Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation
Abstract
1. Introduction
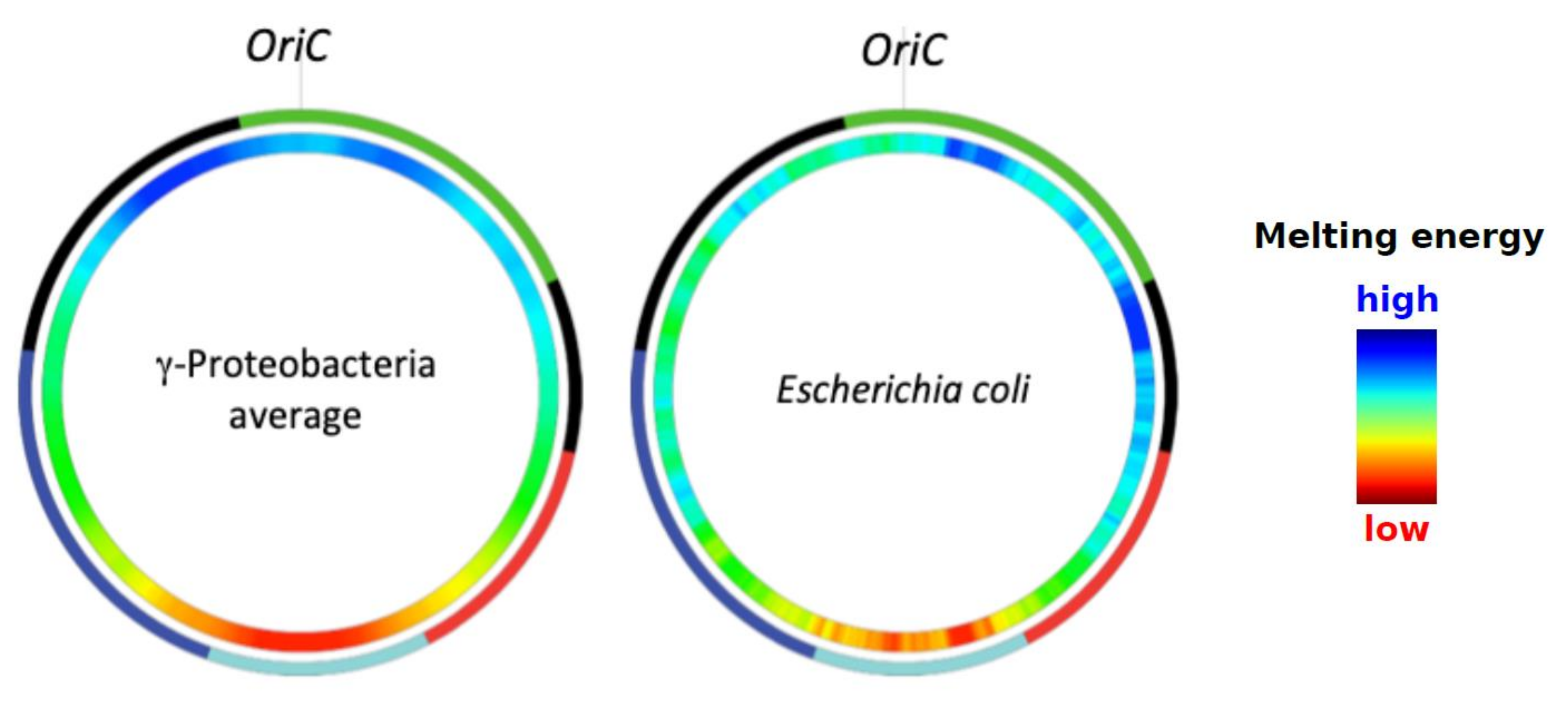
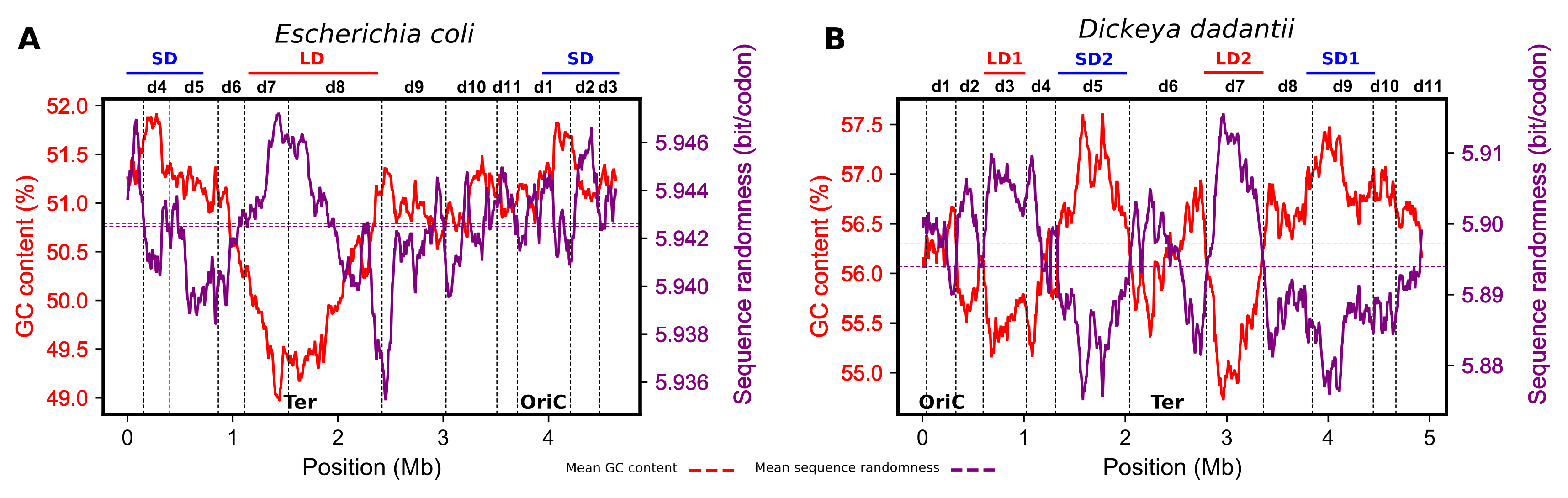
2. Model of Regulation of Gene Expression during the E. coli Growth Cycle—A Brief Overview
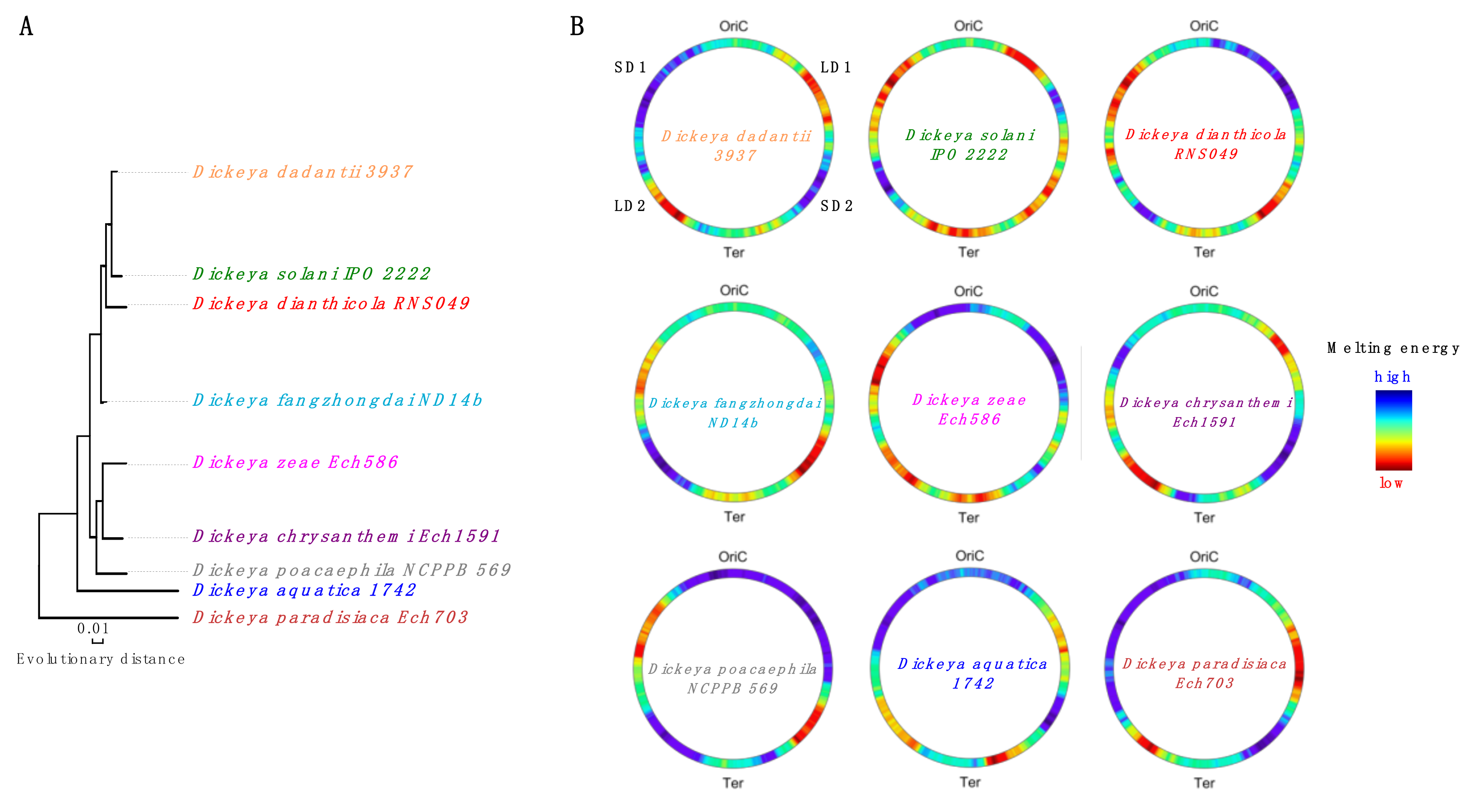
3. Model of Regulation of Gene Expression during the Pathogenic Growth of D. dadantii
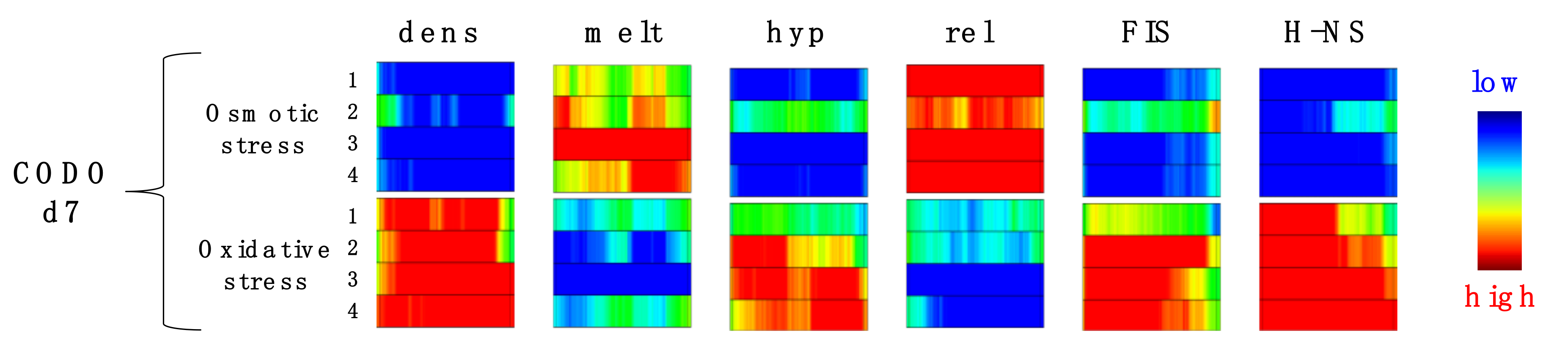
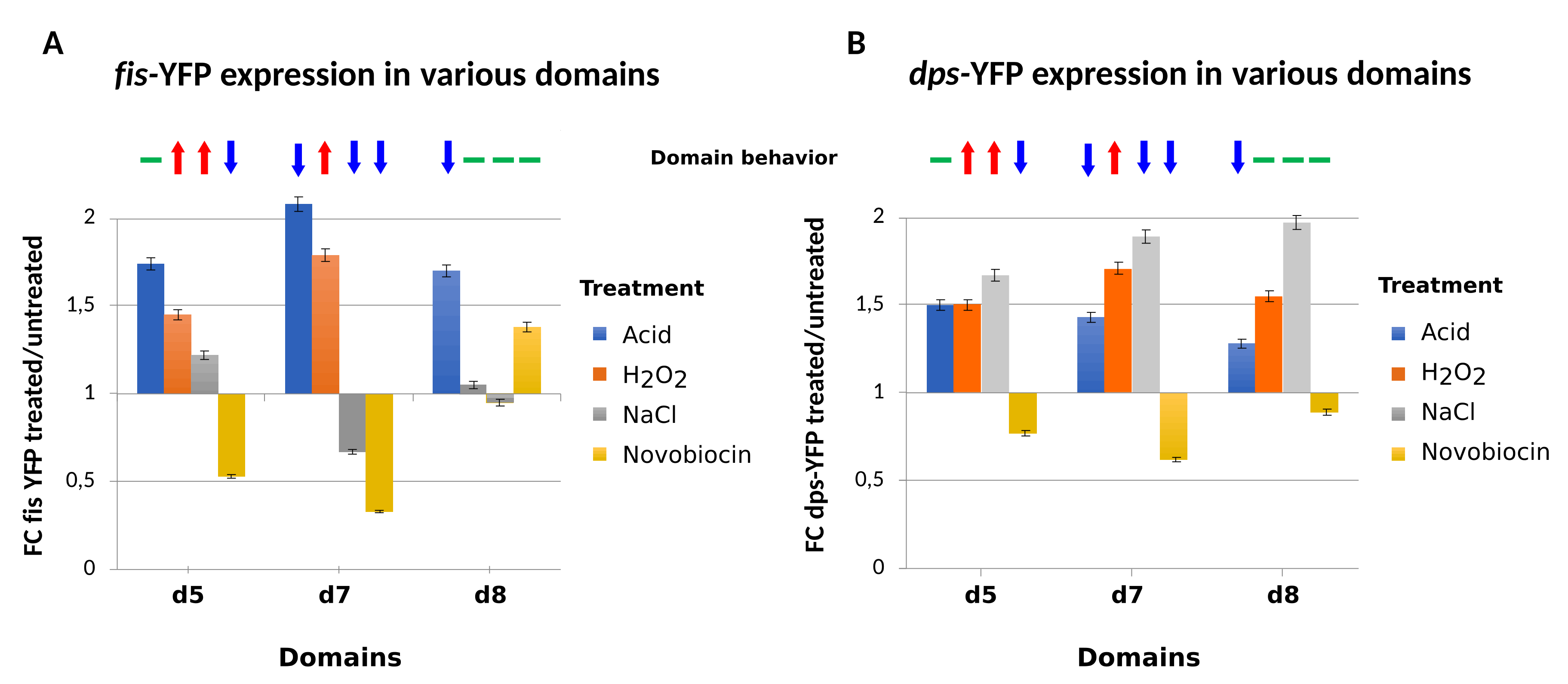
4. Testing the Dynamical Behavior of CODOs by Reporter Constructs.
5. Conclusions
Funding
Conflicts of Interest
References
- Reverchon, S.; Nasser, W. Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 2013, 5, 622–636. [Google Scholar] [PubMed]
- Wade, J.T.; Grainger, D.C. Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014, 12, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.; Gerganova, V.; Berger, P.; Rapiteanu, R.; Lisicovas, V.; Dobrindt, U. Genes on a Wire: The Nucleoid-Associated Protein HU Insulates Transcription Units in Escherichia coli. Sci. Rep. 2016, 6, 31512. [Google Scholar] [CrossRef] [PubMed]
- Junier, I.; Rivoire, O. Conserved Units of Co-Expression in Bacterial Genomes: An Evolutionary Insight into Transcriptional Regulation. PLoS ONE 2016, 11, e0155740. [Google Scholar] [CrossRef] [PubMed]
- Junier, I.; Fremont, P.; Rivoire, O. Universal and idiosyncratic characteristic lengths in bacterial genomes. Phys. Biol. 2018, 15, 035001. [Google Scholar] [CrossRef] [PubMed]
- Yus, E.; Llorens-Rico, V.; Martinez, S.; Gallo, C.; Eilers, H.; Blotz, C.; Stulke, J.; Lluch-Senar, M.; Serrano, L. Determination of the Gene Regulatory Network of a Genome-Reduced Bacterium Highlights Alternative Regulation Independent of Transcription Factors. Cell Syst. 2019, 9, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Sellars, L.E.; Busby, S.J.; Lee, D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014, 42, 11383–11392. [Google Scholar] [CrossRef]
- Brambilla, E.; Sclavi, B. Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 Genes Genomes Genet. 2015, 5, 605–614. [Google Scholar] [CrossRef]
- Scholz, S.A.; Diao, R.; Wolfe, M.B.; Fivenson, E.M.; Lin, X.N.; Freddolino, P.L. High-Resolution Mapping of the Escherichia coli Chromosome Reveals Positions of High and Low Transcription. Cell Syst. 2019, 8, 212–225. [Google Scholar] [CrossRef]
- Hatfield, G.W.; Benham, C.J. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002, 36, 175–203. [Google Scholar] [CrossRef]
