The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx
Abstract
1. Introduction
- Viruses sharing a common node in the phylogenetic tree.
- Monophyletic grouping with a bootstrap value of ≥60 at the clade-defining node (after 1000 neighbor-joining bootstrap replicates).
- Average percentage pairwise nucleotide distances between and within clades of >1.5% and <1.5%, respectively.
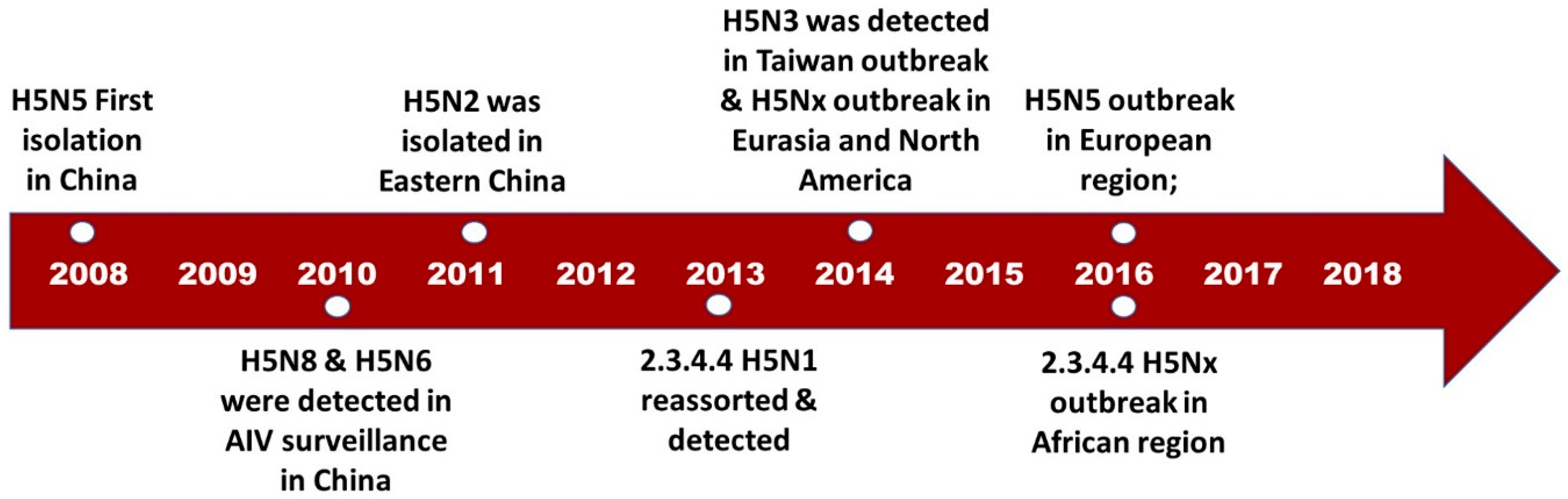
2. Emergence of H5Nx
3. Evolution of H5Nx Subtypes
3.1. H5N5
3.2. H5N8
3.3. H5N6
3.4. H5N2
3.5. H5N3
3.6. H5N1
3.7. H5Nx Provisional Grouping
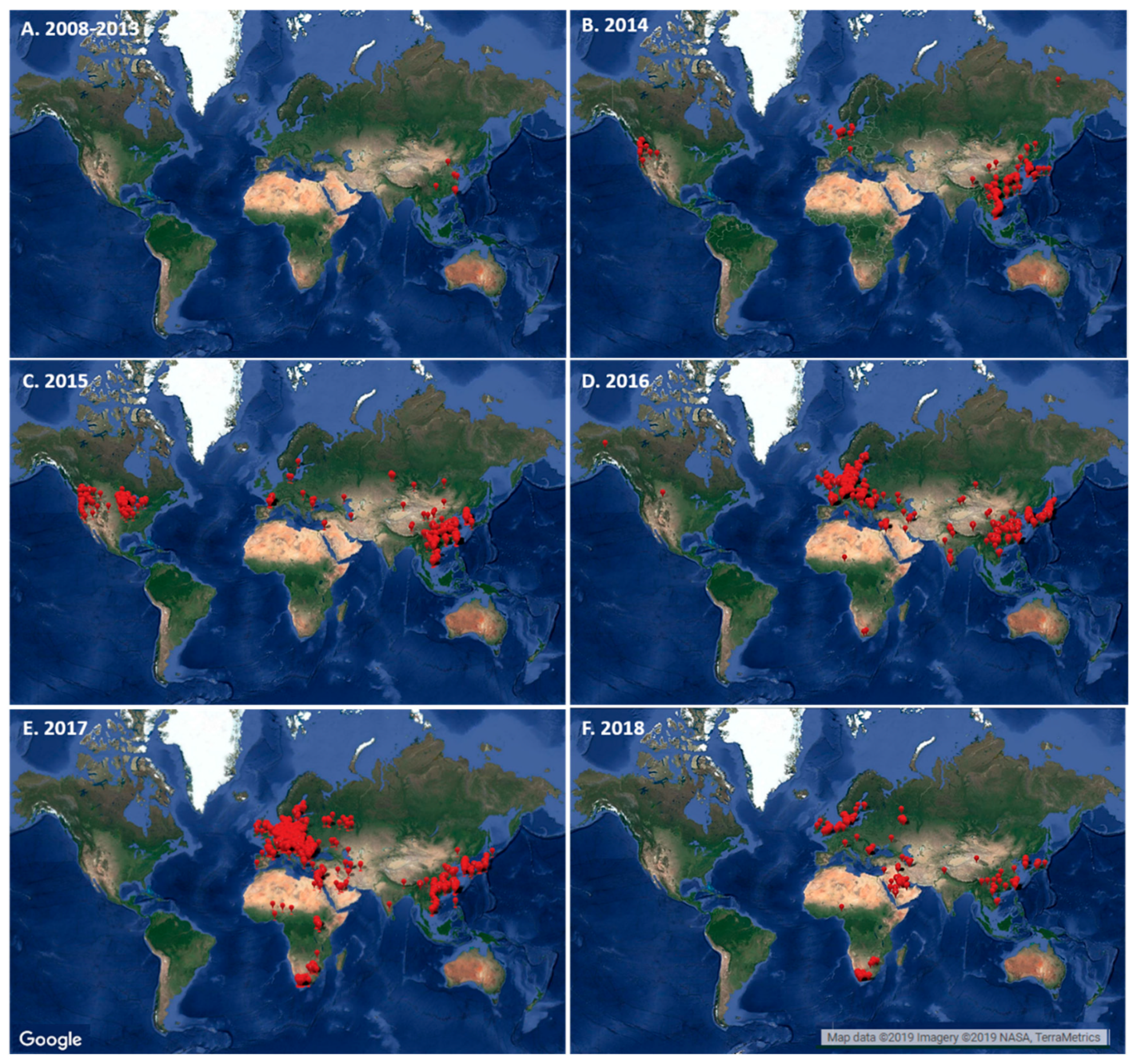
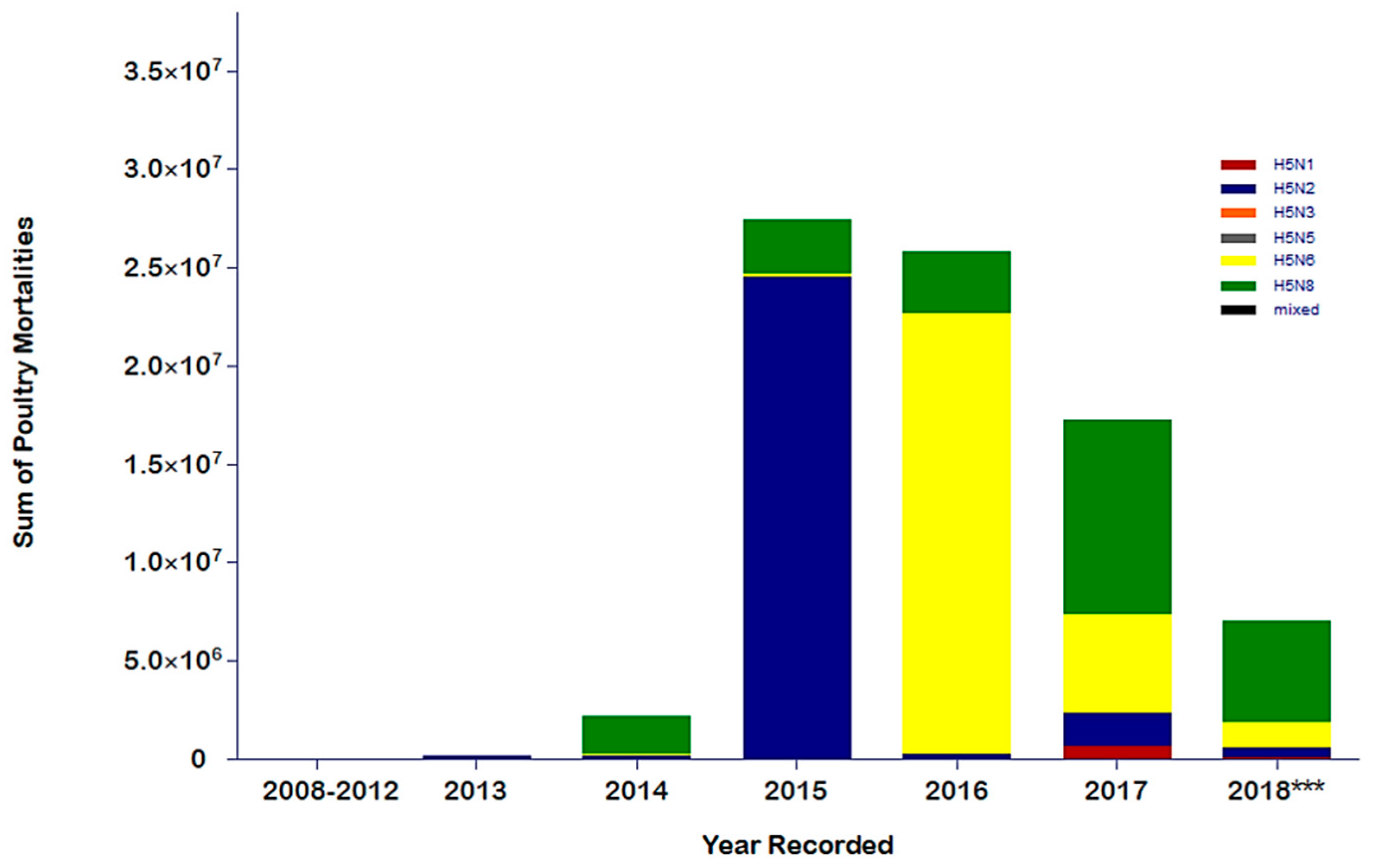
4. Evaluating the Decennary Distribution of H5Nx
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, M.S.; Artois, J.; Robinson, T.P.; Linard, C.; Chaiban, C.; Xenarios, I.; Engler, R.; Liechti, R.; Kuznetsov, D.; Xiao, X.; et al. Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation. eLife 2016, 5. [Google Scholar] [CrossRef]
- Bender, C.; Hall, H.; Huang, J.; Klimov, A.; Cox, N.; Hay, A.; Gregory, V.; Cameron, K.; Lim, W.; Subbarao, K. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997-1998. Virology 1999, 254, 115–123. [Google Scholar] [CrossRef]
