monerawela, C.; Bond, U. Brewing up a storm: The genomes of lager yeasts and how they evolved. Biotechnol. Adv. 2017, 35, 512–519. [Google Scholar] [CrossRef]
- Monerawela, C.; Bond, U. The hybrid genomes of Saccharomyces pastorianus: A current perspective. Yeast 2018, 35, 39–50. [Google Scholar] [CrossRef]
- Libkind, D.; Hittinger, C.T.; Valerio, E.; Goncalves, C.; Dover, J.; Johnston, M.; Goncalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef]
- Bing, J.; Han, P.J.; Liu, W.Q.; Wang, Q.M.; Bai, F.Y. Evidence for a Far East Asian origin of lager beer yeast. Curr. Biol. 2014, 24, R380–R381. [Google Scholar] [CrossRef]
- Baker, E.P.; Peris, D.; Moriarty, R.V.; Li, X.C.; Fay, J.C.; Hittinger, C.T. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 2019, 5, eaav1869. [Google Scholar] [CrossRef] [PubMed]
- Gorter de Vries, A.R.; de Groot, P.A.; van den Broek, M.; Daran, J.G. CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb. Cell Fact. 2017, 16, 222. [Google Scholar] [CrossRef] [PubMed]
- Duong, C.T.; Strack, L.; Futschik, M.; Katou, Y.; Nakao, Y.; Fujimura, T.; Shirahige, K.; Kodama, Y.; Nevoigt, E. Identification of Sc-type ILV6 as a target to reduce diacetyl formation in lager brewers’ yeast. Metab. Eng. 2011, 13, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Murakami, N.; Miyoshi, S.; Yokoyama, R.; Hoshida, H.; Akada, R.; Ogata, T. Construction of a URA3 deletion strain from the allotetraploid bottom-fermenting yeast Saccharomyces pastorianus. Yeast 2012, 29, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Bolat, I.; Romagnoli, G.; Zhu, F.; Pronk, J.T.; Daran, J.M. Functional analysis and transcriptional regulation of two orthologs of ARO10, encoding broad-substrate-specificity 2-oxo-acid decarboxylases, in the brewing yeast Saccharomyces pastorianus CBS1483. FEMS Yeast Res 2013, 13, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Wendland, J. PCR-based methods facilitate targeted gene manipulations and cloning procedures. Curr. Genet. 2003, 44, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wach, A.; Brachat, A.; Pohlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Alberti-Segui, C.; Rebischung, C.; Philippsen, P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 1997, 13, 1065–1075. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353, aaf1420. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, E.; VanderSluis, B.; Wang, W.; Tan, G.; Deshpande, R.; Chen, Y.; Usaj, M.; Balint, A.; Mattiazzi Usaj, M.; van Leeuwen, J.; et al. Systematic analysis of complex genetic interactions. Science 2018, 360, eaao1729. [Google Scholar] [CrossRef] [PubMed]
- Bahler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; McKenzie, A., 3rd; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Gerami-Nejad, M.; Berman, J.; Gale, C.A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 2001, 18, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Gerami-Nejad, M.; Hausauer, D.; McClellan, M.; Berman, J.; Gale, C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 2004, 21, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Gerami-Nejad, M.; Dulmage, K.; Berman, J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast 2009, 26, 399–406. [Google Scholar] [CrossRef]
- Gola, S.; Martin, R.; Walther, A.; Dunkler, A.; Wendland, J. New modules for PCR-based gene targeting in Candida albicans: Rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 2003, 20, 1339–1347. [Google Scholar] [CrossRef]
- Wendland, J.; Ayad-Durieux, Y.; Knechtle, P.; Rebischung, C.; Philippsen, P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 2000, 242, 381–391. [Google Scholar] [CrossRef]
- Amelina, H.; Moiseeva, V.; Collopy, L.C.; Pearson, S.R.; Armstrong, C.A.; Tomita, K. Sequential and counter-selectable cassettes for fission yeast. BMC Biotechnol. 2016, 16, 76. [Google Scholar] [CrossRef]
- Kooistra, R.; Hooykaas, P.J.; Steensma, H.Y. Efficient gene targeting in Kluyveromyces lactis. Yeast 2004, 21, 781–792. [Google Scholar] [CrossRef]