- Blot, N.; Mavathur, R.; Geertz, M.; Travers, A.; Muskhelishvili, G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006, 7, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Marr, C.; Geertz, M.; Hutt, M.T.; Muskhelishvili, G. Dissecting the logical types of network control in gene expression profiles. BMC Syst. Biol. 2008, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, N.; Hutt, M.T.; Stoyan, H.; Stoyan, D. Ranges of control in the transcriptional regulation of Escherichia coli. BMC Syst. Biol. 2009, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, N.; Geertz, M.; Muskhelishvili, G.; Hutt, M.T. Analog regulation of metabolic demand. BMC Syst. Biol. 2011, 5, 40. [Google Scholar] [CrossRef]
- Berthoumieux, S.; de Jong, H.; Baptist, G.; Pinel, C.; Ranquet, C.; Ropers, D.; Geiselmann, J. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol. Syst. Biol. 2013, 9, 634. [Google Scholar] [CrossRef]
- Meyer, S.; Beslon, G. Torsion-mediated interaction between adjacent genes. PLoS Comput. Biol. 2014, 10, e1003785. [Google Scholar] [CrossRef]
- Sobetzko, P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016, 44, 1514–1524. [Google Scholar] [CrossRef]
- El Houdaigui, B.; Forquet, R.; Hindre, T.; Schneider, D.; Nasser, W.; Reverchon, S.; Meyer, S. Bacterial genome architecture shapes global transcriptional regulation by DNA supercoiling. Nucleic Acids Res. 2019, 47, 5648–5657. [Google Scholar] [CrossRef]
- Jeong, K.S.; Ahn, J.; Khodursky, A.B. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004, 5, R86. [Google Scholar] [CrossRef]
- Peter, B.J.; Arsuaga, J.; Breier, A.M.; Khodursky, A.B.; Brown, P.O.; Cozzarelli, N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004, 5, R87. [Google Scholar] [CrossRef]
- Berger, M.; Farcas, A.; Geertz, M.; Zhelyazkova, P.; Brix, K.; Travers, A.; Muskhelishvili, G. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010, 11, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Ferrandiz, M.J.; Martin-Galiano, A.J.; Schvartzman, J.B.; de la Campa, A.G. The genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 2010, 38, 3570–3581. [Google Scholar] [CrossRef] [PubMed]
- Sobetzko, P.; Glinkowska, M.; Travers, A.; Muskhelishvili, G. DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol. Biosyst. 2013, 9, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sobetzko, P.; Nasser, W.; Reverchon, S.; Muskhelishvili, G. Chromosomal “stress-response” domains govern the spatiotemporal expression of the bacterial virulence program. MBio 2015, 6, e00353-15. [Google Scholar] [CrossRef]
- Meyer, S.; Reverchon, S.; Nasser, W.; Muskhelishvili, G. Chromosomal organization of transcription: In a nutshell. Curr. Genet. 2018, 64, 555–565. [Google Scholar] [CrossRef]
