DNA Damage Response Pathways in Dinoflagellates
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
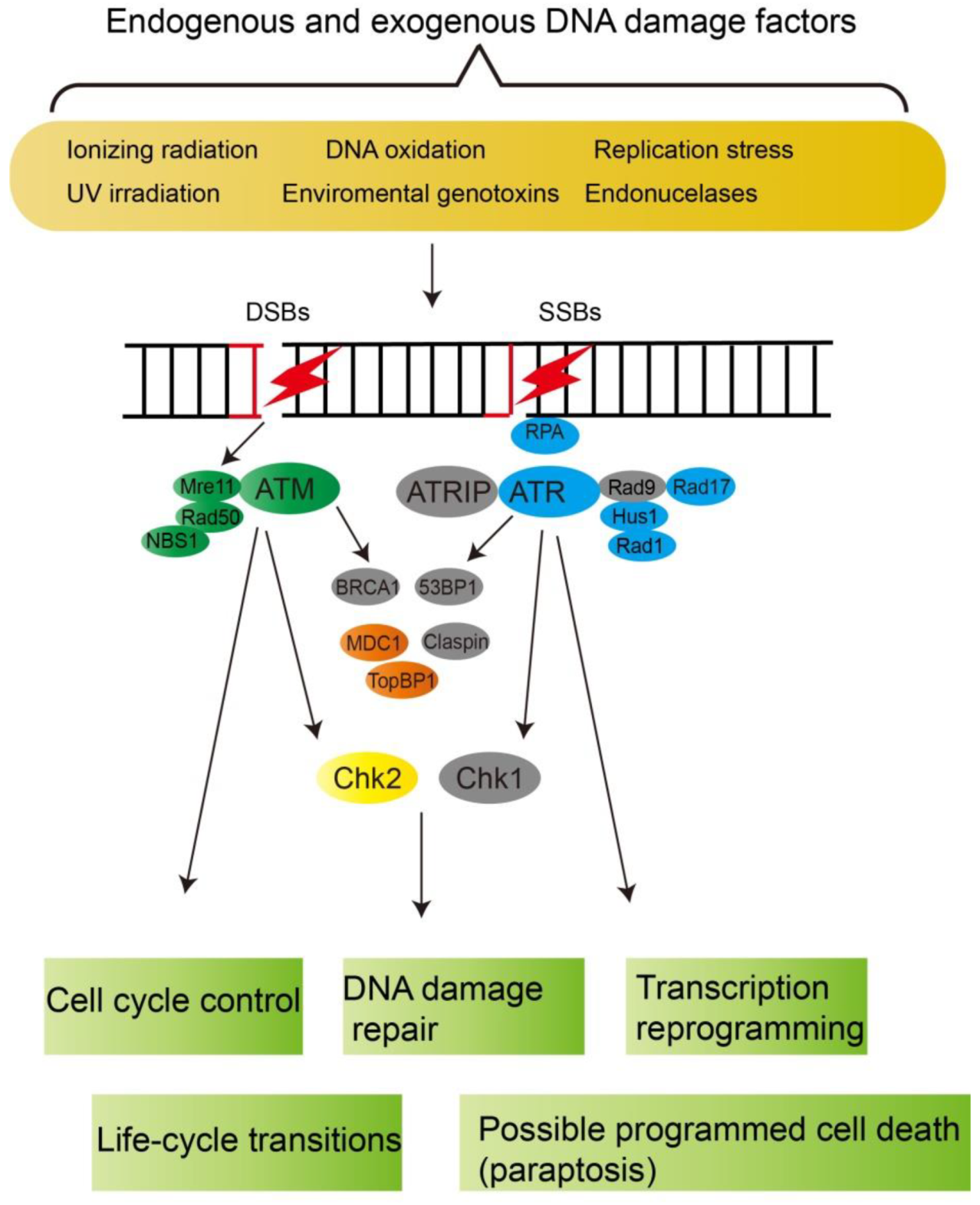
3.1. DNA Damage Checkpoint Signaling Networks
3.2. DNA Repair Pathways
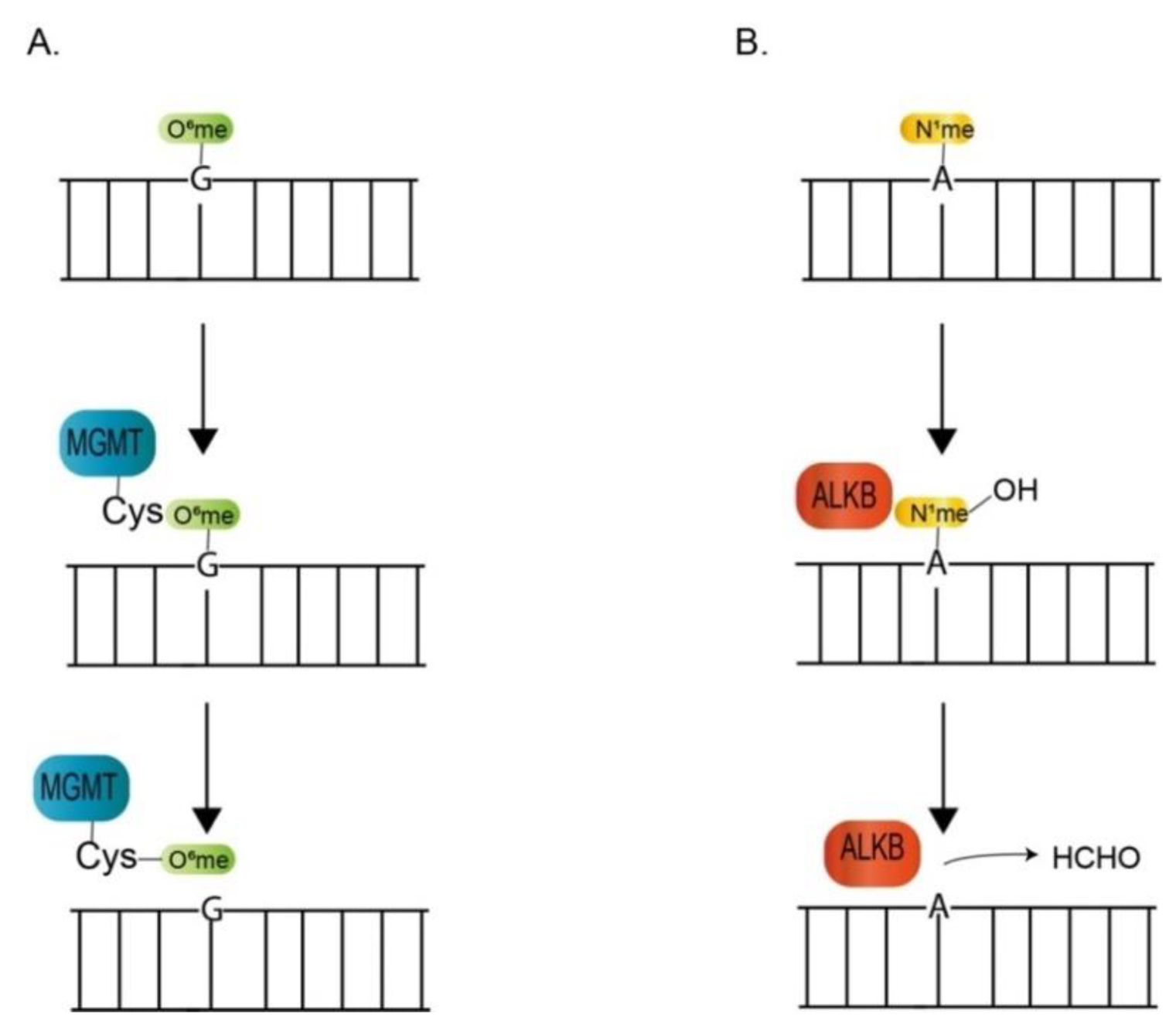
3.2.1. Direct Reversal of DNA Lesion
3.2.2. Photoreactivation
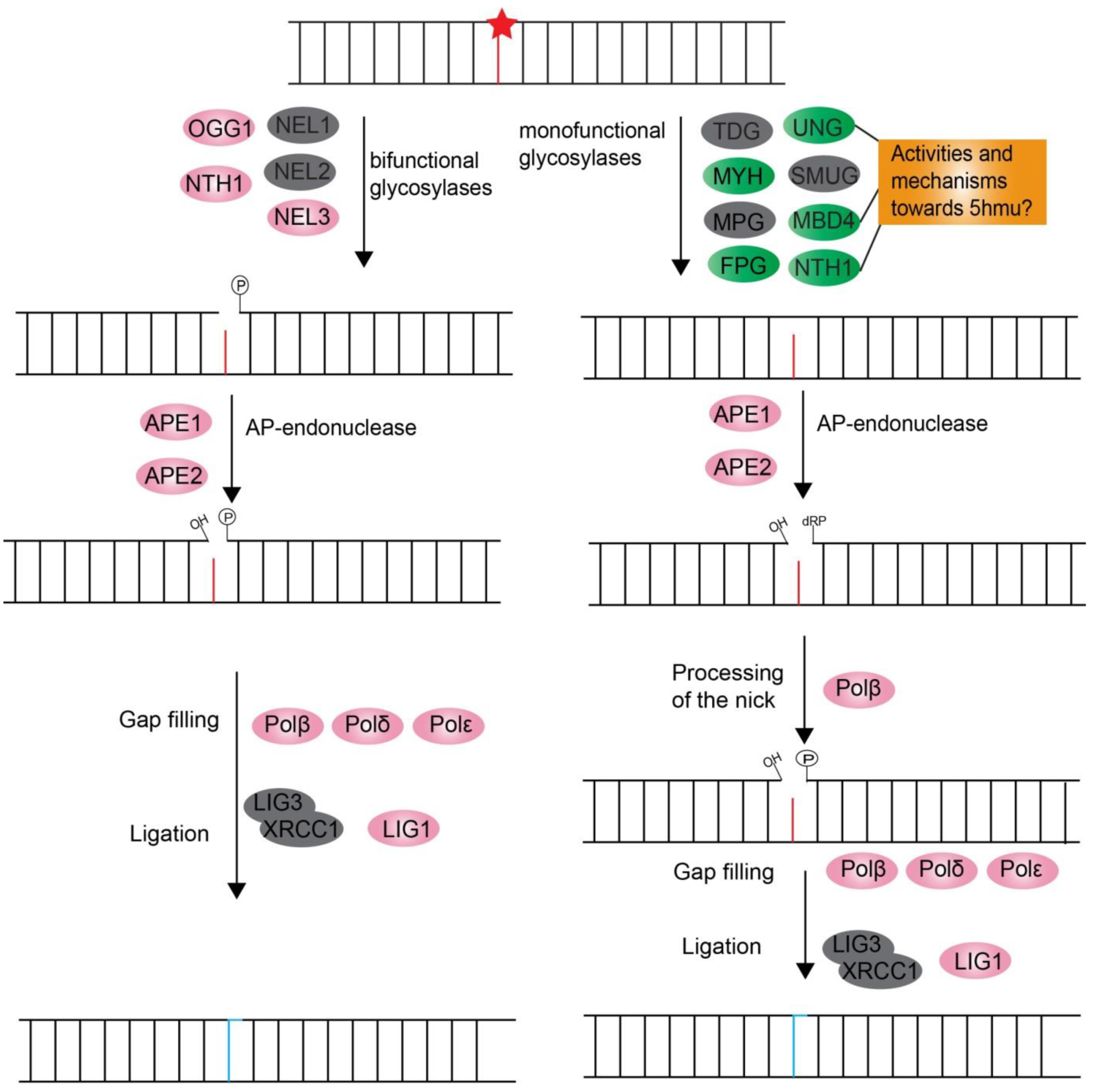
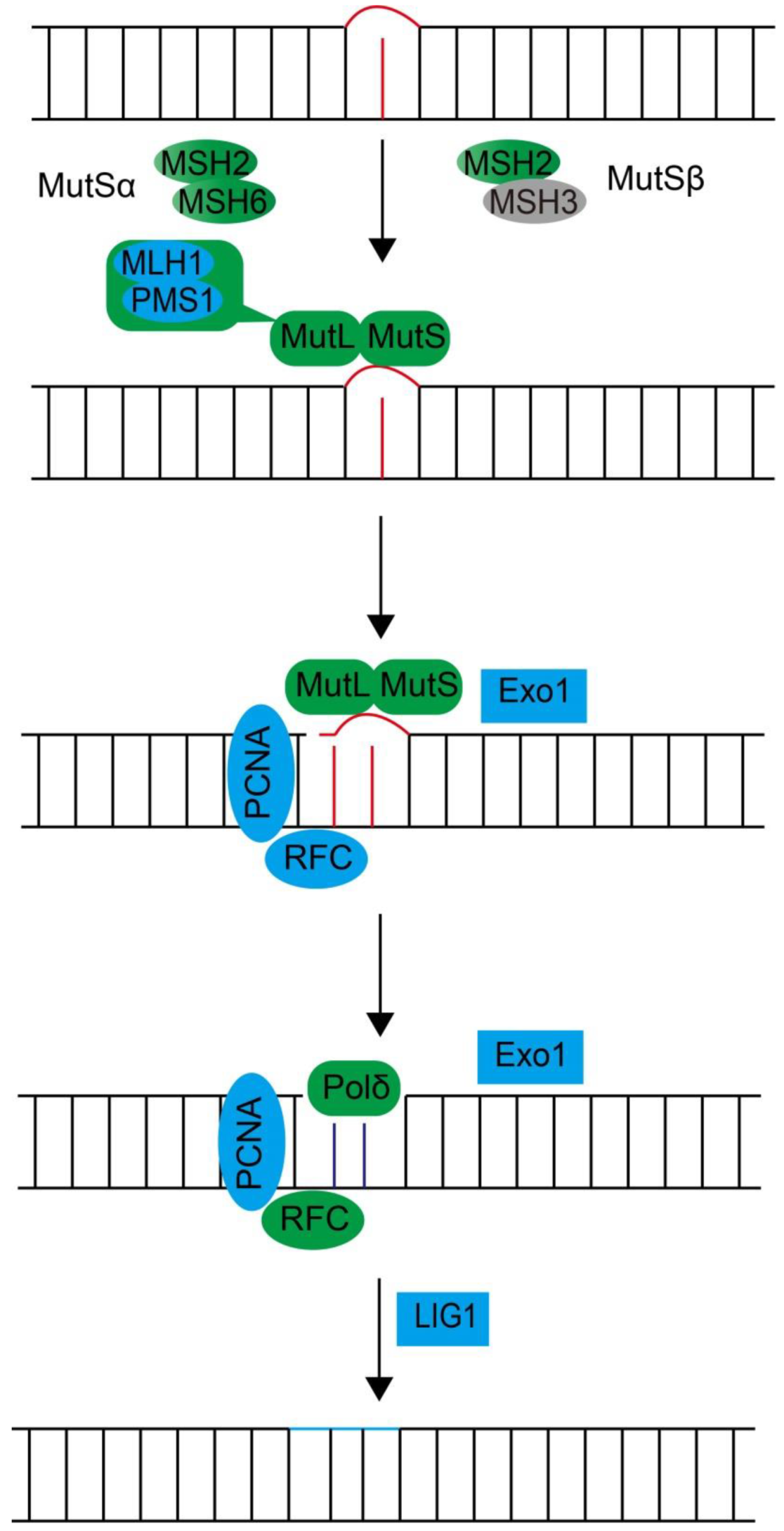
3.2.3. Three Excision Repair Pathways, Nucleotide Excision Repair (NER), Base Excision Repair (BER), and DNA Mismatch Repair (MMR), Confer Single-Strand DNA Damage Repair Through Excision-Coupled Resynthesis
Base Excision Repair (BER)
Nucleotide Excision Repair
Mismatch Repair
3.2.4. DNA Double-Strand Breaks Repair
Non-Homologous End Joining
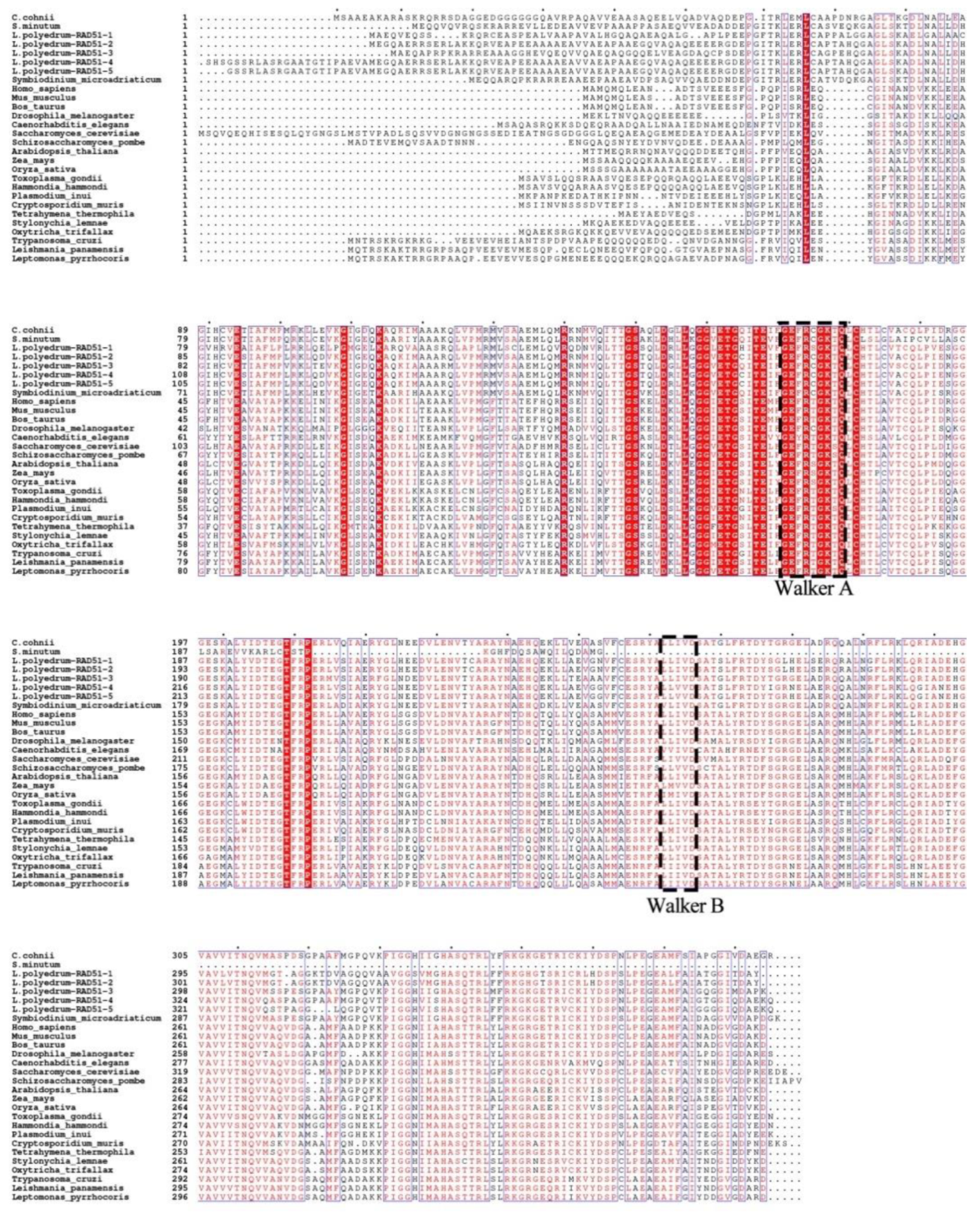
Homologous Recombination
3.2.5. Translesion Synthesis
3.2.6. DNA Interstrand Crosslinks Repair
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Matthiessen, J.; Schreck, M. Dinoflagellates. In Encyclopedia of Marine Geosciences; Harff, J., Meschede, M., Petersen, S., Thiede, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 1–7. [Google Scholar]
- Saldarriaga, J.F.; Taylor, F.J.R. Dinoflagellata. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J., Corliss, J.O., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–54. [Google Scholar]
- Anderson, D.M.; Cembella, A.D.; Hallegraeff, G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 2011, 4, 143–176. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Anderson, D.M.; et al. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef] [PubMed]
- Franklin, D.J.; Berges, J.A. Mortality in cultures of the dinoflagellate Amphidinium carterae during culture senescence and darkness. Proc. R. Soc. Lond. B Biol. Sci. 2004, 271, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.Y. Architectural organization of dinoflagellate liquid crystalline chromosomes. Microorganisms 2019, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Yeung, P.K.; Wong, J.T. Inhibition of cell proliferation by mechanical agitation involves transient cell cycle arrest at G1 phase in dinoflagellates. Protoplasma 2003, 220, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Lesser, M.P. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 1997, 16, 187–192. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral reefs under rapid climate change and ocean acidification. Science 2007, 318, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Ekelund, N.G.A. Effects of UV-B radiation on growth and motility of four phytoplankton species. Physiol. Plant. 1990, 78, 590–594. [Google Scholar] [CrossRef]
- Ekelund, N.G.A. The effects of UV-B radiation on dinoflagellates. J. Plant Physiol. 1991, 138, 274–278. [Google Scholar] [CrossRef]
- Banaszak, A.T.; Trench, R.K. Effects of ultraviolet (UV) radiation on marine microalgal-invertebrate symbioses. I. Response of the algal symbionts in culture an in hospite. J. Exp. Mar. Biol. Ecol. 1995, 194, 213–232. [Google Scholar] [CrossRef]
- Dodge, J. Effects of ultra-violet light on the survival and nuclear division of a Dinoflagellate. Protoplasma 1965, 59, 485–493. [Google Scholar] [CrossRef]
- Lessard, E.J. The trophic role of heterotrophic dinoflagellates in diverse marine environments. Mar. Microb. Food Webs. 1991, 5, 49–58. [Google Scholar]
- Schnepf, E.; Elbrächter, M. Nutritional strategies in dinoflagellates: A review with emphasis on cell biological aspects. Eur. J. Protistol. 1992, 28, 3–24. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. J. Phycol. 2010, 46, 220–235. [Google Scholar] [CrossRef]
- Gao, K.; Zhang, Y.; Häder, D.-P. Individual and interactive effects of ocean acidification, global warming, and UV radiation on phytoplankton. J. Appl. Phycol. 2018, 30, 743–759. [Google Scholar] [CrossRef]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Takeuchi, T.; Hisata, K.; Tanaka, M.; Fujiwara, M.; et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef]
- Chan, W.S.; Kwok, A.C.M.; Wong, J.T.Y. Knockdown of dinoflagellate cellulose synthase CesA1 resulted in malformed intracellular cellulosic thecal plates and severely impeded cyst-to-swarmer transition. Front. Microbiol. 2019, 10, 546. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling with a user-friendly interface. BioRXiv. 2018, 289660. [Google Scholar]
- Tamura, K.; Peterson, D.; Stecher, G.; Peterson, N.; Kumar, S.; Nei, M. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell. 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. CSH Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- So, S.; Davis, A.J.; Chen, D.J. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 2009, 187, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; Van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Savic, V.; Yin, B.; Maas, N.L.; Bredemeyer, A.L.; Carpenter, A.C.; Helmink, B.A.; Yang-Iott, K.S.; Sleckman, B.P.; Bassing, C.H. Formation of dynamic γ-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell 2009, 34, 298–310. [Google Scholar] [CrossRef]
- Kumagai, A.; Lee, J.; Yoo, H.Y.; Dunphy, W.G. TopBP1 activates the ATR-ATRIP complex. Cell 2006, 124, 943–955. [Google Scholar] [CrossRef]
- Lee, J.; Kumagai, A.; Dunphy, W.G. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007, 282, 28036–28044. [Google Scholar] [CrossRef]
- Delacroix, S.; Wagner, J.M.; Kobayashi, M.; Yamamoto, K.-i.; Karnitz, L.M. The Rad9–Hus1–Rad1 (9–1–1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007, 21, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Alveal, C.; Calonge, T.M.; O’Connell, M.J. Regulation of Chk1. Cell. Div. 2009, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, R.D.; Cimprich, K.A. The ATR pathway: Fine-tuning the fork. DNA Repair 2007, 6, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Besteiro, M.A.G.; Gottifredi, V. The fork and the kinase: A DNA replication tale from a CHK1 perspective. Mutat. Res. 2015, 763, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Zannini, L.; Delia, D.; Buscemi, G. CHK2 kinase in the DNA damage response and beyond. J. Mol. Cell Biol. 2014, 6, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.B.; Cools, T.; de Veylder, L. Mechanisms used by plants to cope with DNA damage. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Reviews: Palo Alto, CA, USA, 2016; Volume 67, pp. 439–462. [Google Scholar]