- Wesolowski-Louvel, M. An efficient method to optimize Kluyveromyces lactis gene targeting. FEMS Yeast Res. 2011, 11, 509–513. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kretzschmar, A.; Otto, C.; Holz, M.; Werner, S.; Hubner, L.; Barth, G. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 2013, 59, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, Y.; Suzuki, K.; Ishii, C.; Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2004, 101, 12248–12253. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, Y.S.; Cairns, T.C.; Chaudhari, Y.K.; Usher, J.; Talbot, N.J.; Studholme, D.J.; Csukai, M.; Haynes, K. Exploitation of sulfonylurea resistance marker and non-homologous end joining mutants for functional analysis in Zymoseptoria tritici. Fungal Genet. Biol. 2015, 79, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Arras, S.D.; Fraser, J.A. Chemical Inhibitors of Non-Homologous End Joining Increase Targeted Construct Integration in Cryptococcus neoformans. PLoS ONE 2016, 11, e0163049. [Google Scholar] [CrossRef] [PubMed]
- Yanez, R.J.; Porter, A.C. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999, 6, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, O.G.; Lanzov, V.A.; Ogawa, H.; Filatov, M.V. Overexpression of bacterial RecA protein stimulates homologous recombination in somatic mammalian cells. Mutat. Res. 2000, 459, 65–71. [Google Scholar] [CrossRef]
- Liu, L.; Maguire, K.K.; Kmiec, E.B. Genetic re-engineering of Saccharomyces cerevisiae RAD51 leads to a significant increase in the frequency of gene repair in vivo. Nucleic Acids Res. 2004, 32, 2093–2101. [Google Scholar] [CrossRef][Green Version]
- Thompson, J.R.; Register, E.; Curotto, J.; Kurtz, M.; Kelly, R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 1998, 14, 565–571. [Google Scholar] [CrossRef]
- Suga, M.; Hatakeyama, T. High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast 2001, 18, 1015–1021. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [PubMed]
- Dunkler, A.; Wendland, J. Use of MET3 promoters for regulated gene expression in Ashbya gossypii. Curr. Genet. 2007, 52, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Junker, K.; Chailyan, A.; Hesselbart, A.; Forster, J.; Wendland, J. Multi-omics characterization of the necrotrophic mycoparasite Saccharomycopsis schoenii. PLoS Pathog. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. PCR-based gene targeting in Candida albicans. Nat. Protoc. 2008, 3, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Carroll, E.; Trinh, T.N.; Son, H.; Lee, Y.W.; Seo, J.A. Comprehensive analysis of fungal diversity and enzyme activity in nuruk, a Korean fermenting starter, for acquiring useful fungi. J. Microbiol. 2017, 55, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Farh, M.E.; Cho, Y.; Lim, J.Y.; Seo, J.A. A diversity study of Saccharomycopsis fibuligera in rice wine starter nuruk, reveals the evolutionary process associated with its interspecies hybrid. J. Microbiol. 2017, 55, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Uhl, M.A.; Johnson, A.D. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 2001, 147, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Greig, D. Reproductive isolation in Saccharomyces. Heredity 2009, 102, 39–44. [Google Scholar] [CrossRef]
- Sipiczki, M. Yeast two- and three-species hybrids and high-sugar fermentation. Microb. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Leducq, J.B.; Charron, G.; Diss, G.; Gagnon-Arsenault, I.; Dube, A.K.; Landry, C.R. Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genet. 2012, 8, e1003161. [Google Scholar] [CrossRef]
- Piatkowska, E.M.; Naseeb, S.; Knight, D.; Delneri, D. Chimeric protein complexes in hybrid species generate novel phenotypes. PLoS Genet. 2013, 9, e1003836. [Google Scholar] [CrossRef]
- Pfliegler, W.P.; Antunovics, Z.; Sipiczki, M. Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res 2012, 12, 703–718. [Google Scholar] [CrossRef]
- Barnes, G.; Rio, D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 867–872. [Google Scholar] [CrossRef]
- Polotnianka, R.M.; Li, J.; Lustig, A.J. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 1998, 8, 831–834. [Google Scholar] [CrossRef]