- Reverchon, S.; Nasser, W.; Sobetzko, P.; Muskhelishvili, G. Rethinking the bacterial genetic regulation. Biochem. Anal. Biochem. 2015, 4, 1. [Google Scholar] [CrossRef]
- Booker, B.M.; Deng, S.; Higgins, N.P. DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2010, 78, 1348–1364. [Google Scholar] [CrossRef]
- Le, T.B.; Imakaev, M.V.; Mirny, L.A.; Laub, M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013, 342, 731–734. [Google Scholar] [CrossRef]
- Fritsche, M.; Li, S.; Heermann, D.W.; Wiggins, P.A. A model for Escherichia coli chromosome packaging supports transcription factor-induced DNA domain formation. Nucleic Acids Res. 2012, 40, 972–980. [Google Scholar] [CrossRef]
- Dame, R.T.; Kalmykowa, O.J.; Grainger, D.C. Chromosomal macrodomains and associated proteins: Implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 2011, 7, e1002123. [Google Scholar] [CrossRef]
- Scolari, V.F.; Bassetti, B.; Sclavi, B.; Lagomarsino, M.C. Gene clusters reflecting macrodomain structure respond to nucleoid perturbations. Mol. Biosyst. 2011, 7, 878–888. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lioy, V.S.; Cournac, A.; Marbouty, M.; Duigou, S.; Mozziconacci, J.; Espeli, O.; Boccard, F.; Koszul, R. Multiscale Structuring of the E. coli Chromosome by Nucleoid-Associated and Condensin Proteins. Cell 2018, 172, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Kaper, J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef] [PubMed]
- Bouyioukos, C.; Reverchon, S.; Kepes, F. From multiple pathogenicity islands to a unique organized pathogenicity archipelago. Sci. Rep. 2016, 6, 27978. [Google Scholar] [CrossRef]
- Crooke, E.; Hwang, D.S.; Skarstad, K.; Thony, B.; Kornberg, A.E. coli minichromosome replication: Regulation of initiation at oriC. Res. Microbiol. 1991, 142, 127–130. [Google Scholar] [CrossRef]
- Gille, H.; Egan, J.B.; Roth, A.; Messer, W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991, 19, 4167–4172. [Google Scholar] [CrossRef]
- Margulies, C.; Kaguni, J.M. The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 1998, 26, 5170–5175. [Google Scholar] [CrossRef][Green Version]
- Ryan, V.T.; Grimwade, J.E.; Nievera, C.J.; Leonard, A.C. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 2002, 46, 113–124. [Google Scholar] [CrossRef]
- Ryan, V.T.; Grimwade, J.E.; Camara, J.E.; Crooke, E.; Leonard, A.C. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 2004, 51, 1347–1359. [Google Scholar] [CrossRef]