- Stracker, T.H.; Usui, T.; Petrini, J.H.J. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair 2009, 8, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.; Daoust, P.; Benribague, S. A transcriptome-based perspective of cell cycle regulation in dinoflagellates. Protist 2016, 167, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Wang, B.; Bignell, C.R.; Taylor, A.M.R.; Elledge, S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003, 421, 961–966. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Ho, B.N.; Singh, A.T.K. The evolution, functions and applications of the breast cancer genes BRCA1 and BRCA2. Cancer Genom. Proteom. 2017, 14, 293–298. [Google Scholar]
- Bachvaroff, T.R.; Gornik, S.G.; Concepcion, G.T.; Waller, R.F.; Mendez, G.S.; Lippmeier, J.C.; Delwiche, C.F. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogenet. Evol. 2014, 70, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.D.; Pittman, D.L. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006, 19, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 2012, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Duguid, E.M.; He, C. Direct reversal of DNA alkylation damage. Chem. Rev. 2006, 106, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.C.; Lindahl, T. Repair of alkylated DNA: Recent advances. DNA Repair 2007, 6, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.Ø.; Rognes, T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003, 154, 531–538. [Google Scholar] [CrossRef]
- Yang, C.-G.; Garcia, K.; He, C. Damage detection and base flipping in direct DNA alkylation repair. ChemBioChem 2009, 10, 417–423. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Rae, P. Hydroxymethyluracil in eukaryote DNA: A natural feature of the pyrrophyta (dinoflagellates). Science 1976, 194, 1062–1064. [Google Scholar] [CrossRef]
- Rae, P.M.M.; Steele, R.E. Modified bases in the DNAs of unicellular eukaryotes: An examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems 1978, 10, 37–53. [Google Scholar] [CrossRef]
- Heyn, H.; Esteller, M. An adenine code for DNA: A second life for N6-methyladenine. Cell 2015, 161, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Doré, L.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA methylation on N6-adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416. [Google Scholar] [CrossRef]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N6-methyladenine DNA modification in the human genome. Mol. Cell 2018, 71, 306–318. [Google Scholar] [CrossRef]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef]
- Lynch, M.; Lucas-Lledó, J.I. Evolution of mutation rates: Phylogenomic analysis of the photolyase/cryptochrome family. Mol. Biol. Evol. 2009, 26, 1143–1153. [Google Scholar]
- Dany, A.-L.; Douki, T.; Triantaphylides, C.; Cadet, J. Repair of the main UV-induced thymine dimeric lesions within Arabidopsis thaliana DNA: Evidence for the major involvement of photoreactivation pathways. J. Photochem. Photobiol. B 2001, 65, 127–135. [Google Scholar] [CrossRef]
- Britt, A.B. Repair of DNA damage induced by solar UV. Photosynth. Res. 2004, 81, 105–112. [Google Scholar] [CrossRef]
- Sancar, A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003, 103, 2203–2238. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, A.E.; Annunziata, R.; Jaubert, M.; Bouly, J.-P.; Falciatore, A. Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 2015, 172, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Brettel, K.; Byrdin, M. Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 2010, 20, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Zhong, D. Dynamics and mechanisms of DNA repair by photolyase. Phys. Chem. Chem. Phys. 2015, 17, 11933–11949. [Google Scholar] [CrossRef] [PubMed]
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700. [Google Scholar] [CrossRef]
- Wang, J.; Du, X.; Pan, W.; Wang, X.; Wu, W. Photoactivation of the cryptochrome/photolyase superfamily. J. Photochem. Photobiol. C 2015, 22, 84–102. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; Zhong, D. Photolyase: Dynamics and mechanisms of repair of sun-induced DNA damage. Photochem. Photobiol. 2017, 93, 78–92. [Google Scholar] [CrossRef]
- Sung, J.-S.; Demple, B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006, 273, 1620–1629. [Google Scholar] [CrossRef]
- Whitaker, A.M.; Schaich, M.A.; Smith, M.R.; Flynn, T.S.; Freudenthal, B.D. Base excision repair of oxidative DNA damage: From mechanism to disease. Front. Biosci. 2017, 22, 1493–1522. [Google Scholar]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-excision repair of oxidative DNA damage. Nature 2007, 447, 941. [Google Scholar] [CrossRef] [PubMed]
- Baute, J.; Depicker, A. Base excision repair and its role in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 239–276. [Google Scholar] [CrossRef] [PubMed]
- Papaluca, A.; Wagner, J.R.; Saragovi, H.U.; Ramotar, D. UNG-1 and APN-1 are the major enzymes to efficiently repair 5-hydroxymethyluracil DNA lesions in C. elegans. Sci. Rep. 2018, 8, 6860. [Google Scholar] [CrossRef] [PubMed]
- Olinski, R.; Starczak, M.; Gackowski, D. Enigmatic 5-hydroxymethyluracil: Oxidatively modified base, epigenetic mark or both? Mutat. Res. 2016, 767, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Sjolund, A.B.; Senejani, A.G.; Sweasy, J.B. MBD4 and TDG: Multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013, 743–744, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.I.; Nakamura, N.; Doi, T.; Sugiyama, H.; Yamamoto, K.; Yonei, S.; Zhang, Q.M. Recombinant Schizosaccharomyces pombe Nth1 protein exhibits DNA glycosylase activities for 8-oxo-7,8-dihydroguanine and thymine residues oxidized in the methyl group. J. Radiat. Res. 2007, 48, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Rae, P.M.M. 5-Hydroxymethyluracil in the DNA of a Dinoflagellate. Proc. Natl. Acad. Sci. USA 1973, 70, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Lindahl, T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef]
- Robertson, A.B.; Klungland, A.; Rognes, T.; Leiros, I. Base excision repair: The long and short of it. Cell. Mol. Life Sci. 2009, 66, 981–993. [Google Scholar] [CrossRef]
- Simsek, D.; Jasin, M. DNA ligase III: A spotty presence in eukaryotes, but an essential function where tested. Cell Cycle 2011, 10, 3636–3644. [Google Scholar] [CrossRef]
- Córdoba-Cañero, D.; Roldán-Arjona, T.; Ariza, R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011, 68, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Prakash, L. Nucleotide excision repair in yeast. Mutat. Res. 2000, 451, 13–24. [Google Scholar] [CrossRef]
- Wood, R.D.; Araujo, S.J.; Ariza, R.R.; Batty, D.P.; Biggerstaff, M.; Evans, E.; Gaillard, P.H.; Gunz, D.; Köberle, B.; Kuraoka, I.; et al. DNA damage recognition and nucleotide excision repair in mammalian cells. CSH Symp. Quant. Biol. 2000, 65, 173–182. [Google Scholar] [CrossRef]