- Mertens, S.; Gallone, B.; Steensels, J.; Herrera-Malaver, B.; Cortebeek, J.; Nolmans, R.; Saels, V.; Vyas, V.K.; Verstrepen, K.J. Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts. PLoS ONE 2019, 14, e0209124. [Google Scholar] [CrossRef]



| Strain Number | Feature/Genotype | Source |
|---|---|---|
| B003 | Saccharomyces cerevisiae BY4741 MATa his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| B237 | Weihenstephan WS34/70 lager yeast | Lab collection |
| B256 | WS34/70; pRAD51 (HYG3, CEN6/ARSH4) | This study |
| B257 | WS34/70; pHYG3 (CEN6/ARSH4) | This study |
| G001 | WS34/70; pRAD51; ade2::YES1 | This study |
| G002 | WS34/70; pRAD51; hsp104::YES1 | This study |
| Strain Number | Feature/Genotype | Source |
|---|---|---|
| E008 | pRS415 | [41] |
| E025 | pRS417-AgTEF1p-LacZ-GEN3 | [42] |
| E026 | pGEM-AsTEF1p | This study |
| E054 | pUC57-HYG3 | GenScript |
| E065 | pRS417-AsTEF1p-LacZ | This study |
| E066 | pYES1 | This study |
| E068 | pYES2 | This study |
| E088 | pGEM-YES1 | This study |
| E120 | pGEM-ScRAD51 | This study |
| E150 | pRS-AsTEF1p-ScRAD51-HYG3-GEN3 | This study |
| E160 | pRS-AsoTEF1p-ScRAD51-HYG3-LEU2 | This study |
| Primer Number | Primer Name | Sequence 5′→3′ * |
|---|---|---|
| 106 | 5′-AsTEF1p | GTCCAGAATAACATCAAATC |
| 107 | 3′-AsTEF1p | CTATAAAAAATGTTAGTATGGAG |
| 108 | 5′-AsTEFp-pRS | CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGCTCGAGTCCAGAATAACATCAAATC |
| 109 | 3′-AsTEFp-lacZ | CAATCTTTGGATCGTTTAAATAAGTTTGAATTTTTTCAGTCATGTTCTATAAAAAATGTTAGTATGGAG |
| 226 | P3-pRS415-AsTEF1p | TGTAAAACGACGGCCAGTGAGCGCGCGTAATACGACTCACTATAGGAAGCTTCGTACGCTGCAGGTCGGATCCCCCGGGGGCGCGCCGTCCAGAATAACATCAAATC |
| 227 | P4-AsTEF1p-kanR | GTTGGAGTTCAAACGTGGTCTGGAAACGTGAGTCTTTTCCTTACCCTATAAAAAATGTTAGTATGGAG |
| 228 | P5-kan-ORF | GGTAAGGAAAAGACTCACGTTTCCA |
| 229 | P6-kanR-pRS415 | GGAAACAGCTATGACCATGATTACGCCAAGCGCGCAATTAACCCTTCTGATATCATCGATGAATTCGAGCTCGTTTAAACATTGGTAATAG |
| 230 | P7-5′-HYG3 | CTGACTTTTGTCTTGTTATGGACTCCATACTAACATTTTTTATAGAAAAAACCAGAATTGACTGCTACTTC |
| 231 | P8-3′-HYG3 | CTGATATCATCGATGAATTCGAGCTCGTTTAAACATTGGTAATAGGACCACCTTTGATTGTAAATAG |
| 253 | 5-HYG3+AD | GGCGCGCCAGATCTAGCCTCCTCAGAGAAAATTGCACAAAAAAAAGGAAGCTTCGTACGCTGCAGGTC |
| 254 | 3-HYG3+AD | TTACGCCAAGCGCGCAATTAACCCTCACTAAAGGGAACAAAAGCTGACCACCTTTGATTGTAAATAG |
| 258 | 5-ScRAD51+AD | CTGACTTTTGTCTTGTTATGGACTCCATACTAACATTTTTTATAGTCTCAAGTTCAAGAACAACATATATCAG |
| 259 | 3-ScRAD51+AD | CTTTTTTTTGTGCAATTTTCTCTGAGGAGGCTAGATCTGGCGCGCCGAAAAATACATATATTTCATGGGTGACAG |
| 260 | 5-ScRAD51 | TCTCAAGTTCAAGAACAACATATATCAG |
| 261 | 3-ScRAD51 | GAAAAATACATATATTTCATGGGTGACAG |
| 344 | S1-LEU2 | GGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTAAAGTGCAATTCTTTTTCC |
| 345 | S2-LEU2 | CTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCGGTCGAGGAGAACTTC |
| 355 | G2-YES1 | GAATGAATCTACTGGTTTGG |
| 356 | G3-YES1 | GTGTCGGTATCGCAGAC |
| 369 | S1-ADE2 | CCTACTATAACATTCAAGAAAAACAAGAAAACCGGACAAAACAATCAAGTGTCCAGAATAACATCAAATC |
| 371 | S2-ADE2 | TATATCATTTTATATTATTTGCTGTGCAAGTATATCAATAAACTTATATATAATAAATTATTTTTATTGTTG |
| 373 | G1-ADE2 | GACTCTTGTTGCAGGGCT |
| 389 | G4-ADE2 | GTGATGCATTGAGCCGCC |
| 390 | S1-HSP104 | TATATTACTGATTCTTGTTCGAAAGTTTTTAAAAATCACACTATATTAAAGTCCAGAATAACATCAAATC |
| 391 | S2-HSP104 | AACAAAGAAAAAAGAAATCAACTACACGTACCATAAAATATACAGAATATTAATAAATTATTTTTATTGTTG |
| 392 | G1-HSP104 | CCCGTATTCTAATAATGGACC |
| 393 | G4-HSP104 | CAAACTTATGCAACCTGCCAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardi, B.; Kayacan, Y.; Akan, M.; Wendland, J. Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast. Microorganisms 2019, 7, 192. https://doi.org/10.3390/microorganisms7070192
Bernardi B, Kayacan Y, Akan M, Wendland J. Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast. Microorganisms. 2019; 7(7):192. https://doi.org/10.3390/microorganisms7070192
Chicago/Turabian StyleBernardi, Beatrice, Yeseren Kayacan, Madina Akan, and Jürgen Wendland. 2019. "Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast" Microorganisms 7, no. 7: 192. https://doi.org/10.3390/microorganisms7070192
APA StyleBernardi, B., Kayacan, Y., Akan, M., & Wendland, J. (2019). Overexpression of RAD51 Enables PCR-Based Gene Targeting in Lager Yeast. Microorganisms, 7(7), 192. https://doi.org/10.3390/microorganisms7070192