- Chodavarapu, S.; Gomez, R.; Vicente, M.; Kaguni, J.M. Escherichia coli Dps interacts with DnaA protein to impede initiation: A model of adaptive mutation. Mol. Microbiol. 2008, 67, 1331–1346. [Google Scholar] [CrossRef]
- Magnan, D.; Bates, D. Regulation of DNA Replication Initiation by Chromosome Structure. J. Bacteriol. 2015, 197, 3370–3377. [Google Scholar] [CrossRef] [PubMed]
- Wolanski, M.; Donczew, R.; Zawilak-Pawlik, A.; Zakrzewska-Czerwinska, J. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2014, 5, 735. [Google Scholar] [PubMed]
- Van Workum, M.; van Dooren, S.J.; Oldenburg, N.; Molenaar, D.; Jensen, P.R.; Snoep, J.L.; Westerhoff, H.V. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol. Microbiol. 1996, 20, 351–360. [Google Scholar] [CrossRef]
- Mukherjee, K.; Nagai, H.; Shimamoto, N.; Chatterji, D. GroEL is involved in activation of Escherichia coli RNA polymerase devoid of the omega subunit in vivo. Eur. J. Biochem. 1999, 266, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Minakhin, L.; Bhagat, S.; Brunning, A.; Campbell, E.A.; Darst, S.A.; Ebright, R.H.; Severinov, K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Oostra, B.A.; van Vliet, A.J.; Ab, G.; Gruber, M. Enhancement of ribosomal ribonucleic acid synthesis by deoxyribonucleic acid gyrase activity in Escherichia coli. J. Bacteriol. 1981, 148, 782–787. [Google Scholar]
- Bosch, L.; Nilsson, L.; Vijgenboom, E.; Verbeek, H. FIS-dependent trans-activation of tRNA and rRNA operons of Escherichia coli. Biochim. Biophys. Acta 1990, 1050, 293–301. [Google Scholar] [CrossRef]
- Ross, W.; Thompson, J.F.; Newlands, J.T.; Gourse, R.L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990, 9, 3733–3742. [Google Scholar] [CrossRef]
- Ohlsen, K.L.; Gralla, J.D. Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli ribosomal RNA rrnB P1 promoter. Mol. Microbiol. 1992, 6, 2243–2251. [Google Scholar] [CrossRef]
- Sander, P.; Langert, W.; Mueller, K. Mechanisms of upstream activation of the rrnD promoter P1 of Escherichia coli. J. Biol. Chem. 1993, 268, 16907–16916. [Google Scholar]
- Appleman, J.A.; Ross, W.; Salomon, J.; Gourse, R.L. Activation of Escherichia coli rRNA transcription by FIS during a growth cycle. J. Bacteriol. 1998, 180, 1525–1532. [Google Scholar] [PubMed]
- Schneider, R.; Travers, A.; Muskhelishvili, G. The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol. Microbiol. 2000, 38, 167–175. [Google Scholar] [CrossRef]
- Zhi, H.; Wang, X.; Cabrera, J.E.; Johnson, R.C.; Jin, D.J. Fis stabilizes the interaction between RNA polymerase and the ribosomal promoter rrnB P1, leading to transcriptional activation. J. Biol. Chem. 2003, 278, 47340–47349. [Google Scholar] [CrossRef]
