Global Transcriptional Response of Three Highly Acid-Tolerant Field Strains of Listeria monocytogenes to HCl Stress
Abstract
1. Introduction
2. Results and Discussion
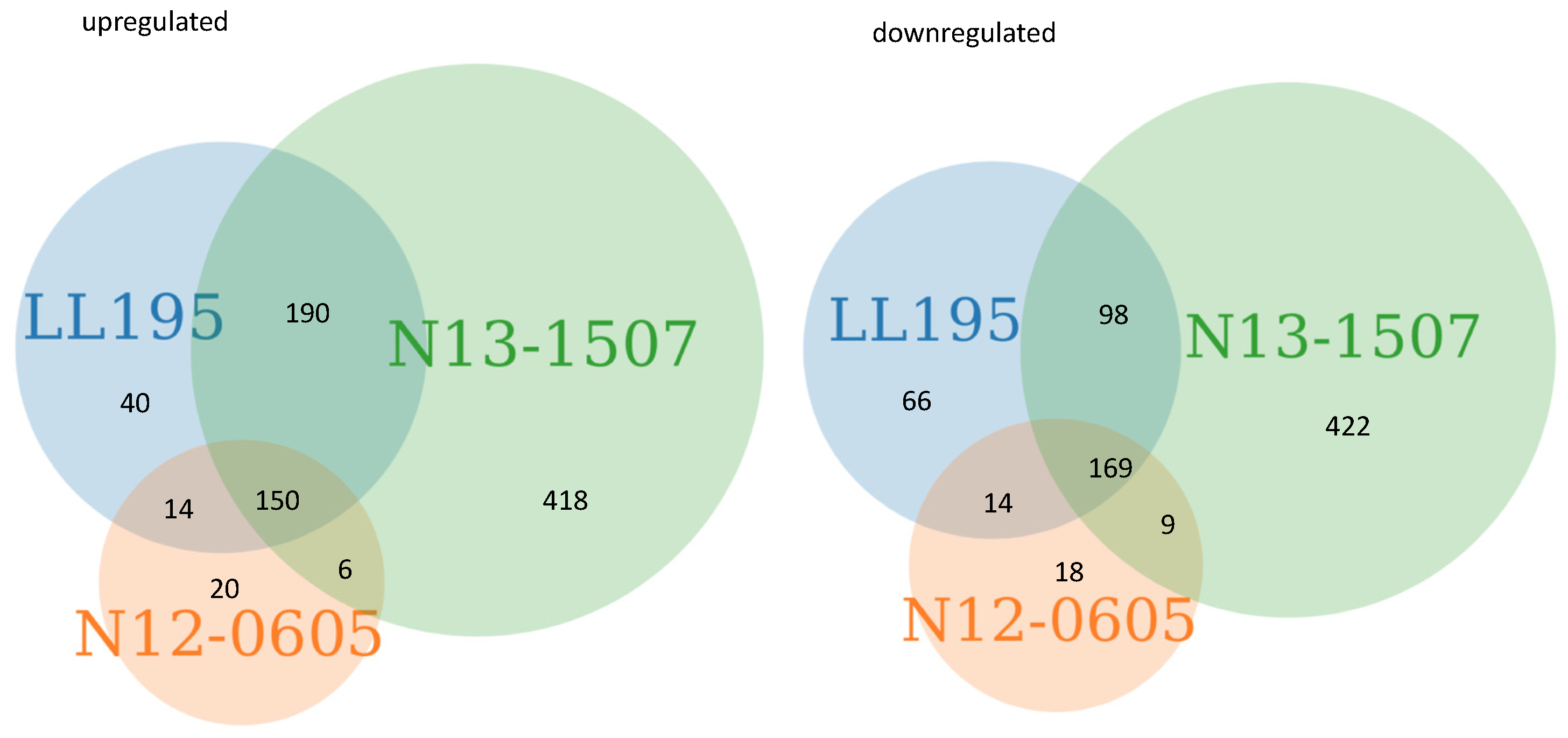
2.1. General Response of 3 Strains to Acid Stress
2.2. Components of the Acid Response in L. monocytogenes were Differentially Expressed after Acid Stress
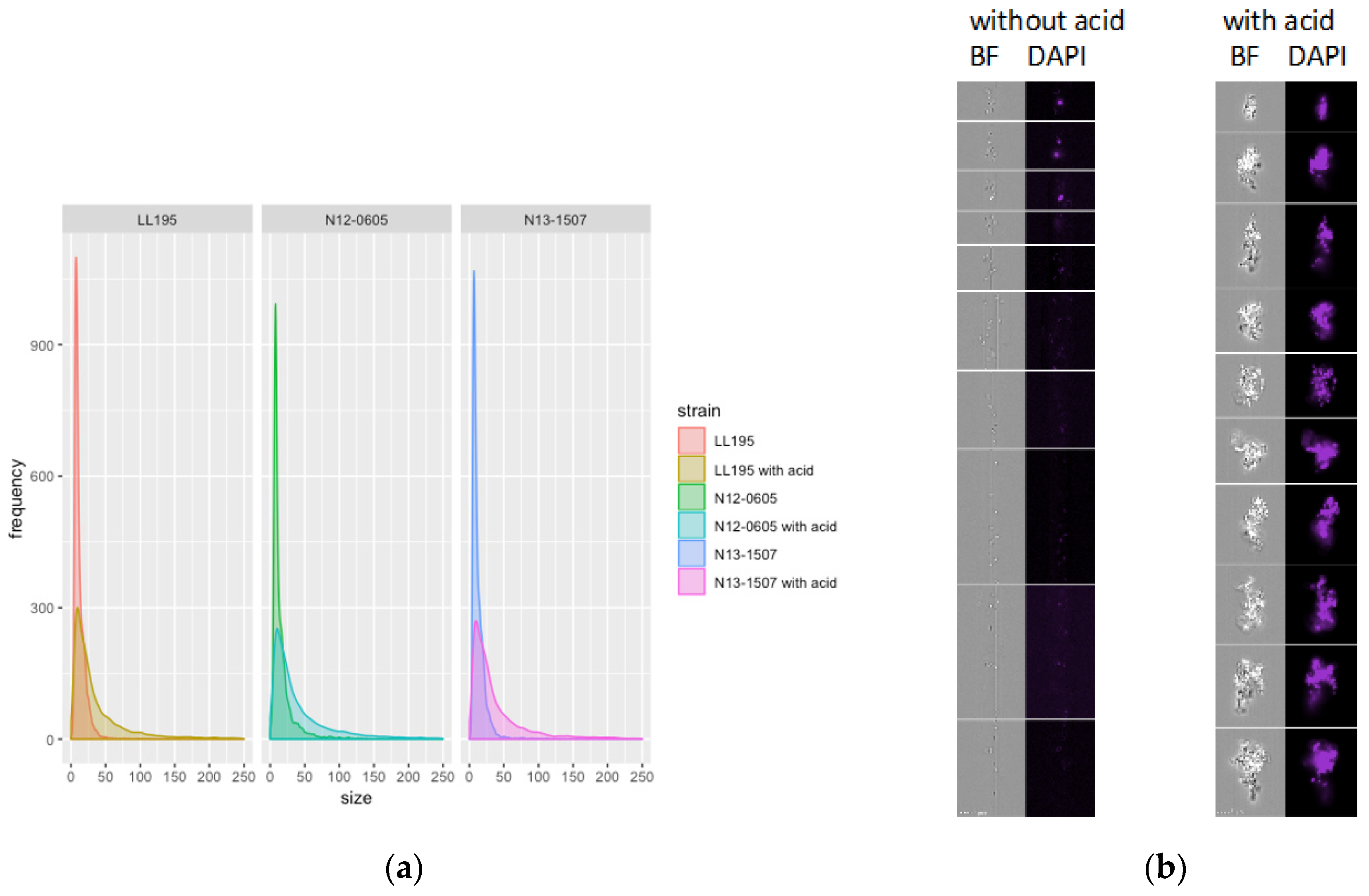
2.3. Genes Involved in Cell Division and Gene Expression were Downregulated after Acid Stress
2.4. Changes in Metabolism after Acid Stress
2.5. Shift in Pyruvate Fermentation
2.6. Genes Involved in Cell Wall Biology and Biofilm Formation were Differentially Expressed after Acid Stress
2.7. Genes Involved in Cellular Respiration were Differentially Expressed after Acid Stress
2.8. Cell Motility Related Genes were Downregulated after Acid Stress
2.9. Early-Stage Virulence Genes were Upregulated after Acid Stress
2.10. Promoter Analysis of Differentially Expressed Genes
3. Materials and Methods
3.1. Bacterial Strains
3.2. Growth Conditions
3.3. RNA Isolation
3.4. rRNA Depletion
3.5. Library Preparation and Sequencing
3.6. RNAseq Analysis
3.7. Morphological Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thakur, M.; Asrani, R.K.; Patial, V. Listeria monocytogenes: A Food-Borne Pathogen. In Foodborne Diseases; Elsevier: Amsterdam, The Netherland, 2018; pp. 157–192. ISBN 9780128114964. [Google Scholar]
- Smith, J.L.; Liu, Y.; Paoli, G.C. How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 2013, 59, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Hill, C.; Gahan, C.G.M. Acid stress responses in Listeria monocytogenes. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherland, 2008; Volume 65, pp. 67–91. ISBN 9780123744296. [Google Scholar]
- O’Byrne, C.P.; Karatzas, K.A.G. The role of Sigma B (σB) in the stress adaptations of Listeria monocytogenes: Overlaps between stress adaptation and virulence. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherland, 2008; pp. 115–140. ISBN 9780123744296. [Google Scholar]
- Raengpradub, S.; Wiedmann, M.; Boor, K.J. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 2008, 74, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, M.; Arvik, T.J.; Hurley, R.J.; Boor, K.J. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 1998, 180, 3650–3656. [Google Scholar] [PubMed]
- Ferreira, A.; O’Byrne, C.P.; Boor, K.J. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 2001, 67, 4454–4457. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef]