- Schärer, O.D. Nucleotide excision repair in eukaryotes. CSH Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Kamileri, I.; Karakasilioti, I.; Garinis, G.A. Nucleotide excision repair: New tricks with old bricks. Trends Genet. 2012, 28, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Mullenders, L.H.F. Solar UV damage to cellular DNA: From mechanisms to biological effects. Photochem. Photobiol. Sci. 2018, 17, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Tatum, D.; Li, S. Nucleotide excision repair in S. cerevisiae. In DNA Repair-On the Pathways to Fixing DNA Damage and Errors; Storici, F., Ed.; IntechOpen: Rijeka, Croatia, 2011; pp. 97–122. [Google Scholar]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26. DNA Repair 2015, 36, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Hanawalt, P.C.; Spivak, G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008, 9, 958. [Google Scholar] [CrossRef]
- Compe, E.; Egly, J.-M. TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012, 13, 343. [Google Scholar] [CrossRef]
- Rimel, J.K.; Taatjes, D.J. The essential and multifunctional TFIIH complex. Protein Sci. 2018, 27, 1018–1037. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair 2016, 44, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Kimzey, A.L.; Masselon, C.D.; Bruce, J.E.; Garner, E.C.; Brown, C.J.; Dunker, A.K.; Smith, R.D.; Ackerman, E.J. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001, 10, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Canturk, F.; Karaman, M.; Selby, C.P.; Kemp, M.G.; Kulaksiz-Erkmen, G.; Hu, J.; Li, W.; Lindsey-Boltz, L.A.; Sancar, A. Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 4706–4710. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Morse, D. Transcription and maturation of mRNA in Dinoflagellates. Microorganisms. 2013, 1, 71–99. [Google Scholar] [CrossRef] [PubMed]
- Genois, M.M.; Paquet, E.R.; Laffitte, M.C.N.; Maity, R.; Rodrigue, A.; Ouellette, M.; Masson, J.Y. DNA repair pathways in Trypanosomatids: From DNA repair to drug resistance. Microbiol. Mol. Biol. Rev. 2014, 78, 40–73. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.X.; Schmidt, T.T.; Kolodner, R.D.; Hombauer, H. New insights into the mechanism of DNA mismatch repair. Chromosoma 2015, 124, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Spies, M.; Fishel, R. Mismatch repair during homologous and homeologous recombination. CSH Perspect. Biol. 2015, 7, a022657. [Google Scholar] [CrossRef]
- Kunz, C.; Saito, Y.; Schär, P. Mismatched repair: Variations on a theme. Cell. Mol. Life Sci. 2009, 66, 1021–1038. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007, 18, 85. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Tarique, M.; Ahmad, M.; Chauhan, M.; Tuteja, R. Genome wide in silico analysis of the mismatch repair components of Plasmodium falciparum and their comparison with human host. Front. Microbiol. 2017, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. CSH Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.B.; Lewis, A.L.; Baldwin, K.K.; Resnick, M.A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 1993, 90, 5613–5617. [Google Scholar] [CrossRef] [PubMed]
- Chiruvella, K.K.; Liang, Z.; Wilson, T.E. Repair of double-strand breaks by end joining. CSH Perspect. Biol. 2013, 5, a012757. [Google Scholar] [CrossRef] [PubMed]
- Emerson, C.H.; Bertuch, A.A. Consider the workhorse: Nonhomologous end-joining in budding yeast. Biochem. Cell Biol. 2016, 94, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Weaver, D.T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997, 16, 6874–6885. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001, 412, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002, 108, 781–794. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Yu, Y.; Riballo, E.; Douglas, P.; Walker, S.A.; Ye, R.; Härer, C.; Marchetti, C.; Morrice, N.; Jeggo, P.A.; et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006, 25, 3880–3889. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kanno, S.I.; Watanabe, R.; Ogiwara, H.; Kohno, T.; Watanabe, G.; Yasui, A.; Lieber, M.R. Polynucleotide Kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3’ exonuclease and can participate in DNA end joining in a biochemical system. J. Biol. Chem. 2011, 286, 36368–36377. [Google Scholar] [CrossRef] [PubMed]
- Grundy, G.J.; Rulten, S.L.; Zeng, Z.; Arribas-Bosacoma, R.; Iles, N.; Manley, K.; Oliver, A.; Caldecott, K.W. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013, 32, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Ahel, I.; Rass, U.; El-Khamisy, S.F.; Katyal, S.; Clements, P.M.; McKinnon, P.J.; Caldecott, K.W.; West, S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006, 443, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Ahel, I.; Rass, U.; El-Khamisy, S.F.; Katyal, S.; Clements, P.M.; McKinnon, P.J.; Caldecott, K.W.; West, S.C. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 2005, 17, 657–670. [Google Scholar]
- Wilson, T.E.; Lieber, M.R. Efficient processing of DNA ends during yeast nonhomologous end joining: Evidence for a DNA polymerase β (pol4)-dependent pathway. J. Biol. Chem. 1999, 274, 23599–23609. [Google Scholar] [CrossRef] [PubMed]
- Bertocci, B.; de Smet, A.; Weill, J.-C.; Reynaud, C.-A. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination In Vivo. Immunity 2006, 25, 31–41. [Google Scholar] [CrossRef]
- Moon, A.F.; Garcia-Diaz, M.; Batra, V.K.; Beard, W.A.; Bebenek, K.; Kunkel, T.A.; Wilson, S.H.; Pedersen, L.C. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair 2007, 6, 1709–1725. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Grawunder, U.; Wilm, M.; Wu, X.; Kulesza, P.; Wilson, T.E.; Mann, M.; Lieber, M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997, 388, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Lu, H.; Tsai, A.G.; Schwarz, K.; Lieber, M.R. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Res. 2007, 35, 5755–5762. [Google Scholar] [CrossRef] [PubMed]
- Modesti, M.; Hesse, J.E.; Gellert, M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999, 18, 2008–2018. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, Y.; Suo, F.; Sun, L.-L.; Zhao, D.; Du, L.-L. Genome-wide screens for sensitivity to ionizing radiation identify the fission yeast nonhomologous end joining factor Xrc4. G3-Genes Genom. Genet. 2014, 4, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants-from models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Symington, L.S.; Fidock, D.A. DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol. Mol. Biol. Rev. 2014, 78, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S. End resection at double-strand breaks: Mechanism and regulation. CSH Perspect. Biol. 2014, 6, a016436. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef]