- Zhang, X.; Bremer, H. Effects of Fis on ribosome synthesis and activity and on rRNA promoter activities in Escherichia coli. J. Mol. Biol. 1996, 259, 27–40. [Google Scholar] [CrossRef]
- Choi, H.S.; Kim, K.S.; Park, J.W.; Jung, Y.H.; Lee, Y. Effects of FIS protein on rnpB transcription in Escherichia coli. Mol. Cells 2005, 19, 239–245. [Google Scholar]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef]
- Yu, H.; Droge, P. Replication-induced supercoiling: A neglected DNA transaction regulator? Trends Biochem. Sci. 2014, 39, 219–220. [Google Scholar] [CrossRef]
- Muskhelishvili, G.; Travers, A. DNA structure and bacterial nucleoid-associated proteins. In Bacterial Gene Regulation and Transcriptional Networks; Babu, M.M., Ed.; Horizon Scientific Press: Norwich, UK, 2013. [Google Scholar]
- Yang, Y.; Ames, G.F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc. Natl. Acad. Sci. USA 1988, 85, 8850–8854. [Google Scholar] [CrossRef]
- Sutormin, D.; Rubanova, N.; Logacheva, M.; Ghilarov, D.; Severinov, K. Single-nucleotide-resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome. Nucleic Acids Res. 2019, 47, 1373–1388. [Google Scholar] [CrossRef]
- Sobetzko, P.; Travers, A.; Muskhelishvili, G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. USA 2012, 109, E42–E50. [Google Scholar] [CrossRef]
- Ferrandiz, M.J.; Carreno, D.; Ayora, S.; de la Campa, A.G. HU of Streptococcus pneumoniae Is Essential for the Preservation of DNA Supercoiling. Front. Microbiol. 2018, 9, 493. [Google Scholar] [CrossRef]
- Rochman, M.; Aviv, M.; Glaser, G.; Muskhelishvili, G. Promoter protection by a transcription factor acting as a local topological homeostat. EMBO Rep. 2002, 3, 355–360. [Google Scholar] [CrossRef]
- Muskhelishvili, G.; Travers, A. Transcription factor as a topological homeostat. Front. Biosci. 2003, 8, 279–285. [Google Scholar] [CrossRef]
- Ishihama, A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000, 54, 499–518. [Google Scholar] [CrossRef]
- Typas, A.; Becker, G.; Hengge, R. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 2007, 63, 1296–1306. [Google Scholar] [CrossRef]
- Typas, A.; Hengge, R. Differential ability of sigma(s) and sigma70 of Escherichia coli to utilize promoters containing half or full UP-element sites. Mol. Microbiol. 2005, 55, 250–260. [Google Scholar] [CrossRef]
- Ohniwa, R.L.; Morikawa, K.; Kim, J.; Ohta, T.; Ishihama, A.; Wada, C.; Takeyasu, K. Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J. 2006, 25, 5591–5602. [Google Scholar] [CrossRef]