- Duan, L.; Campitelli, L.; Fan, X.H.; Leung, Y.H.; Vijaykrishna, D.; Zhang, J.X.; Donatelli, I.; Delogu, M.; Li, K.S.; Foni, E.; et al. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: Implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 2007, 81, 7529–7539. [Google Scholar] [CrossRef] [PubMed]
- Hill, N.J.; Hussein, I.T.; Davis, K.R.; Ma, E.J.; Spivey, T.J.; Ramey, A.M.; Puryear, W.B.; Das, S.R.; Halpin, R.A.; Lin, X.; et al. Reassortment of Influenza A Viruses in Wild Birds in Alaska before H5 Clade 2.3.4.4 Outbreaks. Emerg. Infect. Dis. 2017, 23, 654–657. [Google Scholar] [CrossRef]
- Venkatesh, D.; Poen, M.J.; Bestebroer, T.M.; Scheuer, R.D.; Vuong, O.; Chkhaidze, M.; Machablishvili, A.; Mamuchadze, J.; Ninua, L.; Fedorova, N.B.; et al. Avian Influenza Viruses in Wild Birds: Virus Evolution in a Multihost Ecosystem. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. H5Nx Panzootic Bird Flu—Influenza’s Newest Worldwide Evolutionary Tour. Emerging infectious diseases 2017, 23, 340–342. [Google Scholar] [CrossRef]