- Spirek, M.; Altmannova, V.; Zhao, X.; Krejci, L. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar]
- Wright, W.D.; Shah, S.S.; Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Renkawitz, J.; Lademann, C.A.; Jentsch, S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 2014, 15, 369. [Google Scholar] [CrossRef] [PubMed]
- Kaniecki, K.; de Tullio, L.; Greene, E.C. A change of view: Homologous recombination at single-molecule resolution. Nat. Rev. Genet. 2017, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.; Krejci, L.; van Komen, S.; Sehorn, M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003, 278, 42729–42732. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, M.; Kawabata, T.; Nishibori, M. Role of recA/RAD51 family proteins in mammals. Acta Med. Okayama 2005, 59, 1–9. [Google Scholar] [PubMed]
- Miller-Messmer, M.; Kühn, K.; Bichara, M.; le Ret, M.; Imbault, P.; Gualberto, J.M. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012, 159, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Rowan, B.A.; Oldenburg, D.J.; Bendich, A.J. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J. Exp. Bot. 2010, 61, 2575–2588. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Johnson, R.E.; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005, 74, 317–353. [Google Scholar] [CrossRef]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009, 73, 134–154. [Google Scholar] [CrossRef] [PubMed]
- Sale, J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. CSH Perspect. Biol. 2013, 5, a012708. [Google Scholar] [CrossRef]
- Zhao, L.; Washington, M.T. Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes 2017, 8, 24. [Google Scholar] [CrossRef]
- Marians, K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018, 87, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.T.; Washington, M.T. Eukaryotic translesion synthesis: Choosing the right tool for the job. DNA Repair 2018, 71, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.J.; Cimprich, K.A. DNA damage tolerance: When it's OK to make mistakes. Nat. Chem. Biol. 2009, 5, 82. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, S.D.; Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467. [Google Scholar] [CrossRef]
- Shen, X.; Li, L. Mutagenic repair of DNA interstrand crosslinks. Environ. Mol. Mutagen. 2010, 51, 493–499. [Google Scholar] [CrossRef]
- Hashimoto, S.; Anai, H.; Hanada, K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016, 38, 9. [Google Scholar] [CrossRef]
- Muniandy, P.A.; Liu, J.; Majumdar, A.; Liu, S.-t.; Seidman, M.M. DNA interstrand crosslink repair in mammalian cells: Step by step. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 23–49. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337. [Google Scholar] [CrossRef]
- Clauson, C.; Schärer, O.D.; Niedernhofer, L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. CSH Perspect. Biol. 2013, 5, a012732. [Google Scholar] [CrossRef]
- Kottemann, M.C.; Smogorzewska, A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013, 493, 356. [Google Scholar] [CrossRef] [PubMed]
- Dangel, N.J.; Knoll, A.; Puchta, H. MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014, 78, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Daee, D.L.; Ferrari, E.; Longerich, S.; Zheng, X.F.; Xue, X.; Branzei, D.; Sung, P.; Myung, K. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J. Biol. Chem. 2012, 287, 26563–26575. [Google Scholar] [CrossRef] [PubMed]
- Cattell, E.; Sengerová, B.; McHugh, P.J. The SNM1/Pso2 family of ICL repair nucleases: From yeast to man. Environ. Mol. Mutagen. 2010, 51, 635–645. [Google Scholar] [CrossRef]
- Munari, F.M.; Guecheva, T.N.; Bonatto, D.; Henriques, J.A.P. New features on Pso2 protein family in DNA interstrand cross-link repair and in the maintenance of genomic integrity in Saccharomyces cerevisiae. Fungal Genet. Biol. 2013, 60, 122–132. [Google Scholar] [CrossRef][Green Version]
- van Attikum, H.; Gasser, S.M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009, 19, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Vidanes, G.M.; Bonilla, C.Y.; Toczyski, D.P. Complicated tails: Histone modifications and the DNA damage response. Cell 2005, 121, 973–976. [Google Scholar] [CrossRef] [PubMed]

| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Upstream factors | ||||
| ATR | Unigene24416_All (2e-022) | symbB.v1.2.040344.t1 (2e-067) | Lp_Unigene54547_All (9e-118) | apical checkpoint serine/threonine kinase |
| Unigene40907_All (1e-087) | symbB.v1.2.040393.t1 (1e-018) | |||
| ATRIP | * | * | * | ATR-interacting protein, necessary for checkpoint signaling upon DNA damage |
| ATM | Unigene1664_All (3e-091) | symbB.v1.2.029032.t1 (1e-087) | Lp_Unigene23700_All (6e-089) | apical checkpoint serine/threonine-protein kinase |
| Rad17 | Unigene30410_All (1e-015) | symbB.v1.2.000094.t1 (7e-008) | Lp_Unigene58859_All (4e-018) | involved in ATR-dependent checkpoint activation |
| Rad1 | CL2244.Contig1_All (2e-024) | - | Lp_Unigene59067_All (6e-006) | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| CL2244.Contig2_All (2e-029) | ||||
| Hus1 | Unigene57737_All (1e-008) | symbB.v1.2.041619.t1 (3e-014) | Lp_Unigene75437_All (2e-015) | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| Rad9 | * | * | * | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| Mediators | ||||
| BRCA1 | * | * | * | E3 ubiquitin-protein ligase activity for formation of poly-ubiquitin chains |
| 53BP1 | * | * | * | p53-binding protein 1, binds damaged DNA |
| MDC1 | Unigene15380_All (9e-007) | * | Lp_CL4955.Contig1_All (5e-009) | mediator of DNA damage checkpoint, recruitment of repair proteins to DNA damage foci |
| Lp_CL4955.Contig2_All ((5e-009)) | ||||
| Claspin | * | * | * | upstream regulator of Chk1 |
| TopBP1 | * | * | Lp_Unigene64350_All (1e-006) | topoisomerase II beta interacting protein, ATR activator |
| Effectors | ||||
| Chk1 | * | * | * | serine/threonine-protein kinase |
| Chk2 | Unigene61604_All (3e-056) | symbB.v1.2.014603.t1 (4e-043) | Lp_CL13254.Contig1_All (2e-052) | serine/threonine-protein kinase |
| Unigene13287_All (2e-054) | symbB.v1.2.000403.t1 (3e-028) | Lp_Unigene47534_All (3e-041) | ||
| Unigene68813_All (2e-045) | Lp_CL1544.Contig1_All (6e-049) | |||
| Unigene52773_All (3e-048) | ||||
| Unigene5075_All (1e-041) | ||||
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| direct reversal | ||||
| MGMT | * | * | Lp_Unigene68135_All (1e-009) | O6-methylguanine DNA methyltransferase |
| Lp_Unigene71190_All (5e-007) | ||||