- Pul, U.; Wurm, R.; Lux, B.; Meltzer, M.; Menzel, A.; Wagner, R. LRP and H-NS--cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol. Microbiol. 2005, 58, 864–876. [Google Scholar] [CrossRef]
- Lal, A.; Dhar, A.; Trostel, A.; Kouzine, F.; Seshasayee, A.S.; Adhya, S. Genome scale patterns of supercoiling in a bacterial chromosome. Nat. Commun. 2016, 7, 11055. [Google Scholar] [CrossRef]
- Nigatu, D.; Henkel, W.; Sobetzko, P.; Muskhelishvili, G. Relationship between digital information and thermodynamic stability in bacterial genomes. EURASIP J. Bioinform. Syst. Biol. 2016, 2016, 4. [Google Scholar] [CrossRef]
- Brinza, L.; Calevro, F.; Charles, H. Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera. BMC Genom. 2013, 14, 73. [Google Scholar] [CrossRef]
- Muskhelishvili, G.; Travers, A. Order from the order: How a spatiotemporal genetic program is encoded in a 2D genetic map of the bacterial chromosome. J. Mol. Microbiol. Biotechnol. 2014, 24, 332–343. [Google Scholar] [CrossRef]
- Jiang, X.; Zghidi-Abouzid, O.; Oger-Desfeux, C.; Hommais, F.; Greliche, N.; Muskhelishvili, G.; Nasser, W.; Reverchon, S. Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ. Microbiol. 2016, 18, 3651–3672. [Google Scholar] [CrossRef] [PubMed]
- Reverchon, S.; Muskhelisvili, G.; Nasser, W. Virulence Program of a Bacterial Plant Pathogen: The Dickeya Model. Prog. Mol. Biol. Transl. Sci. 2016, 142, 51–92. [Google Scholar] [PubMed]
- Duprey, A.; Taib, N.; Leonard, S.; Garin, T.; Flandrois, J.P.; Nasser, W.; Brochier-Armanet, C.; Reverchon, S. The phytopathogenic nature of Dickeya aquatica 174/2 and the dynamic early evolution of Dickeya pathogenicity. Environ. Microbiol. 2019, 21, 2809–2835. [Google Scholar] [CrossRef] [PubMed]
- SantaLucia, J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Ricci, V.; Whitehead, R.; Patel, M.; Fookes, M.; Ivens, A.; Piddock, L.J. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio 2013, 4, e00273-12. [Google Scholar] [CrossRef]
- Szafran, M.J.; Gongerowska, M.; Malecki, T.; Elliot, M.; Jakimowicz, D. Transcriptional Response of Streptomyces coelicolor to Rapid Chromosome Relaxation or Long-Term Supercoiling Imbalance. Front. Microbiol. 2019, 10, 1605. [Google Scholar] [CrossRef]
- Ferrandiz, M.J.; Arnanz, C.; Martin-Galiano, A.J.; Rodriguez-Martin, C.; de la Campa, A.G. Role of global and local topology in the regulation of gene expression in Streptococcus pneumoniae. PLoS ONE 2014, 9, e101574. [Google Scholar] [CrossRef]