- Abram, F.; Starr, E.; Karatzas, K.A.G.; Matlawska-Wasowska, K.; Boyd, A.; Wiedmann, M.; Boor, K.J.; Connally, D.; O’Byrne, C.P. Identification of Components of the Sigma B Regulon in Listeria monocytogenes That Contribute to Acid and Salt Tolerance. Appl. Environ. Microbiol. 2008, 74, 6848–6858. [Google Scholar] [CrossRef]
- Wemekamp-Kamphuis, H.H.; Wouters, J.A.; de Leeuw, P.P.L.A.; Hain, T.; Chakraborty, T.; Abee, T. Identification of Sigma Factor B-Controlled Genes and Their Impact on Acid Stress, High Hydrostatic Pressure, and Freeze Survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 2004, 70, 3457–3466. [Google Scholar] [CrossRef]
- Cotter, P.D.; Gahan, C.G.M.; Hill, C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 2001, 40, 465–475. [Google Scholar] [CrossRef]
- van der Veen, S.; Abee, T. Contribution of Listeria monocytogenes RecA to acid and bile survival and invasion of human intestinal Caco-2 cells. Int. J. Med. Microbiol. 2011, 301, 334–340. [Google Scholar] [CrossRef]
- van der Veen, S.; van Schalkwijk, S.; Molenaar, D.; de Vos, W.M.; Abee, T.; Wells-Bennik, M.H.J. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 2010, 156, 374–384. [Google Scholar] [CrossRef]
- Cotter, P.D.; Emerson, N.; Gahan, C.G.; Hill, C. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 1999, 181, 6840–6843. [Google Scholar] [PubMed]
- Soares, C.A.; Knuckley, B. Mechanistic studies of the agmatine deiminase from Listeria monocytogenes. Biochem. J. 2016, 473, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.R.; Benjamin, M.M. Factors controlling acid tolerance of Listeria monocytogenes: Effects of nisin and other ionophores. Appl. Environ. Microbiol. 1997, 63, 4123–4126. [Google Scholar] [PubMed]
- Bécavin, C.; Bouchier, C.; Lechat, P.; Archambaud, C.; Creno, S.; Gouin, E.; Wu, Z.; Kühbacher, A.; Brisse, S.; Pucciarelli, M.G.; et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio 2014, 5, e00969-14. [Google Scholar] [CrossRef]
- Lundén, J.; Tolvanen, R.; Korkeala, H. Acid and heat tolerance of persistent and nonpersistent Listeria monocytogenes food plant strains. Lett. Appl. Microbiol. 2007, 46, 276–280. [Google Scholar] [CrossRef]