| ALKB/APH | Unigene40709_All (5e-018) | symbB.v1.2.000566.t1 (1e-006) | Lp_Unigene17309_All (9e-015) | nucleic acid dioxygenase involved in alkylation damage |
| symbB.v1.2.012827.t1 (5e-017) | ||||
| photoreactivation | ||||
| 6-4 photolyase | CL2432.Contig2_All (2e-113) | symbB.v1.2.031430.t1 (3e-051) | Lp_Unigene48358_All (9e-128) | involved in repair of (6-4) pyrimidine–pyrimidine induced by UV irradiation |
| CL6843.Contig1_All (8e-119) | symbB.v1.2.033063.t1 (5e-114) | Lp_Unigene59149_All (2e-121) | ||
| CPD photolyase | Unigene1971_All (2e-151) | symbB.v1.2.000806.t1 (3e-022) | Lp_Unigene17168_All (3e-015) | involved in repair of cyclobutane-pyrimidine dimer (CPD) induced by UV irradiation |
| CL5100.Contig1_All (1e-80) | symbB.v1.2.012126.t1 (6e-027) | Lp_Unigene75562_All (9e-024) | ||
| symbB.v1.2.023060.t1 (5e-028) | Lp_Unigene21665_All (3e-013) | |||
| symbB.v1.2.037491.t1 (4e-038) | Lp_Unigene43660_All (3e-012) | |||
| symbB.v1.2.001247.t1 (7e-041) | Lp_Unigene4570_All (2e-069) | |||
| symbB.v1.2.030395.t1 (2e-043) | Lp_Unigene41185_All (3e-021) | |||
| symbB.v1.2.030396.t1 (3e-057) | Lp_Unigene47312_All (2e-074) | |||
| symbB.v1.2.030397.t1 (6e-022) | Lp_Unigene58611_All (3e-155) | |||
| symbB.v1.2.031429.t1 (4e-010) | Lp_Unigene63301_All (2e-020) | |||
| symbB.v1.2.011563.t1 (4e-035) | ||||
| cryptochrome DASH | Unigene5536_All (5e-47) | symbB.v1.2.014092.t1 (9e-017) | Lp_Unigene16311_All (3e-108) | removal of cyclobutane-pyrimidine dimers (CPD) in ssDNA |
| Unigene41133_All (4e-110) | symbB.v1.2.021362.t1 (8e-068) | Lp_Unigene17251_All (3e-068) | ||
| Unigene49976_All (5e-69) | symbB.v1.2.025343.t1 (1e-102) | Lp_Unigene20031_All (4e-074) | ||
| symbB.v1.2.028883.t1 (1e-043) | Lp_Unigene25347_All (9e-078) | |||
| symbB.v1.2.027391.t1 (7e-046) | Lp_Unigene63655_All (2e-075) | |||
| symbB.v1.2.021130.t1 (1e-084) | Lp_Unigene67177_All (9e-101) | |||
| symbB.v1.2.030740.t1 (1e-062) | Lp_CL14399.Contig1_All (2e-097) | |||
| symbB.v1.2.029072.t1 (1e-035) | Lp_CL14399.Contig2_All (3e-097) | |||
| symbB.v1.2.015001.t1 (3e-080) | Lp_Unigene7953_All (1e-092) | |||
| symbB.v1.2.006488.t1 (1e-084) | Lp_Unigene24084_All (4e-065) | |||
| symbB.v1.2.007315.t1 (2e-026) | Lp_Unigene39631_All (3e-071) | |||
| Lp_Unigene51086_All (1e-032) | ||||
| Lp_Unigene54913_All (6e-065) | ||||
| Lp_Unigene6860_All (8e-012) | ||||
| Lp_Unigene8874_All (6e-079) | ||||
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| UNG | Unigene44697_All (2e-059) | symbB.v1.2.022126.t1 (9e-069) | Lp_Unigene44751_All (3e-078) | uracil-DNA glycosylases; removes uracil |
| Unigene70580_All (6e-071) | symbB.v1.2.022126.t2 (3e-070) | Lp_Unigene47003_All (4e-071) | ||
| symbB.v1.2.031173.t1 (6e-052) | ||||
| SMUG1 | * | * | * | single-strand selective mono-functional uracil-DNA glycosylase |
| MBD4 | Unigene51401_All (7e-008) | symbB.v1.2.021865.t1 (2e-014) | Lp_Unigene54655_All (3e-019) | DNA N-glycosylase involved in removal of thymine mismatch |
| TDG | * | * | * | thymine DNA glycosylase; removes thymine mismatch, uracil |
| OGG1 | Unigene85095_All (4e-019) | symbB.v1.2.040865.t1 (5e-011) | Lp_Unigene28278_All (1e-054) | 8-oxoG DNA glycosylase |
| Unigene82000_All (1e-013) | Lp_Unigene40876_All (4e-054) | |||
| MYH | Unigene67393_All (4e-036) | symbB.v1.2.014787.t1 (4e-067) | Lp_Unigene17300_All (4e-078) | adenine DNA glycosylase; removes A opposite G |
| NTH1 | Unigene62006_All (2e-034) | symbB.v1.2.007780.t1 (4e-050) | Lp_CL9519.Contig1_All (5e-006) | Bi-functional DNA N-glycosylase; removal of oxidized pyrimidines, formamidopyrimidines, 5-formyluracil |
| Unigene64896_All (2e-045) | symbB.v1.2.038247.t1 (2e-037) | Lp_Unigene32526_All (1e-061) | ||
| Lp_Unigene58890_All (6e-053) | ||||
| MPG/Mag1 | * | symbB.v1.2.024316.t1 (4e-007) | Lp_Unigene52978_All (4e-013) | DNA-3-methyladenine glycosylase |
| FPG | Unigene42128_All (6e-049) | symbB.v1.2.002485.t1 (2e-071) | Lp_Unigene4975_All (3e-064) | formamidopyrimidine-DNA glycosylase |
| CL7240.Contig3_All (3e-048) | Lp_Unigene48155_All (6e-008) | |||
| NEIL1 | * | * | * | DNA glycosylases; removal of oxidized pyrimidines, thymine glycol |
| NEIL2 | * | * | * | DNA glycosylases; removal of oxidized pyrimidines |
| NEIL3 | Unigene48672_All (1e-011) | symbB.v1.2.022143.t1 (5e-018) | Lp_Unigene13451_All (5e-014) | DNA glycosylases; removal of oxidized pyrimidines |
| APE1 | Unigene9067_All (1e-033) | symbB.v1.2.023749.t1 (4e-039) | Lp_CL8817.Contig2_All (3e-011) | apurinic/apyrimidinic (AP) endonuclease |
| Unigene17453_All (6e-006) | symbB.v1.2.024175.t1 (4e-012) | Lp_CL8837.Contig2_All (1e-036) | ||
| Unigene34000_All (2e-012) | symbB.v1.2.033468.t1 (1e-035) | Lp_Unigene2242_All (5e-025) | ||
| Unigene73707_All (2e-011) | symbB.v1.2.033468.t2 (1e-035) | Lp_Unigene33584_All (7e-035) | ||
| Unigene41862_All (3e-037) | ||||
| Unigene73755_All (1e-032) | ||||
| APE2 | CL7855.Contig3_All (1e-014) | symbB.v1.2.021745.t1 (1e-009) | Lp_Unigene55979_All (4e-008) | apurinic/apyrimidinic (AP) endonuclease |
| Lp_Unigene48320_All (7e-010) | ||||
| LIG1 | CL2462.Contig1_All (7e-151) | symbB.v1.2.029028.t1 (3e-011) | Lp_CL8189.Contig1_All (9e-022) | DNA ligase required for long-patch BER |
| Unigene56781_All (2e-051) | symbB.v1.2.029030.t1 (1e-055) | Lp_CL8189.Contig2_All (6e-043) | ||
| symbB.v1.2.007861.t1 (7e-021) | Lp_CL13983.Contig1_All (8e-060) | |||
| symbB.v1.2.007862.t3 (3e-094) | Lp_CL13983.Contig2_All (1e-059) | |||
| symbB.v1.2.007862.t4 (1e-011) | Lp_Unigene18447_All (6e-020) | |||
| symbB.v1.2.007862.t5 (5e-081) | Lp_Unigene44117_All (2e-154) | |||
| symbB.v1.2.007862.t6 (4e-081) | ||||
| XRCC1 | * | * | * | interacts with PARP, LIG3, and Polβ |
| LIG3 | * | * | * | DNA ligase required for short-patch BER |
| PKNP | CL5342.Contig1_All (5e-029) | Lp_Unigene21188_All (3e-025) | DNA 3′-phosphatase 5′-kinase required for restoration of 5′-phosphate and 3′-hydroxyl termini | |
| CL5342.Contig2_All (2e-029) | Lp_Unigene67913_All (4e-012) | |||
| CL5342.Contig3_All (3e-029) | Lp_Unigene67503_All (8e-027) | |||
| FEN1 | Unigene36893_All (6e-102) | symbB.v1.2.012846.t1 (2e-067) | Lp_Unigene74714_All (3e-108) | structure-specific nuclease |
| symbB.v1.2.005353.t1 (1e-047) | ||||
| symbB.v1.2.017794.t1 (9e-099) | ||||
| PCNA | CL2939.Contig1_All (1e-075) | symbB.v1.2.024689.t1 (1e-069) | Lp_CL16467.Contig1_All (3e-051) | loading platform of PolD1 and PolE1 |
| CL2939.Contig2_All (3e-075) | symbB.v1.2.005740.t1 (1e-072) | Lp_CL16467.Contig2_All (5e-072) | ||
| CL2939.Contig3_All (5e-076) | symbB.v1.2.003346.t1 (1e-036) | Lp_CL16467.Contig3_All (2e-039) | ||
| CL2939.Contig4_All (1e-075) | Lp_CL16467.Contig5_All (4e-051) | |||
| CL2939.Contig5_All (1e-075) | Lp_CL16467.Contig6_All (3e-056) | |||