- Sutherland, L.; Cairney, J.; Elmore, M.J.; Booth, I.R.; Higgins, C.F. Osmotic regulation of transcription: Induction of the proU betaine transport gene is dependent on accumulation of intracellular potassium. J. Bacteriol. 1986, 168, 805–814. [Google Scholar] [CrossRef]
- Dinnbier, U.; Limpinsel, E.; Schmid, R.; Bakker, E.P. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 1988, 150, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.K.; Gorin, A.A.; Lu, X.J.; Hock, L.M.; Zhurkin, V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 11163–11168. [Google Scholar] [CrossRef] [PubMed]
- Brogaard, K.; Xi, L.; Wang, J.P.; Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 2012, 486, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Cagliero, C.; Izard, J.; Chen, Y.; Zhou, Y.N.; Heinz, W.F.; Schneider, T.D.; Jin, D.J. Density of sigma70 promoter-like sites in the intergenic regions dictates the redistribution of RNA polymerase during osmotic stress in Escherichia coli. Nucleic Acids Res. 2019, 47, 3970–3985. [Google Scholar] [CrossRef]
- Schneider, R.; Travers, A.; Muskhelishvili, G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol. Microbiol. 1997, 26, 519–530. [Google Scholar] [CrossRef]
- Liu, G.; Ma, Q.; Xu, Y. Physical properties of DNA may direct the binding of nucleoid-associated proteins along the E. coli genome. Math. Biosci. 2018, 301, 50–58. [Google Scholar] [CrossRef]
- Ouafa, Z.A.; Reverchon, S.; Lautier, T.; Muskhelishvili, G.; Nasser, W. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 2012, 40, 4306–4319. [Google Scholar] [CrossRef]
- Duprey, A.; Muskhelishvili, G.; Reverchon, S.; Nasser, W. Temporal control of Dickeya dadantii main virulence gene expression by growth phase-dependent alteration of regulatory nucleoprotein complexes. Biochim. Biophys. Acta 2016, 1859, 1470–1480. [Google Scholar] [CrossRef]
- Japaridze, A.; Muskhelishvili, G.; Benedetti, F.; Gavriilidou, A.F.; Zenobi, R.; De Los Rios, P.; Longo, G.; Dietler, G. Hyperplectonemes: A Higher Order Compact and Dynamic DNA Self-Organization. Nano Lett. 2017, 17, 1938–1948. [Google Scholar] [CrossRef]
- Dorman, C.J. H-NS, the genome sentinel. Nat. Rev. Microbiol. 2007, 5, 157–161. [Google Scholar] [CrossRef]
- Japaridze, A.; Renevey, S.; Sobetzko, P.; Stoliar, L.; Nasser, W.; Dietler, G.; Muskhelishvili, G. Spatial organization of DNA sequences directs the assembly of bacterial chromatin by a nucleoid-associated protein. J. Biol. Chem. 2017, 292, 7607–7618. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, H.; Kenney, L.J.; Yan, J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010, 24, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, R.A.; Vreede, J.; Qin, L.; Moolenaar, G.F.; Hofmann, A.; Goosen, N.; Dame, R.T. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. Elife 2017, 6, e27369. [Google Scholar] [CrossRef] [PubMed]