- Skandamis, P.N.; Gounadaki, A.S.; Geornaras, I.; Sofos, J.N. Adaptive acid tolerance response of Listeria monocytogenes strains under planktonic and immobilized growth conditions. Int. J. Food Microbiol. 2012, 159, 160–166. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Ainsworth, A.J.; Austin, F.W. Comparative assessment of acid, alkali and salt tolerance in Listeria monocytogenes virulent and avirulent strains. FEMS Microbiol. Lett. 2005, 243, 373–378. [Google Scholar] [CrossRef]
- Lianou, A.; Stopforth, J.D.; Yoon, Y.; Wiedmann, M.; Sofos, J.N. Growth and Stress Resistance Variation in Culture Broth among Listeria monocytogenes Strains of Various Serotypes and Origins. J. Food Prot. 2006, 69, 2640–2647. [Google Scholar] [CrossRef]
- Horlbog, J.A.; Kent, D.; Stephan, R.; Guldimann, C. Surviving host—And food relevant stresses: Phenotype of L. monocytogenes strains isolated from food and clinical sources. Sci. Rep. 2018, 8, 12931. [Google Scholar] [CrossRef]
- Ondrusch, N.; Kreft, J. Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS ONE 2011, 6, e16151. [Google Scholar] [CrossRef]
- Utratna, M.; Cosgrave, E.; Baustian, C.; Ceredig, R.; O’Byrne, C. Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioengineered 2012, 3, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Guldimann, C.; Guariglia-Oropeza, V.; Harrand, S.; Kent, D.; Boor, K.J.; Wiedmann, M. Stochastic and Differential Activation of σB and PrfA in Listeria monocytogenes at the Single Cell Level under Different Environmental Stress Conditions. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Sue, D.; O’Byrne, C.P.; Boor, K.J. Role of Listeria monocytogenes sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 2003, 69, 2692–2698. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Mazel, D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 2014, 38, 1126–1145. [Google Scholar] [CrossRef]
- Cheng, C.; Chen, J.; Fang, C.; Xia, Y.; Shan, Y.; Liu, Y.; Wen, G.; Song, H.; Fang, W. Listeria monocytogenes aguA1, but Not aguA2, Encodes a Functional Agmatine Deiminase. J. Biol. Chem. 2013, 288, 26606–26615. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Carvalho, A.L.; Turner, D.L.; Fonseca, L.L.; Solopova, A.; Catarino, T.; Kuipers, O.P.; Voit, E.O.; Neves, A.R.; Santos, H. Metabolic and Transcriptional Analysis of Acid Stress in Lactococcus lactis, with a Focus on the Kinetics of Lactic Acid Pools. PLoS ONE 2013, 8, e68470. [Google Scholar] [CrossRef]
- Fujita, Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009, 73, 245–259. [Google Scholar] [CrossRef]
- Ramström, H.; Sanglier, S.; Leize-Wagner, E.; Philippe, C.; Van Dorsselaer, A.; Haiech, J. Properties and Regulation of the Bifunctional Enzyme HPr Kinase/Phosphatase in Bacillus subtilis. J. Biol. Chem. 2003, 278, 1174–1185. [Google Scholar] [CrossRef]