- Lee, Y.N.; Lee, E.K.; Song, B.M.; Heo, G.B.; Woo, S.H.; Cheon, S.H.; Lee, Y.J. Evaluation of the zoonotic potential of multiple subgroups of clade 2.3.4.4 influenza A (H5N8) virus. Virology 2018, 516, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Bertran, K.; Kwon, J.H.; Swayne, D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017, 18, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Bahl, J.; Torchetti, M.K.; Killian, M.L.; Ip, H.S.; DeLiberto, T.J.; Swayne, D.E. Highly Pathogenic Avian Influenza Viruses and Generation of Novel Reassortants, United States, 2014-2015. Emerg. Infect. Dis 2016, 22, 1283–1285. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Zou, S.M.; Zhang, Y.; Bai, T.; Gao, R.B.; Zhang, X.; Wu, J.; Shu, Y.L. A novel reassortant H2N3 influenza virus isolated from China. Biomed. Environ. Sci. 2014, 27, 240–249. [Google Scholar] [CrossRef]
- Liu, C.G.; Liu, M.; Liu, F.; Lv, R.; Liu, D.F.; Qu, L.D.; Zhang, Y. Emerging multiple reassortant H5N5 avian influenza viruses in ducks, China, 2008. Vet. Microbiol. 2013, 167, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Highly pathogenic H5 avian influenza in 2016 and 2017—Observations and future perspectives. Focus On 2017. Available online: http://www.fao.org/3/a-i8068e.pdf (accessed on 24 May 2019).