- Joyeux, M. Role of Salt Valency in the Switch of H-NS Proteins between DNA-Bridging and DNA-Stiffening Modes. Biophys. J. 2018, 114, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Will, W.R.; Whitham, P.J.; Reid, P.J.; Fang, F.C. Modulation of H-NS transcriptional silencing by magnesium. Nucleic Acids Res. 2018, 46, 5717–5725. [Google Scholar] [CrossRef] [PubMed]
- Rafiei, N.; Cordova, M.; Navarre, W.W.; Milstein, J.N. Growth Phase Dependent Chromosome Condensation and H-NS Protein Redistribution in Escherichia coli Under Osmotic Stress. J. Bacteriol. 2019, 201, e00469-19. [Google Scholar] [CrossRef]
- Lang, B.; Blot, N.; Bouffartigues, E.; Buckle, M.; Geertz, M.; Gualerzi, C.O.; Mavathur, R.; Muskhelishvili, G.; Pon, C.L.; Rimsky, S.; et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007, 35, 6330–6337. [Google Scholar] [CrossRef]
- Maurer, S.; Fritz, J.; Muskhelishvili, G. A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J. Mol. Biol. 2009, 387, 1261–1276. [Google Scholar] [CrossRef]
- Price, M.N.; Deutschbauer, A.M.; Skerker, J.M.; Wetmore, K.M.; Ruths, T.; Mar, J.S.; Kuehl, J.V.; Shao, W.; Arkin, A.P. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol. Syst. Biol. 2013, 9, 660. [Google Scholar] [CrossRef]
- Helmann, T.C.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. USA 2019, 116, 18900–18910. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muskhelishvili, G.; Forquet, R.; Reverchon, S.; Meyer, S.; Nasser, W. Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation. Microorganisms 2019, 7, 694. https://doi.org/10.3390/microorganisms7120694
Muskhelishvili G, Forquet R, Reverchon S, Meyer S, Nasser W. Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation. Microorganisms. 2019; 7(12):694. https://doi.org/10.3390/microorganisms7120694
Chicago/Turabian StyleMuskhelishvili, Georgi, Raphaël Forquet, Sylvie Reverchon, Sam Meyer, and William Nasser. 2019. "Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation" Microorganisms 7, no. 12: 694. https://doi.org/10.3390/microorganisms7120694
APA StyleMuskhelishvili, G., Forquet, R., Reverchon, S., Meyer, S., & Nasser, W. (2019). Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation. Microorganisms, 7(12), 694. https://doi.org/10.3390/microorganisms7120694