- Warner, J.B.; Lolkema, J.S. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Romick; Fleming Acetoin production as an indicator of growth and metabolic inhibition of Listeria monocytogenes. J. Appl. Microbiol. 1998, 84, 18–24. [CrossRef] [PubMed]
- Tsau, J.L.; Guffanti, A.A.; Montville, T.J. Conversion of Pyruvate to Acetoin Helps to Maintain pH Homeostasis in Lactobacillus plantarum. Appl. Environ. Microbiol. 1992, 58, 891–894. [Google Scholar]
- Wilks, J.C.; Kitko, R.D.; Cleeton, S.H.; Lee, G.E.; Ugwu, C.S.; Jones, B.D.; BonDurant, S.S.; Slonczewski, J.L. Acid and Base Stress and Transcriptomic Responses in Bacillus subtilis. Appl. Environ. Microbiol. 2009, 75, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; Lee Chang, K.J.; Pinfold, T.; Ross, T. Transcriptomic and phenotypic responses of Listeria monocytogenes strains possessing different growth efficiencies under acidic conditions. Appl. Environ. Microbiol. 2010, 76, 4836–4850. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Wiedmann, M.; Bergholz, T.M. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl. Environ. Microbiol. 2011, 77, 5294–5306. [Google Scholar] [CrossRef]
- Phillips; Humphrey; Lappin-Scott Chilling invokes different morphologies in two Salmonella Enteritidis PT4 strains. J. Appl. Microbiol. 1998, 84, 820–826. [CrossRef]
- Isom, L.L.; Khambatta, Z.S.; Moulf, J.L.; Akers, D.F.; Martin, S.E. Filament Formation in Listeria monocytogenes. J. Food Prot. 1995, 58, 1031–1033. [Google Scholar] [CrossRef]
- Everis, L.; Betts, G. pH stress can cause cell elongation in Bacillus and Clostridium species: A research note. Food Control 2001, 12, 53–56. [Google Scholar] [CrossRef]
- Kawai, Y.; Moriya, S.; Ogasawara, N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol. Microbiol. 2003, 47, 1113–1122. [Google Scholar] [CrossRef]
- Molle, V.; Fujita, M.; Jensen, S.T.; Eichenberger, P.; González-Pastor, J.E.; Liu, J.S.; Losick, R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003, 50, 1683–1701. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Makita, Y.; de Hoon, M.; Nakai, K. DBTBS: A database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008, 36, D93–D96. [Google Scholar] [CrossRef] [PubMed]