- WHO/OIE/FAO, H.N.E.W.G. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect. Dis. 2008, 14. [Google Scholar] [CrossRef]
- WHO-OIE-FAO, H.E.W.G. Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: Divergence of clade 2.2 viruses. Influenza other Respir. Viruses 2009, 3, 59–62. [Google Scholar] [CrossRef]
- WHO/OIE/FAO, H.N.E.W.G. Continued evolution of highly pathogenic avian influenza A (H5N1): Updated nomenclature. Influenza Other Respir. Viruses 2011, 6, 1–5. [Google Scholar] [CrossRef]
- WHO/OIE/FAO, H.N.E.W.G. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir. Viruses 2014, 8, 384–388. [Google Scholar] [CrossRef]
- Claes, F.; Morzaria, S.P.; Donis, R.O. Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr. Opin. Virol. 2016, 16, 158–163. [Google Scholar] [CrossRef]
- Gu, M.; Zhao, G.; Zhao, K.; Zhong, L.; Huang, J.; Wan, H.; Wang, X.; Liu, W.; Liu, H.; Peng, D.; et al. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg. Infect. Dis. 2013, 19, 2021–2024. [Google Scholar] [CrossRef]
- Neumann, G.; Chen, H.; Gao, G.F.; Shu, Y.; Kawaoka, Y. H5N1 influenza viruses: Outbreaks and biological properties. Cell Res. 2010, 20, 51–61. [Google Scholar] [CrossRef]
- Ducatez, M.; Sonnberg, S.; Crumpton, J.C.; Rubrum, A.; Phommachanh, P.; Douangngeun, B.; Peiris, M.; Guan, Y.; Webster, R.; Webby, R. Highly pathogenic avian influenza H5N1 clade 2.3.2.1 and clade 2.3.4 viruses do not induce a clade-specific phenotype in mallard ducks. J. Gen. Virol. 2017, 98, 1232–1244. [Google Scholar] [CrossRef]
- Saito, T.; Tanikawa, T.; Uchida, Y.; Takemae, N.; Kanehira, K.; Tsunekuni, R. Intracontinental and intercontinental dissemination of Asian H5 highly pathogenic avian influenza virus (clade 2.3.4.4) in the winter of 2014-2015. Rev. Med Virol. 2015, 25, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, R.A.; Naughtin, M.J.; Horm, S.V.; San, S.; Buchy, P. A(H5N1) Virus Evolution in South East Asia. Viruses 2009, 1, 335–361. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Chen, Q.; Wang, Q.; Chen, J.; Jin, T.; Wong, G.; Quan, C.; Liu, J.; Wu, J.; Yin, R.; et al. Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China. Cell Host Microbe 2016, 20, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Liu, H.; Xiong, C.; Di, L.; Shi, W.; Li, M.; Liu, S.; Chen, J.; Chen, G.; Li, Y.; et al. Novel avian influenza A (H5N6) viruses isolated in migratory waterfowl before the first human case reported in China, 2014. Sci. Rep. 2016, 6, 29888. [Google Scholar] [CrossRef]
- Yang, H.; Carney, P.J.; Mishin, V.P.; Guo, Z.; Chang, J.C.; Wentworth, D.E.; Gubareva, L.V.; Stevens, J. Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses. J. Virol. 2016, 90, 5770–5784. [Google Scholar] [CrossRef]
- Bodewes, R.; Kuiken, T. Changing Role of Wild Birds in the Epidemiology of Avian Influenza A Viruses. Adv. Virus Res. 2018, 100, 279–307. [Google Scholar] [CrossRef]
- Chang, C.F.; King, C.C.; Wan, C.H.; Chang, Y.C.; Chan, T.C.; David Lee, C.C.; Chou, P.H.; Li, Z.R.; Li, Y.T.; Tseng, T.J.; et al. Lessons from the Largest Epidemic of Avian Influenza Viruses in Taiwan, 2015. Avian Dis. 2016, 60, 156–171. [Google Scholar] [CrossRef]
- El-Shesheny, R.; Barman, S.; Feeroz, M.M.; Hasan, M.K.; Jones-Engel, L.; Franks, J.; Turner, J.; Seiler, P.; Walker, D.; Friedman, K.; et al. Genesis of Influenza A(H5N8) Viruses. Emerg Infect. Dis. 2017, 23, 1368–1371. [Google Scholar] [CrossRef]