| CL2939.Contig6_All (5e-076) | Lp_CL16467.Contig7_All (6e-057) | |||
| Lp_CL16467.Contig9_All (9e-058) | ||||
| Lp_Unigene31367_All (2e-075) | ||||
| Lp_Unigene31369_All (3e-072) | ||||
| Lp_CL9592.Contig1_All (1e-008) | ||||
| Lp_CL15065.Contig1_All (1e-011) | ||||
| Lp_CL15065.Contig2_All (1e-010) | ||||
| Lp_CL15065.Contig4_All (5e-010) | ||||
| Lp_Unigene7276_All (2e-033) | ||||
| Lp_Unigene31368_All (7e-029) | ||||
| Lp_Unigene31370_All (1e-027) | ||||
| Lp_Unigene31371_All (2e-022) | ||||
| PolB | Unigene65173_All (8e-055) | symbB.v1.2.005179.t1 (8e-051) | Lp_Unigene74664_All (5e-061) | DNA polymerase β/beta |
| Unigene70894_All (1e-009) | ||||
| POLE1 | Unigene29376_All (0.0) | symbB.v1.2.023008.t1 (4e-015) | Lp_CL7119.Contig1_All (0.0) | DNA polymerase ε/epsilon catalytic subunit |
| Unigene37482_All (3e-126) | symbB.v1.2.023011.t1 (3e-019) | Lp_CL7119.Contig2_All (0.0) | ||
| symbB.v1.2.017036.t1 (6e-006) | Lp_Unigene71337_All (5e-029) | |||
| symbB.v1.2.001887.t1 (7e-131) | ||||
| symbB.v1.2.001887.t2 (2e-130) | ||||
| symbB.v1.2.001887.t3 (3e-048) | ||||
| symbB.v1.2.001889.t1 (1e-042) | ||||
| POLD1 | Unigene4585_All (3e-050) | symbB.v1.2.029507.t1 (1e-045) | Lp_Unigene28445_All (5e-082) | DNA polymerase δ/delta catalytic subunit |
| Unigene44692_All (3e-029) | symbB.v1.2.037870.t1 (2e-089) | Lp_Unigene37178_All (3e-071) | ||
| Unigene50187_All (8e-055) | symbB.v1.2.036929.t1 (3e-144) | Lp_Unigene55814_All (1e-019) | ||
| Unigene61374_All (0.0) | symbB.v1.2.036930.t1 (3e-118) | Lp_Unigene70233_All (0.0) | ||
| Unigene77552_All (2e-148) | symbB.v1.2.033323.t1 (2e-014) | Lp_Unigene71779_All (0.0) | ||
| PARP1 | CL1934.Contig3_All (1e-015) | symbB.v1.2.024122.t1 (6e-011) | Lp_CL11756.Contig2_All (2e-006) | poly(ADP-Ribose) polymerase-1; necessary for recruitment of other DNA-repairing enzymes |
| Unigene13204_All (1e-027) | symbB.v1.2.018837.t1 (5e-048) | Lp_CL11756.Contig3_All (2e-007) | ||
| Unigene82709_All (1e-015) | Lp_Unigene29089_All (6e-048) | |||
| PARP2 | Unigene9060_All (7e-006) | symbB.v1.2.001263.t1 (4e-022) | Lp_CL1803.Contig2_All (5e-028) | poly(ADP-Ribose) polymerase-2; necessary for recruitment of other DNA-repairing enzymes |
| Unigene45362_All (7e-017) | Lp_Unigene35603_All (2e-045) | |||
| Lp_Unigene36239_All (5e-019) | ||||
| Lp_Unigene54635_All (2e-142) | ||||
| PARP3 | Unigene53685_All (3e-016) | symbB.v1.2.040205.t1 (2e-035) | Lp_Unigene5203_All (6e-032) | poly(ADP-Ribose) polymerase-3; necessary for recruitment of other DNA-repairing enzymes |
| symbB.v1.2.034465.t1 (1e-015) | ||||
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| DDB1 | * | symbB.v1.2.027839.t1 (2e-058) | Lp_Unigene56329_All (2e-161) | component of UV-damaged DNA-binding protein complex |
| DDB2/XPE | * | * | * | component of UV-damaged DNA-binding protein complex |
| XPC/Rad4 | Unigene32549_All (8e-015) | symbB.v1.2.001527.t1 (2e-009) | Lp_Unigene33147_All (3e-017) | binds damaged DNA as XPC complex |
| Rad23 | Unigene21346_All (2e-006) | symbB.v1.2.023245.t1 (2e-022) | Lp_CL13062.Contig2_All (2e-007) | binds damaged DNA as XPC complex |
| Unigene68760_All (3e-019) | symbB.v1.2.021833.t1 (6e-017) | Lp_Unigene20125_All (6e-010) | ||
| symbB.v1.2.010422.t1 (6e-009) | Lp_Unigene72427_All (2e-012) | |||
| Lp_CL7915.Contig1_All (7e-009) | ||||
| Lp_CL7915.Contig2_All (8e-009) | ||||
| Lp_CL13062.Contig1_All (2e-007) | ||||
| Lp_Unigene36792_All (8e-006) | ||||
| Lp_Unigene50304_All (3e-006) | ||||
| CETN2 | Unigene29893_All (5e-049) | symbB.v1.2.018116.t1 (5e-043) | Lp_CL4243.Contig1_All (1e-043) | component of the XPC complex |
| Unigene38411_All (2e-047) | Lp_CL4243.Contig2_All (7e-043) | |||
| Lp_CL4243.Contig3_All (2e-043) | ||||
| ERCC8/CSA/Rad28 | Unigene20816_All (1e-009) | symbB.v1.2.020387.t1 (2e-010) | * | required for transcription-coupled nucleotide excision repair |
| symbB.v1.2.020387.t2 (2e-010) | ||||
| ERCC-6/Rad26/CSB | Unigene349_All (7e-166) | symbB.v1.2.032523.t1 (5e-084) | Lp_CL6060.Contig1_All (1e-038) | required for transcription-coupled excision repair |
| Unigene1702_All (8e-056) | symbB.v1.2.026497.t1 (7e-057) | Lp_CL6060.Contig2_All (2e-036) | ||
| CL1148.Contig1_All (1e-037) | symbB.v1.2.019527.t1 (1e-161) | Lp_CL12386.Contig2_All (8e-123) | ||
| Unigene61410_All (2e-041) | symbB.v1.2.019505.t1 (1e-033) | Lp_CL12386.Contig1_All (7e-123) | ||
| symbB.v1.2.019507.t1 (2e-024) | Lp_CL14187.Contig1_All (6e-157) | |||
| symbB.v1.2.026498.t1 (3e-021) | Lp_CL14187.Contig2_All (7e-157) | |||
| Lp_Unigene36232_All (2e-038) | ||||
| XPA | * | * | * | binds to DNA damage foci in pre-incision complex |
| RPA1 | Unigene13507_All (3e-041) | symbB.v1.2.000388.t1 (7e-036) | Lp_CL2032.Contig1_All (4e-047) | subunits of heterotrimeric replication protein A complex |
| Unigene40788_All (9e-018) | symbB.v1.2.016776.t1 (3e-023) | Lp_CL2032.Contig3_All (2e-048) | ||
| Unigene69752_All (7e-034) | symbB.v1.2.010569.t1 (9e-043) | Lp_CL2032.Contig4_All (2e-048) | ||
| Unigene72956_All (1e-043) | symbB.v1.2.015898.t1 (2e-051) | Lp_CL2227.Contig4_All (1e-054) | ||
| Lp_CL2227.Contig5_All (2e-057) | ||||
| Lp_CL2227.Contig6_All (2e-060) | ||||
| Lp_CL2227.Contig8_All (4e-058) | ||||
| Lp_CL2227.Contig9_All (1e-047) | ||||
| Lp_Unigene5237_All (3e-048) | ||||
| Lp_CL2032.Contig2_All (6e-047) | ||||
| Lp_CL2227.Contig1_All (3e-044) | ||||
| Lp_CL2227.Contig2_All (2e-042) | ||||
| Lp_CL2227.Contig3_All (2e-037) | ||||
| Lp_CL2227.Contig7_All (3e-045) | ||||
| Lp_CL14409.Contig1_All (2e-045) | ||||
| Lp_CL14409.Contig2_All (2e-046) | ||||
| Lp_CL14409.Contig3_All (2e-046) | ||||
| Lp_Unigene27814_All (2e-045) | ||||
| RPA2 | Unigene14481_All (5e-010) | symbB.v1.2.033139.t1 (3e-007) | Lp_CL6472.Contig2_All (1e-007) | subunits of heterotrimeric replication protein A complex |
| Lp_CL6472.Contig5_All (7e-008) | ||||
| Lp_CL6472.Contig6_All (6e-007) | ||||
| Lp_CL9761.Contig1_All (2e-008) | ||||
| Lp_CL9761.Contig2_All (1e-008) | ||||
| Lp_Unigene6245_All (8e-008) | ||||
| Lp_Unigene13916_All (9e-009) | ||||
| RPA3 | * | * | * | subunits of heterotrimeric replication protein A complex |
| RFC1 | CL4680.Contig1_All (1e-076) | symbB.v1.2.031425.t1 (2e-037) | Lp_CL3410.Contig1_All (2e-050) | required for strand displacement and DNA synthesis |
| CL4680.Contig2_All (2e-023) | symbB.v1.2.031427.t1 (9e-013) | Lp_CL3410.Contig3_All (1e-084) | ||
| CL4680.Contig3_All (2e-076) | symbB.v1.2.000210.t1 (1e-073) | Lp_CL3410.Contig5_All (2e-006) | ||
| Unigene45646_All (2e-052) | symbB.v1.2.025385.t1 (3e-054) | Lp_CL3410.Contig6_All (3e-016) | ||
| Unigene77812_All (3e-089) | Lp_Unigene33492_All (2e-008) | |||
| Lp_Unigene35510_All (6e-040) | ||||