- Fadda, D.; Pischedda, C.; Caldara, F.; Whalen, M.B.; Anderluzzi, D.; Domenici, E.; Massidda, O. Characterization of divIVA and Other Genes Located in the Chromosomal Region Downstream of the dcw Cluster in Streptococcus pneumoniae. J. Bacteriol. 2003, 185, 6209–6214. [Google Scholar] [CrossRef] [PubMed]
- Challan Belval, S.; Gal, L.; Margiewes, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. Assessment of the Roles of LuxS, S-Ribosyl Homocysteine, and Autoinducer 2 in Cell Attachment during Biofilm Formation by Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 2006, 72, 2644–2650. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, S.; Abee, T. Dependence of Continuous-Flow Biofilm Formation by Listeria monocytogenes EGD-e on SOS Response Factor YneA. Appl. Environ. Microbiol. 2010, 76, 1992–1995. [Google Scholar] [CrossRef] [PubMed]
- Renier, S.; Hébraud, M.; Desvaux, M. Molecular biology of surface colonization by Listeria monocytogenes: An additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 2011, 13, 835–850. [Google Scholar] [CrossRef]
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189. [Google Scholar] [CrossRef]
- Morgan, R.; Kohn, S.; Hwang, S.-H.; Hassett, D.J.; Sauer, K. BdlA, a Chemotaxis Regulator Essential for Biofilm Dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7335–7343. [Google Scholar] [CrossRef]
- Briandet, R.; Leriche, V.; Carpentier, B.; Bellon-Fontain, M.-N. Effects of the growth procedure on the surface hydrophobicity of Listeria monocytogenes cells and their adhesion to stainless steel. J. Food Prot. 1999, 62, 994–998. [Google Scholar] [CrossRef]
- Barbosa, J.; Borges, S.; Camilo, R.; Magalhães, R.; Ferreira, V.; Santos, I.; Silva, J.; Almeida, G.; Teixeira, P. Biofilm Formation among Clinical and Food Isolates of Listeria monocytogenes. Int. J. Microbiol. 2013, 2013, 524975. [Google Scholar] [CrossRef]
- Simon, J.; van Spanning, R.J.M.; Richardson, D.J. The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim. Biophys. Acta Bioenerg. 2008, 1777, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Magalon, A.; Fedor, J.G.; Walburger, A.; Weiner, J.H. Molybdenum enzymes in bacteria and their maturation. Coord. Chem. Rev. 2011, 255, 1159–1178. [Google Scholar] [CrossRef]
- Hennon, S.W.; Soman, R.; Zhu, L.; Dalbey, R.E. YidC/Alb3/Oxa1 family of insertases. J. Biol. Chem. 2015, 290, 14866–14874. [Google Scholar] [CrossRef] [PubMed]