- Hu, T.; Song, J.; Zhang, W.; Zhao, H.; Duan, B.; Liu, Q.; Zeng, W.; Qiu, W.; Chen, G.; Zhang, Y.; et al. Emergence of novel clade 2.3.4 influenza A (H5N1) virus subgroups in Yunnan Province, China. Infect. Genet. Evol. 2015, 33, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Negovetich, N.J.; Forrest, H.L.; Webster, R.G. Ducks: The “Trojan horses” of H5N1 influenza. Influenza Other Respir. Viruses 2009, 3, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Lycett, S.J.; Bodewes, R.; Pohlmann, A.; Banks, J.; Bányai, K.; Boni, M.F.; Bouwstra, R.; Breed, A.C.; Brown, I.H.; Chen, H. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 2016, 354, 213–217. [Google Scholar]
- Yang, L.; Zhu, W.; Li, X.; Bo, H.; Zhang, Y.; Zou, S.; Gao, R.; Dong, J.; Zhao, X.; Chen, W.; et al. Genesis and Dissemination of Highly Pathogenic H5N6 Avian Influenza Viruses. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Z.; Li, H.; Wang, X.; Li, B.; Ren, X.; Zeng, Z.; Zhang, X.; Liu, S.; Hu, P.; et al. Biological Characterizations of H5Nx Avian Influenza Viruses Embodying Different Neuraminidases. Front. Microbiol. 2017, 8, 1084. [Google Scholar] [CrossRef]
- Bertran, K.; Lee, D.H.; Pantin-Jackwood, M.J.; Spackman, E.; Balzli, C.; Suarez, D.L.; Swayne, D.E. Pathobiology of Clade 2.3.4.4 H5Nx High-Pathogenicity Avian Influenza Virus Infections in Minor Gallinaceous Poultry Supports Early Backyard Flock Introductions in the Western United States in 2014-2015. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Smith, G.J.; Donis, R.O. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir. Viruses 2015, 9, 271–276. [Google Scholar] [CrossRef]
- Zhao, Z.; Guo, Z.; Zhang, C.; Liu, L.; Chen, L.; Wang, Z.; Fu, Y.; Li, J.; Shao, H.; Luo, Q.; et al. Avian Influenza H5N6 Viruses Exhibit Differing Pathogenicities and Transmissibilities in Mammals. Sci. Rep. 2017, 7, 16280. [Google Scholar] [CrossRef]
- Xu, W.; Dai, Y.; Hua, C.; Wang, Q.; Zou, P.; Deng, Q.; Jiang, S.; Lu, L. Genomic signature analysis of the recently emerged highly pathogenic A(H5N8) avian influenza virus: Implying an evolutionary trend for bird-to-human transmission. Microbes Infect. 2017, 19, 597–604. [Google Scholar] [CrossRef]
- Fan, S.; Zhou, L.; Wu, D.; Gao, X.; Pei, E.; Wang, T.; Gao, Y.; Xia, X. A novel highly pathogenic H5N8 avian influenza virus isolated from a wild duck in China. Influenza Other Respir. Viruses 2014, 8, 646–653. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Kang, H.-M.; Lee, E.-K.; Song, B.-M.; Jeong, J.; Kwon, Y.-K.; Kim, H.-R.; Lee, K.-J.; Hong, M.-S.; Jang, I. Novel reassortant influenza A (H5N8) viruses, South Korea, 2014. Emerg. Infect. Dis. 2014, 20, 1087. [Google Scholar] [CrossRef]
- Ma, L.; Jin, T.; Wang, H.; Liu, H.; Wang, R.; Li, Y.; Yang, G.; Xiong, Y.; Chen, J.; Zhang, J.; et al. Two reassortant types of highly pathogenic H5N8 avian influenza virus from wild birds in Central China in 2016. Emerg. Microbes Infect. 2018, 7, 14. [Google Scholar] [CrossRef]
- Ma, M.J.; Chen, S.H.; Wang, G.L.; Zhao, T.; Qian, Y.H.; Wu, M.N.; Liu, Y.; Gray, G.C.; Lu, B.; Cao, W.C. Novel Highly Pathogenic Avian H5 Influenza A Viruses in Live Poultry Markets, Wuxi City, China, 2013-2014. Open Forum Infect. Dis. 2016, 3, ofw054. [Google Scholar] [CrossRef]
- Song, Y.; Cui, J.; Song, H.; Ye, J.; Zhao, Z.; Wu, S.; Xu, C.; Jiao, P.; Liao, M. New reassortant H5N8 highly pathogenic avian influenza virus from waterfowl in Southern China. Front. Microbiol. 2015, 6, 1170. [Google Scholar] [CrossRef] [PubMed]