| Lp_Unigene43360_All (3e-049) | ||||
| TFIIH complex | catalyzes DNA helix unwinding in pre-incision complex | |||
| GTF2H1 | * | * | * | core TFIIH subunit p62 |
| GTF2H2 | Unigene58152_All (5e-034) | * | Lp_Unigene48571_All (2e-051) | core TFIIH subunit p44 |
| GTF2H3 | * | symbB.v1.2.011909.t1 (2e-007) | Lp_Unigene75735_All (6e-014) | core TFIIH subunit p34 |
| GTF2H4 | Unigene45546_All (2e-030) | symbB.v1.2.036961.t2 (2e-016) | Lp_Unigene63874_All (5e-022) | core TFIIH subunit p52 |
| GTF2H5 | * | * | * | core TFIIH subunit p8 |
| ERCC3/XPB | Unigene17412_All (1e-177) | symbB.v1.2.000409.t1 (1e-150) | Lp_Unigene15679_All (7e-142) | core subunits of TFIIH complex, 3′ to 5′ DNA helicase |
| ERCC2/XPD/Rad3 | Unigene61441_All (0.0) | symbB.v1.2.043770.t1 (3e-017) | Lp_Unigene51894_All (0.0) | core subunits of TFIIH complex, 5′ to 3′ DNA helicase |
| symbB.v1.2.003111.t1 (5e-085) | ||||
| symbB.v1.2.003112.t1 (8e-033) | ||||
| symbB.v1.2.003109.t1 (2e-056) | ||||
| CDK7 | Unigene65253_All (6e-042) | * | Lp_CL3145.Contig1_All (3e-076) | subunit of kinase module of TFIIH complex |
| Unigene66528_All (1e-019) | Lp_CL3145.Contig2_All (4e-079) | |||
| Lp_CL3145.Contig3_All (3e-079) | ||||
| MNAT1 | Unigene73596_All (2e-014) | symbB.v1.2.023842.t1 (3e-018) | Lp_Unigene66408_All (2e-017) | subunit of kinase module of TFIIH complex |
| CCNH | * | * | Lp_Unigene28603_All (7e-010) | subunit of kinase module of TFIIH complex |
| ERCC1 | Unigene34115_All (1e-045) | symbB.v1.2.018882.t1 (1e-038) | Lp_Unigene75127_All (5e-053) | 5′ incision DNA-binding subunit of TFIIH complex |
| XPF/ERCC4 | Unigene21562_All (2e-035) | symbB.v1.2.008218.t1 (1e-056) | Lp_Unigene48147_All (2e-057) | 5′ incision catalytic subunit of TFIIH complex |
| Lp_Unigene63025_All (4e-049) | ||||
| XPG/ERCC5 | Unigene8568_All (2e-031) | symbB.v1.2.006008.t1 (4e-025) | Lp_Unigene10768_All (2e-023) | 3′ incision subunits of TFIIH complex |
| Unigene57291_All (2e-024) | symbB.v1.2.006011.t1 (2e-026) | Lp_Unigene47869_All (4e-033) | ||
| symbB.v1.2.006013.t1 (3e-008) | ||||
| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| MLH1 | Unigene48309_All (7e-103) | symbB.v1.2.021543.t1 (3e-111) | Lp_Unigene17816_All (2e-116) | MutL homolog, form heterodimer with PMS1, MLH2 and MLH3 |
| Unigene56264_All (6e-020) | ||||
| MLH3 | * | symbB.v1.2.038425.t1 (7e-014) | Lp_Unigene24248_All (7e-036) | MutL homolog, form heterodimer with MLH1 |
| PMS1 | CL7093.Contig1_All (1e-042) | symbB.v1.2.037608.t1 (1e-051) | Lp_CL13190.Contig3_All (4e-049) | MutL homolog, form heterodimer with MLH1, part of the DNA mismatch repair (MMR) complex |
| CL7093.Contig2_All (9e-044) | symbB.v1.2.037606.t1 (2e-014) | Lp_Unigene124_All (2e-049) | ||
| CL7093.Contig3_All (3e-041) | Lp_Unigene48112_All (9e-040) | |||
| MSH1 | Unigene56453_All (2e-010) | symbB.v1.2.040100.t1 (2e-012) | Lp_Unigene39523_All (1e-006) | MutS homolog, required for repair of mitochondrial DNA |
| Unigene41842_All (3e-007) | symbB.v1.2.011567.t1 (4e-019) | |||
| MSH2 | Unigene8950_All (3e-133) | symbB.v1.2.002929.t1 (1e-135) | Lp_Unigene75744_All (1e-138) | MutS homolog 2, form heterodimers with MSH3 or MSH6; component of the DNA mismatch repair (MMR) complex |
| symbB.v1.2.002929.t2 (2e-061) | ||||
| MSH3 | * | * | * | MutS homolog 3 |
| MSH6 | Unigene36848_All (2e-072) | symbB.v1.2.026874.t1 (1e-056) | Lp_CL3961.Contig1_All (6e-086) | MutS homolog 6 |
| Unigene62212_All (4e-011) | symbB.v1.2.038785.t1 (5e-012) | Lp_CL3961.Contig2_All (7e-007) | ||
| Unigene69195_All (3e-023) | symbB.v1.2.039566.t1 (1e-014) | Lp_CL3961.Contig3_All (5e-100) | ||
| Lp_CL3961.Contig4_All (2e-100) | ||||
| Lp_CL3961.Contig5_All (2e-082) | ||||
| Lp_CL3961.Contig6_All (7e-101) | ||||
| MSH4 | * | symbB.v1.2.013503.t1 (7e-060) | Lp_Unigene9260_All (8e-065) | MutS homolog 4, specific for meiosis |
| MSH5 | Unigene89588_All (4e-019) | symbB.v1.2.033801.t1 (7e-079) | Lp_Unigene70169_All (1e-039) | MutS homolog 5, specific for meiosis, formation of heterodimer with MSH4 |
| Lp_Unigene63036_All (2e-030) | ||||
| EXO1 | Unigene52745_All (3e-035) | symbB.v1.2.019143.t1 (1e-043) | Lp_CL3557.Contig1_All (9e-048) | 5′ exonuclease |
| CL691.Contig1_All (7e-037) | Lp_Unigene36569_All (2e-037) | |||
| Lp_Unigene71445_All (9e-056) | ||||
| Lp_Unigene77209_All (1e-007) | ||||
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Ku70 | Unigene61676_All (3e-062) | symbB.v1.2.013334.t1 (2e-048) | Lp_Unigene63133_All (8e-069) | components of Ku heterodimer, which binds to damaged DNA ends |
| Ku80 | Unigene21225_All (8e-012) | symbB.v1.2.017317.t1 (2e-033) | Lp_Unigene60142_All (3e-041) | components of Ku heterodimer, which binds to damaged DNA ends |
| DNA-PK/PRKDC | Unigene41976_All (4e-076) | symbB.v1.2.025517.t1 (2e-071) | Lp_Unigene62602_All (1e-075) | catalytic subunit of DNA-dependent serine/threonine-protein kinase |
| Unigene56338_All (3e-006) | ||||
| Unigene72430_All (3e-006) | ||||
| LIG4 | Unigene30802_All (5e-018) | symbB.v1.2.033586.t1 (1e-055) | Lp_Unigene8556_All (7e-100) | DNA ligase which forms complex with XRCC4 and responsible for the NHEJ ligation |
| Unigene57597_All (3e-023) | symbB.v1.2.033587.t1 (2e-022) | |||
| Unigene73919_All (3e-029) | ||||
| XRCC4 | * | * | * | forms complex with LIG4 and responsible for the NHEJ ligation |
| XLF/Nej1 | * | * | * | end-joining factor; serve as bridge for XRCC4-LIG4 complex |
| Artemis | * | * | * | nuclease for DNA ends processing |
| APTX | Unigene85214_All (2e-008) | symbB.v1.2.019417.t1 (2e-018) | * | resolve of DNA single-strand interruptions |
| APLF | * | symbB.v1.2.012028.t1 (7e-006) | * | auxiliary factor for DNA end-joining |
| PolL | Unigene72723_All (3e-018) | symbB.v1.2.039928.t1 (8e-031) | Lp_Unigene32155_All (3e-036) | DNA polymerase λ/lambda, involved in NHEJ of nuclear DNA |
| symbB.v1.2.007119.t1 (2e-037) | Lp_Unigene47083_All (3e-040) | |||
| PolM | * | * | * | DNA polymerase μ/mu |
| Pol4 | * | * | * | DNA polymerase IV |
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| RAD51 | Unigene44686_All (1e-133) | symbB.v1.2.024814.t1 (4e-084) | Lp_CL13972.Contig1_All (8e-120) | recombinase, eukaryote RecA homologue |
| Lp_CL13972.Contig3_All (6e-123) | ||||
| Lp_CL13972.Contig4_All (6e-126) | ||||
| Lp_CL13972.Contig5_All (2e-124) | ||||
| Lp_Unigene24767_All (2e-110) | ||||
| DMC1 | Unigene64853_All (9e-113) | symbB.v1.2.000608.t1 (2e-109) | Lp_Unigene20110_All (1e-118) | RAD51 homologue specific for meiosis |
| symbB.v1.2.008353.t1 (3e-109) | ||||
| RecA | * | symbB.v1.2.015650.t1 (2e-063) | Lp_Unigene51570_All (4e-093) | bacterial recombinase |
| symbB.v1.2.028306.t1 (5e-016) | ||||