- Celebi, N.; Yi, L.; Facey, S.J.; Kuhn, A.; Dalbey, R.E. Membrane Biogenesis of Subunit II of Cytochrome bo Oxidase: Contrasting Requirements for Insertion of N-terminal and C-terminal Domains. J. Mol. Biol. 2006, 357, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Mols, M.; van Kranenburg, R.; Tempelaars, M.H.; van Schaik, W.; Moezelaar, R.; Abee, T. Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int. J. Food Microbiol. 2010, 137, 13–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maurer, L.M.; Yohannes, E.; Bondurant, S.S.; Radmacher, M.; Slonczewski, J.L. pH Regulates Genes for Flagellar Motility, Catabolism, and Oxidative Stress in Escherichia coli K-12. J. Bacteriol. 2005, 187, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Olesen, I.; Vogensen, F.K.; Jespersen, L. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog. Dis. 2009, 6, 669–680. [Google Scholar] [CrossRef]
- Tessema, G.T.; Møretrø, T.; Snipen, L.; Heir, E.; Holck, A.; Naterstad, K.; Axelsson, L. Microarray-based transcriptome of Listeria monocytogenes adapted to sublethal concentrations of acetic acid, lactic acid, and hydrochloric acid. Can. J. Microbiol. 2012, 58, 1112–1123. [Google Scholar] [CrossRef]
- Hajam, I.A.; Dar, P.A.; Shahnawaz, I.; Jaume, J.C.; Lee, J.H. Bacterial flagellin—A potent immunomodulatory agent. Exp. Mol. Med. 2017, 49, e373. [Google Scholar] [CrossRef]
- Martínez-García, E.; Nikel, P.I.; Chavarría, M.; de Lorenzo, V. The metabolic cost of flagellar motion in P seudomonas putida KT2440. Environ. Microbiol. 2014, 16, 291–303. [Google Scholar] [CrossRef]
- O’Neil, H.S.; Marquis, H. Listeria monocytogenes Flagella Are Used for Motility, Not as Adhesins, to Increase Host Cell Invasion. Infect. Immun. 2006, 74, 6675–6681. [Google Scholar]
- Guariglia-Oropeza, V.; Orsi, R.H.; Guldimann, C.; Wiedmann, M.; Boor, K.J. The Listeria monocytogenes Bile Stimulon under Acidic Conditions Is Characterized by Strain-Specific Patterns and the Upregulation of Motility, Cell Wall Modification Functions, and the PrfA Regulon. Front. Microbiol. 2018, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Scortti, M.; Monzó, H.J.; Lacharme-Lora, L.; Lewis, D.A.; Vázquez-Boland, J.A. The PrfA virulence regulon. Microbes Infect. 2007, 9, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P.; Vicente, M.F.; Mengaud, J.; Baquero, F.; Perez-Diaz, J.C.; Berche, P. Listeriolysin O is essential for virulence of Listeria monocytogenes: Direct evidence obtained by gene complementation. Infect. Immun. 1989, 57, 3629–3636. [Google Scholar] [PubMed]
- Gaillard, J.-L.; Berche, P.; Frehel, C.; Gouln, E.; Cossart, P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 1991, 65, 1127–1141. [Google Scholar] [CrossRef]