- Thanh, H.D.; Tran, V.T.; Nguyen, D.T.; Hung, V.K.; Kim, W. Novel reassortant H5N6 highly pathogenic influenza A viruses in Vietnamese quail outbreaks. Comp. Immunol. Microbiol. Infect. Dis 2018, 56, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Novel Highly Pathogenic Avian Influenza A(H5N6) Viruses in the Netherlands. Emerg. Infect. Dis. 2018, 24, 1393. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Gu, M.; Zhong, L.; Duan, Z.; Zhang, Y.; Zhu, Y.; Zhao, G.; Zhao, M.; Chen, Z.; Hu, S.; et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet. Microbiol. 2013, 163, 351–357. [Google Scholar] [CrossRef]
- Zhou, L.C.; Liu, J.; Pei, E.L.; Xue, W.J.; Lyu, J.M.; Cai, Y.T.; Wu, D.; Wu, W.; Liu, Y.Y.; Jin, H.Y.; et al. Novel Avian Influenza A(H5N8) Viruses in Migratory Birds, China, 2013-2014. Emerg. Infect. Dis. 2016, 22, 1121–1123. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Guo, X.; Li, S.; Yang, Y.; Jin, M. Complete genome sequence of a novel natural recombinant H5N5 influenza virus from ducks in central China. J. Virol. 2012, 86, 13878. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Vries, E.; Guo, H.; Dai, M.; Rottier, P.J.; van Kuppeveld, F.J.; de Haan, C.A. Rapid Emergence of Highly Pathogenic Avian Influenza Subtypes from a Subtype H5N1 Hemagglutinin Variant. Emerg. Infect. Dis. 2015, 21, 842–846. [Google Scholar] [CrossRef]
- Guo, H.; de Vries, E.; McBride, R.; Dekkers, J.; Peng, W.; Bouwman, K.M.; Nycholat, C.; Verheije, M.H.; Paulson, J.C.; van Kuppeveld, F.J.; et al. Highly Pathogenic Influenza A(H5Nx) Viruses with Altered H5 Receptor-Binding Specificity. Emerg. Infect. Dis 2017, 23, 220–231. [Google Scholar] [CrossRef]
- Sun, H.; Pu, J.; Hu, J.; Liu, L.; Xu, G.; Gao, G.F.; Liu, X.; Liu, J. Characterization of clade 2.3.4.4 highly pathogenic H5 avian influenza viruses in ducks and chickens. Vet. Microbiol. 2016, 182, 116–122. [Google Scholar] [CrossRef]
- Kang, Y.; Liu, L.; Feng, M.; Yuan, R.; Huang, C.; Tan, Y.; Gao, P.; Xiang, D.; Zhao, X.; Li, Y.; et al. Highly pathogenic H5N6 influenza A viruses recovered from wild birds in Guangdong, southern China, 2014-2015. Sci. Rep. 2017, 7, 44410. [Google Scholar] [CrossRef]
- Gu, M.; Liu, W.; Cao, Y.; Peng, D.; Wang, X.; Wan, H.; Zhao, G.; Xu, Q.; Zhang, W.; Song, Q.; et al. Novel Reassortant Highly Pathogenic Avian Influenza (H5N5) Viruses in Domestic Ducks, China. Emerg. Infect. Dis. 2011, 17, 1060–1063. [Google Scholar] [CrossRef]
- Wu, H.; Peng, X.; Xu, L.; Jin, C.; Cheng, L.; Lu, X.; Xie, T.; Yao, H.; Wu, N. Novel reassortant influenza A(H5N8) viruses in domestic ducks, eastern China. Emerg. Infect. Dis. 2014, 20, 1315–1318. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.; Monne, I.; Mulatti, P.; Zecchin, B.; Bonfanti, L.; Ormelli, S.; Milani, A.; Cecchettin, K.; Lemey, P.; Moreno, A.; et al. Genetic Diversity of Highly Pathogenic Avian Influenza A(H5N8/H5N5) Viruses in Italy, 2016–2017. Emerg. Infect. Dis. 2017, 23, 1543–1547. [Google Scholar] [CrossRef]
- Li, Q.; Wang, X.; Gao, Z.; Sun, Z.; Cui, Z.; Duan, Z.; Li, J.; Gu, M.; Hu, J.; Liu, X. Novel reassortant H5N5 viruses bind to a human-type receptor as a factor in pandemic risk. Vet. Microbiol. 2015, 175, 356–361. [Google Scholar] [CrossRef]
- Animal and Plant Health Agency (APHA), U. Situation assessment following detection and spread of H5N8 HPAI in EU Member States since October 2016. Available online: https://science.vla.gov.uk/flu-lab-net/docs/outbreak-hpai-h5n8-europe.pdf (accessed on 2014 May 2019).
- Disease Events of Avian Influenza 2004–2018. Available online: http://empres-i.fao.org/eipws3g/ (accessed on 28 May 2019).