| RAD51B | Unigene1912_All (2e-016) | * | Lp_Unigene71304_All (1e-023) | RAD51 paralog |
| RAD51C | Unigene41837_All (3e-017) | * | Lp_Unigene66289_All (3e-034) | RAD51 paralog |
| RAD51D | * | * | * | RAD51 paralog |
| XRCC2 | * | symbB.v1.2.008271.t1 (2e-008) | * | DNA break and cross-link repair |
| XRCC3 | * | * | Lp_Unigene71572_All (2e-010) | DNA break and cross-link repair |
| RAD50 | CL6927.Contig1_All (2e-068) | symbB.v1.2.031416.t1 (3e-051) | Lp_Unigene51516_All (1e-065) | part of the MRN complex |
| CL6927.Contig2_All (6e-068) | symbB.v1.2.031416.t2 (3e-051) | |||
| Unigene31993_All (2e-021) | symbB.v1.2.025527.t1 (8e-044) | |||
| Unigene68915_All (7e-019) | ||||
| NBS1 | Unigene77295_All (5e-013) | symbB.v1.2.016565.t1 (3e-014) | Lp_CL4311.Contig2_All (9e-011) | part of the MRN complex |
| MRE11 | Unigene13700_All (8e-117) | symbB.v1.2.022929.t1 (1e-113) | Lp_Unigene63346_All (7e-111) | part of the MRN complex |
| symbB.v1.2.022929.t2 (2e-093) | ||||
| symbB.v1.2.022929.t3 (3e-010) | ||||
| RAD52 | * | * | * | accessory factor for recombination |
| RAD54L | Unigene77564_All (4e-149) | symbB.v1.2.021721.t2 (3e-127) | Lp_CL12947.Contig4_All (4e-148) | involved in recombination |
| symbB.v1.2.012979.t1 (9e-088) | Lp_Unigene46377_All (6e-147) | |||
| symbB.v1.2.021720.t1 (4e-077) | Lp_CL12947.Contig2_All (2e-148) | |||
| symbB.v1.2.021721.t1 (4e-066) | Lp_CL12947.Contig3_All (2e-148) | |||
| symbB.v1.2.021721.t3 (4e-066) | ||||
| RAD54B | * | symbB.v1.2.012978.t1 (6e-024) | Lp_Unigene31872_All (5e-096) | accessory factor for recombination |
| Lp_Unigene981_All (4e-037) | ||||
| BRCA2 | * | symbB.v1.2.003783.t1 (1e-008) | * | involved in recombination |
| DSS1/SHFM1 | * | * | * | BRCA2 accessary factor |
| CtIP | * | symbB.v1.2.018901.t1 (3e-006) | * | involved in DNA end resection |
| BLM/Sgs1 | * | symbB.v1.2.009075.t3 (2e-037) | Lp_Unigene24386_All (1e-018) | 3′-5′ DNA helicase |
| symbB.v1.2.009075.t4 (2e-037) | ||||
| symbB.v1.2.009075.t2 (2e-037) | ||||
| symbB.v1.2.009074.t1 (3e-041) | ||||
| symbB.v1.2.008709.t1 (7e-022) | ||||
| Top3α | Unigene32503_All (2e-163) | symbB.v1.2.026949.t1 (6e-160) | Lp_Unigene1654_All (3e-166) | DNA topoisomerase 3-alpha |
| symbB.v1.2.035328.t1 (2e-021) | Lp_Unigene44680_All (6e-022) | |||
| symbB.v1.2.037631.t1 (2e-006) | Lp_CL12602.Contig1_All (3e-011) | |||
| Lp_Unigene21536_All (4e-012) | ||||
| Lp_Unigene29037_All (1e-011) | ||||
| Lp_Unigene53944_All (3e-006) | ||||
| Lp_Unigene62514_All (3e-012) | ||||
| RMI1 | Unigene5006_All (6e-010) | symbB.v1.2.037665.t1 (1e-006) | Lp_Unigene13746_All (1e-010) | key component of the RMI complex |
| RMI2 | * | * | * | key component of the RMI complex |
| DNA2 | * | * | * | ATP-dependent helicase/nuclease |
| PARI/Srs2 | Unigene16314_All (1e-009) | * | * | inhibit inappropriate homologous recombination |
| MUS81 | Unigene1977_All (6e-016) | symbB.v1.2.018987.t1 (1e-017) | Lp_Unigene50884_All (1e-013) | subunit of structure-specific endonuclease |
| EME1/MMS4 | * | * | * | interaction with Mus81 |
| SLX1 | Unigene40330_All (4e-021) | symbB.v1.2.003012.t1 (1e-020) | Lp_Unigene20489_All (2e-020) | subunit of SLX1-SLX4 structure-specific nuclease |
| symbB.v1.2.009695.t1 (2e-012) | ||||
| SLX4 | * | * | * | subunit of SLX1-SLX4 structure-specific nuclease |
| GEN1/YEN1 | Unigene8182_All (2e-008) | * | Lp_Unigene2100_All (7e-019) | nuclease seperating Holliday junctions |
| Lp_Unigene17855_All (2e-012) | ||||
| SPO11 | Unigene18126_All (3e-012) | symbB.v1.2.038121.t1 (7e-025) | Lp_Unigene20651_All (2e-024) | meiosis specific endonuclease |
| Unigene24373_All (1e-012) | symbB.v1.2.012520.t1 (8e-031) | Lp_Unigene9316_All (2e-038) | ||
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Rev3L/PolZ | Unigene7171_All (1e-017) | symbB.v1.2.025103.t1 (1e-067) | Lp_Unigene29950_All (8e-022) | DNA polymerase ζ/zeta catalytic subunit |
| Unigene32515_All (9e-131) | symbB.v1.2.025103.t2 (3e-067) | Lp_Unigene60556_All (1e-025) | ||
| Unigene56596_All (3e-083) | symbB.v1.2.025103.t3 (2e-067) | Lp_Unigene61611_All (1e-030) | ||
| symbB.v1.2.028081.t1 (7e-091) | Lp_Unigene76685_All (4e-026) | |||
| symbB.v1.2.028084.t1 (1e-024) | Lp_Unigene76716_All (1e-013) | |||
| Lp_Unigene81304_All (2e-008) | ||||
| REV1 | Unigene56396_All (3e-046) | symbB.v1.2.017539.t1 (2e-014) | Lp_Unigene31865_All (3e-008) | non-classical DNA polymerase, dCMP transferase |
| symbB.v1.2.017542.t1 (1e-017) | Lp_Unigene55084_All (5e-053) | |||
| Lp_Unigene62480_All (6e-044) | ||||
| PolH/Rad30 | Unigene678_All (9e-062) | symbB.v1.2.015189.t1 (3e-054) | Lp_Unigene8962_All (3e-049) | DNA polymerase η/eta involved in the DNA repair by translesion synthesis |
| Unigene54870_All (1e-008) | symbB.v1.2.015189.t2 (9e-051) | |||
| symbB.v1.2.017537.t1 (3e-027) | ||||
| PolI/Rad30B | Unigene46925_All (8e-036) | symbB.v1.2.027247.t1 (6e-058) | Lp_Unigene39489_All (1e-056) | error-prone DNA polymerase ι/iota involved in bypass of DNA lesions |
| PolK/DINB1 | Unigene49999_All (1e-044) | symbB.v1.2.024275.t1 (1e-016) | Lp_Unigene16086_All (8e-040) | error-prone DNA polymerase κ/kappa involved in bypass of DNA lesions |
| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| FANCA | * | * | * | core complex member required for interstrand DNA cross-link repair |
| FANCB | * | * | * | ibid |
| FANCC | * | * | * | ibid |
| FANCE | * | * | * | ibid |
| FANCF | * | * | * | ibid |
| FANCG | * | * | * | ibid |
| FANCL | * | * | * | ibid |
| FANCM | * | * | * | lesion recognition and recruitment of the core complex |
| MHF1 | * | * | * | involved in the damage-dependent DNA binding of FANCM |
| MHF2 | * | * | * | involved in the damage-dependent DNA binding of FANCM |
| SNM1 | Unigene68129_All (9e-006) | symbB.v1.2.005478.t1 (5e-046) | Lp_Unigene56381_All (2e-063) | required for interstrand DNA cross-link repair |
| Unigene48769_All (6e-023) | ||||
| SNM1B | * | symbB.v1.2.023872.t2 (1e-024) | Lp_Unigene44216_All (4e-036) | related to SNM1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wong, J.T.Y. DNA Damage Response Pathways in Dinoflagellates. Microorganisms 2019, 7, 191. https://doi.org/10.3390/microorganisms7070191
Li C, Wong JTY. DNA Damage Response Pathways in Dinoflagellates. Microorganisms. 2019; 7(7):191. https://doi.org/10.3390/microorganisms7070191
Chicago/Turabian StyleLi, Chongping, and Joseph Tin Yum Wong. 2019. "DNA Damage Response Pathways in Dinoflagellates" Microorganisms 7, no. 7: 191. https://doi.org/10.3390/microorganisms7070191
APA StyleLi, C., & Wong, J. T. Y. (2019). DNA Damage Response Pathways in Dinoflagellates. Microorganisms, 7(7), 191. https://doi.org/10.3390/microorganisms7070191