- Hall, M.; Grundström, C.; Begum, A.; Lindberg, M.J.; Sauer, U.H.; Almqvist, F.; Johansson, J.; Sauer-Eriksson, A.E. Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc. Natl. Acad. Sci. USA 2016, 113, 14733–14738. [Google Scholar] [CrossRef]
- Cheng, C.; Dong, Z.; Han, X.; Wang, H.; Jiang, L.; Sun, J.; Yang, Y.; Ma, T.; Shao, C.; Wang, X.; et al. Thioredoxin A Is Essential for Motility and Contributes to Host Infection of Listeria monocytogenes via Redox Interactions. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Ollinger, J.; Bowen, B.; Wiedmann, M.; Boor, K.J.; Bergholz, T.M. Listeria monocytogenes B Modulates PrfA-Mediated Virulence Factor Expression. Infect. Immun. 2009, 77, 2113–2124. [Google Scholar] [CrossRef]
- Makariti, I.P.; Printezi, A.; Kapetanakou, A.E.; Zeaki, N.; Skandamis, P.N. Investigating boundaries of survival, growth and expression of genes associated with stress and virulence of Listeria monocytogenes in response to acid and osmotic stress. Food Microbiol. 2015, 45, 231–244. [Google Scholar] [CrossRef]
- Neuhaus, K.; Satorhelyi, P.; Schauer, K.; Scherer, S.; Fuchs, T.M. Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genom. 2013, 14, 285. [Google Scholar] [CrossRef]
- Fuangthong, M.; Helmann, J.D. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 2003, 185, 6348–6357. [Google Scholar] [CrossRef] [PubMed]
- Follmann, M.; Ochrombel, I.; Krämer, R.; Trötschel, C.; Poetsch, A.; Rückert, C.; Hüser, A.; Persicke, M.; Seiferling, D.; Kalinowski, J.; et al. Functional genomics of pH homeostasis in Corynebacterium glutamicum revealed novel links between pH response, oxidative stress, iron homeostasis and methionine synthesis. BMC Genom. 2009, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Jakob, K.; Satorhelyi, P.; Lange, C.; Wendisch, V.F.; Silakowski, B.; Scherer, S.; Neuhaus, K. Gene Expression Analysis of Corynebacterium glutamicum Subjected to Long-Term Lactic Acid Adaptation. J. Bacteriol. 2007, 189, 5582–5590. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carmel-Harel, O.; Storz, G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 2000, 54, 439–461. [Google Scholar] [CrossRef]
- Gray, M.J.; Freitag, N.E.; Boor, K.J. How the Bacterial Pathogen Listeria monocytogenes Mediates the Switch from Environmental Dr. Jekyll to Pathogenic Mr. Hyde. Infect. Immun. 2006, 74, 2505–2512. [Google Scholar] [CrossRef]