- Adlhoch, C.; Gossner, C.; Koch, G.; Brown, I.; Bouwstra, R.; Verdonck, F.; Penttinen, P.; Harder, T. Comparing introduction to Europe of highly pathogenic avian influenza viruses A (H5N8) in 2014 and A (H5N1) in 2005. Eurosurveillance 2014, 19, 20996. [Google Scholar] [CrossRef]
- Lee, D.H.; Sharshov, K.; Swayne, D.E.; Kurskaya, O.; Sobolev, I.; Kabilov, M.; Alekseev, A.; Irza, V.; Shestopalov, A. Novel Reassortant Clade 2.3.4.4 Avian Influenza A(H5N8) Virus in Wild Aquatic Birds, Russia. Emerg. Infect. Dis. 2016, 23, 359–360. [Google Scholar] [CrossRef]
- Kim, Y.I.; Park, S.J.; Kwon, H.I.; Kim, E.H.; Si, Y.J.; Jeong, J.H.; Lee, I.W.; Nguyen, H.D.; Kwon, J.J.; Choi, W.S.; et al. Genetic and phylogenetic characterizations of a novel genotype of highly pathogenic avian influenza (HPAI) H5N8 viruses in 2016/2017 in South Korea. Infect. Genet. Evol. 2017, 53, 56–67. [Google Scholar] [CrossRef]
- Globig, A.; Starick, E.; Homeier, T.; Pohlmann, A.; Grund, C.; Wolf, P.; Zimmermann, A.; Wolf, C.; Heim, D.; Schlößer, H.; et al. Epidemiological and molecular analysis of an outbreak of highly pathogenic avian influenza H5N8 clade 2.3.4.4 in a German zoo: Effective disease control with minimal culling. Transbound Emerg. Dis. 2017, 11. [Google Scholar] [CrossRef]
- Li, M.; Liu, H.; Bi, Y.; Sun, J.; Wong, G.; Liu, D.; Li, L.; Liu, J.; Chen, Q.; Wang, H.; et al. Highly Pathogenic Avian Influenza A(H5N8) Virus in Wild Migratory Birds, Qinghai Lake, China. Emerg. Infect. Dis. 2017, 23, 637–641. [Google Scholar] [CrossRef]
- 2016–2018 Spread of H5N8 highly pathogenic avian influenza (HPAI) in sub-Saharan Africa: Epidemiological and ecological observations. Available online: http://www.fao.org/3/CA1209EN/ca1209en.pdf (accessed on 24 May 2019).
- Kim, Y.I.; Pascua, P.N.; Kwon, H.I.; Lim, G.J.; Kim, E.H.; Yoon, S.W.; Park, S.J.; Kim, S.M.; Choi, E.J.; Si, Y.J.; et al. Pathobiological features of a novel, highly pathogenic avian influenza A(H5N8) virus. Emerg Microbes Infect. 2014, 3, e75. [Google Scholar] [CrossRef]
- Verhagen, J.H.; van der Jeugd, H.P.; Nolet, B.A.; Slaterus, R.; Kharitonov, S.P.; de Vries, P.P.; Vuong, O.; Majoor, F.; Kuiken, T.; Fouchier, R.A. Wild bird surveillance around outbreaks of highly pathogenic avian influenza A(H5N8) virus in the Netherlands, 2014, within the context of global flyways. Eur. Surveill. 2015, 20, 21–32. [Google Scholar] [CrossRef]
- Choi, W.S.; Baek, Y.H.; Kwon, J.J.; Jeong, J.H.; Park, S.J.; Kim, Y.I.; Yoon, S.W.; Hwang, J.; Kim, M.H.; Kim, C.J.; et al. Rapid acquisition of polymorphic virulence markers during adaptation of highly pathogenic avian influenza H5N8 virus in the mouse. Sci Rep. 2017, 7, 40667. [Google Scholar] [CrossRef]
- Takemae, N.; Tsunekuni, R.; Sharshov, K.; Tanikawa, T.; Uchida, Y.; Ito, H.; Soda, K.; Usui, T.; Sobolev, I.; Shestopalov, A.; et al. Five distinct reassortants of H5N6 highly pathogenic avian influenza A viruses affected Japan during the winter of 2016-2017. Virology 2017, 512, 8–20. [Google Scholar] [CrossRef]
- Joob, B.; Viroj, W. H5N6 influenza virus infection, the newest influenza. Asian Pacific J. Trop. BioMed. 2015, 5, 434–437. [Google Scholar] [CrossRef]
- Zhao, G.; Gu, X.; Lu, X.; Pan, J.; Duan, Z.; Zhao, K.; Gu, M.; Liu, Q.; He, L.; Chen, J. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. PLoS ONE 2012, 7, e46183. [Google Scholar] [CrossRef]
- Jiao, P.; Wei, L.; Yuan, R.; Gong, L.; Cao, L.; Song, Y.; Luo, K.; Ren, T.; Liao, M. Complete genome sequence of an H5N2 avian influenza virus isolated from a parrot in Southern China. J. Virol. 2012, 86, 8890–8891. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Chen, L.H.; Chen, Y.P.; Liu, Y.P.; Li, W.C.; Lin, Y.L.; Lee, F. Highly pathogenic avian influenza viruses H5N2, H5N3, and H5N8 in Taiwan in 2015. Vet. Microbiol. 2016, 187, 50–57. [Google Scholar] [CrossRef]
- DeJesus, E.; Costa-Hurtado, M.; Smith, D.; Lee, D.H.; Spackman, E.; Kapczynski, D.R.; Torchetti, M.K.; Killian, M.L.; Suarez, D.L.; Swayne, D.E.; et al. Changes in adaptation of H5N2 highly pathogenic avian influenza H5 clade 2.3.4.4 viruses in chickens and mallards. Virology 2016, 499, 52–64. [Google Scholar] [CrossRef]
- Donatelli, I.; Campitelli, L.; Di Trani, L.; Puzelli, S.; Selli, L.; Fioretti, A.; Alexander, D.J.; Tollis, M.; Krauss, S.; Webster, R.G. Characterization of H5N2 influenza viruses from Italian poultry. J. Gen. Virol. 2001, 82, 623–630. [Google Scholar] [CrossRef]
- Wu, H.; Peng, X.; Xu, L.; Jin, C.; Cheng, L.; Lu, X.; Xie, T.; Yao, H.; Wu, N. Characterization of a novel highly pathogenic H5N2 avian influenza virus isolated from a duck in eastern China. Arch. Virol. 2014, 159, 3377–3383. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.S.; Torchetti, M.K.; Lager, K.M.; Webby, R.J.; Vincent, A.L. Absence of clinical disease and contact transmission of HPAI H5Nx clade 2.3.4.4 from North America in experimentally infected pigs. Influenza Other Respir. Viruses 2017, 11, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Pasick, J.; Berhane, Y.; Joseph, T.; Bowes, V.; Hisanaga, T.; Handel, K.; Alexandersen, S. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Sci. Rep. 2015, 5, 9484. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.H.; Jeong, S.; Lee, D.H.; Swayne, D.E.; Kim, Y.J.; Lee, S.H.; Noh, J.Y.; Erdene-Ochir, T.O.; Jeong, J.H.; Song, C.S. New Reassortant Clade 2.3.4.4b Avian Influenza A(H5N6) Virus in Wild Birds, South Korea, 2017–2018. Emerg. Infect. Dis. 2018, 24, 1953–1955. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.; Starick, E.; Grund, C.; Hoper, D.; Strebelow, G.; Globig, A.; Staubach, C.; Conraths, F.J.; Mettenleiter, T.C.; Harder, T.; et al. Swarm incursions of reassortants of highly pathogenic avian influenza virus strains H5N8 and H5N5, clade 2.3.4.4b, Germany, winter 2016/17. Sci. Rep. 2018, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Si, Y.J.; Kwon, H.I.; Kim, E.H.; Park, S.J.; Robles, N.J.; Nguyen, H.D.; Yu, M.A.; Yu, K.M.; Lee, Y.J.; et al. Pathogenicity and genetic characterisation of a novel reassortant, highly pathogenic avian influenza (HPAI) H5N6 virus isolated in Korea, 2017. Eur. Surveill. 2018, 23. [Google Scholar] [CrossRef]
- OIE. World Animal Health Information Database (WAHIS) Interface: Disease Information. Available online: http://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/statusdetail (accessed on 29 May 2019).
- Shin, J.H.; Woo, C.; Wang, S.J.; Jeong, J.; An, I.J.; Hwang, J.K.; Jo, S.D.; Yu, S.D.; Choi, K.; Chung, H.M.; et al. Prevalence of avian influenza virus in wild birds before and after the HPAI H5N8 outbreak in 2014 in South Korea. J. Microbiol. 2015, 53, 475–480. [Google Scholar] [CrossRef]
| Region | H5Nx Subtype Present |
|---|---|
| Africa | H5N8 |
| Americas | H5N1, H5N2, H5N8 |
| Asia | H5N1, H5N2, H5N3, H5N5, H5N6, H5N8 |
| Europe | H5N1, H5N2, H5N5, H5N6, H5N8 |
| Oceania and Antarctic | none |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antigua, K.J.C.; Choi, W.-S.; Baek, Y.H.; Song, M.-S. The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx. Microorganisms 2019, 7, 156. https://doi.org/10.3390/microorganisms7060156
Antigua KJC, Choi W-S, Baek YH, Song M-S. The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx. Microorganisms. 2019; 7(6):156. https://doi.org/10.3390/microorganisms7060156
Chicago/Turabian StyleAntigua, Khristine Joy C., Won-Suk Choi, Yun Hee Baek, and Min-Suk Song. 2019. "The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx" Microorganisms 7, no. 6: 156. https://doi.org/10.3390/microorganisms7060156
APA StyleAntigua, K. J. C., Choi, W.-S., Baek, Y. H., & Song, M.-S. (2019). The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx. Microorganisms, 7(6), 156. https://doi.org/10.3390/microorganisms7060156