- Camilli, A.; Tilney, L.G.; Portnoy, D.A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993, 8, 143–157. [Google Scholar] [CrossRef]
- Weinmaier, T.; Riesing, M.; Rattei, T.; Bille, J.; Arguedas-Villa, C.; Stephan, R.; Tasara, T. Complete Genome Sequence of Listeria monocytogenes LL195, a Serotype 4b Strain from the 1983–1987 Listeriosis Epidemic in Switzerland. Genome Announc. 2013, 1, e00152-12. [Google Scholar] [CrossRef]
- Edgar, R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Tanner, S.A.; Chassard, C.; Rigozzi, E.; Lacroix, C.; Stevens, M.J.A. Bifidobacterium thermophilum RBL67 impacts on growth and virulence gene expression of Salmonella enterica subsp. enterica serovar Typhimurium. BMC Microbiol. 2016, 16, 46. [Google Scholar]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Kushwaha, M.; Salis, H.M. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun. 2015, 6, 7832. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef]
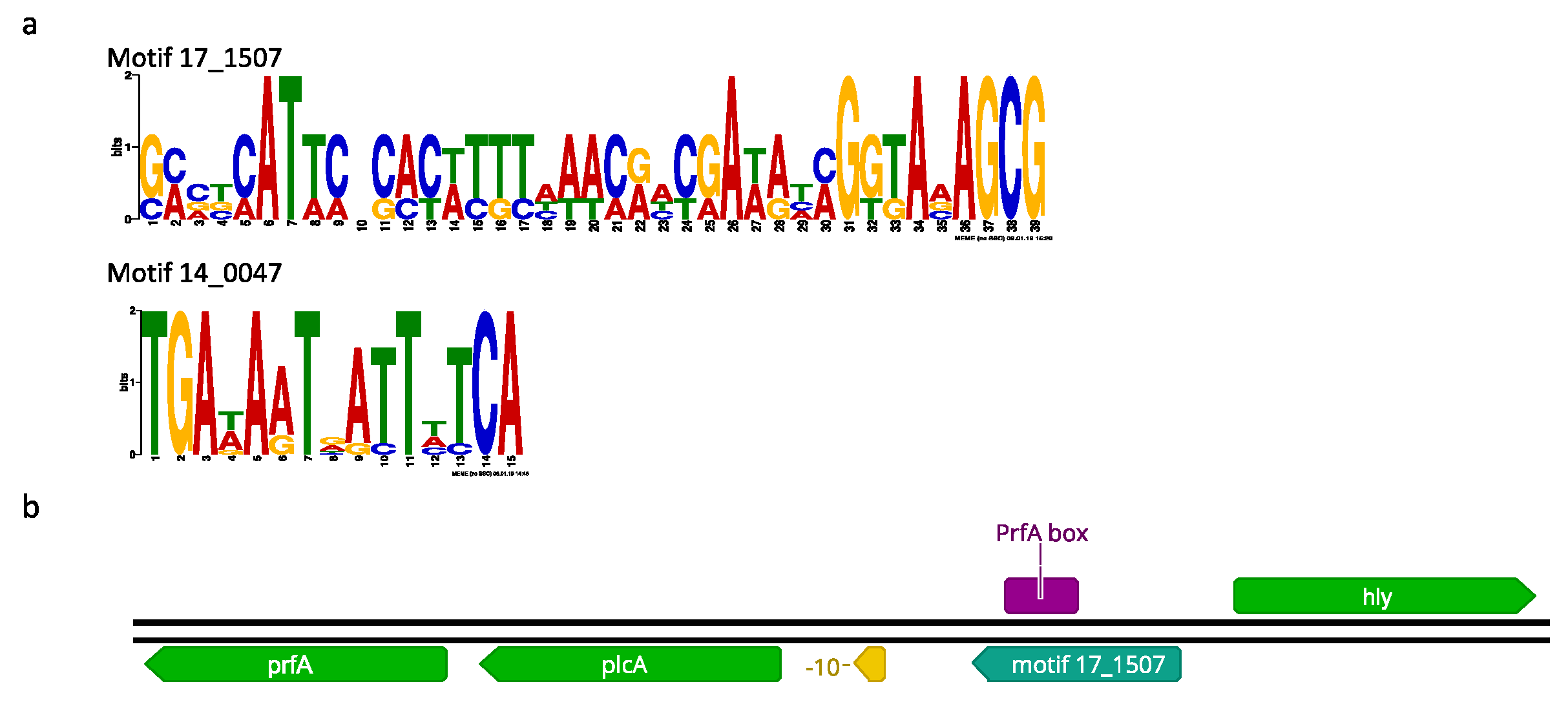
| Motif 17_1507 | |||||||||
| Strain | Motif sequence | Strand 1 | e-value | Product up 2 | Product down 2 | FC up 3 | FDR up 4 | FC down 3 | FDR down 4 |
| LL195 | GAAGAATATCCAAATGTGATTAAAAATATAGTTACACCG | reverse | 9.16E-07 | Cyclic-di-AMP phosphodiesterase GdpP | 50S ribosomal protein L9 | −0.220 | 0.774 | −0.229 | 0.700 |
| LL195 | CGCTTTACCTGCTTCGGCGATTGAATGTCAGTATGAGGC | forward | 2.19E-16 | hypothetical protein | hypothetical protein | 0.969 | 0.057 | 0.769 | 0.143 |
| LL195 | GAGGCATTAACATTTGTTAACGACGATAAAGGGACAGCG | reverse | 1.81E-15 | 1-phosphatidylinositol phosphodiesterase | Listeriolysin O | 0.802 | 0.107 | 3.013 | 0.000 |
| LL195 | CAAGAAATGGCGCTTTTCTTCAACAAAAAAGAGAAAGCA | reverse | 1.18E-07 | RNA polymerase sigma-H factor | Protein translocase subunit SecE | −0.650 | 0.227 | 0.679 | 0.281 |
| LL195 | GCAGAATACACCTATTTAGTAGGAGATAAAGTGAATGCT | reverse | 1.48E-07 | Internalin-A | hypothetical protein | 0.079 | 0.904 | 0.283 | 0.686 |
| LL195 | CAAGAATTAGCAAATGTAAACGACGTAGCAATGGATGAG | reverse | 6.03E-07 | hypothetical protein | Nitrite transporter NirC | 0.414 | 0.462 | 0.355 | 0.668 |
| LL195 | CACTTTAATGATATTAACGAAGAAGATGATTACTGGGGG | forward | 3.67E-07 | hypothetical protein | Glutathione reductase | 1.576 | 0.001 | 1.218 | 0.028 |
| LL195 | ACAGAAATCAGCCACTTAATTAGCGAAACAATGACAGAG | reverse | 7.55E-07 | Putative TrmH family tRNA/rRNA methyltransferase | hypothetical protein | 0.310 | 0.658 | 1.145 | 0.024 |
| LL195 | CTCTCTATCTTCAATGATTTTTGCAAGCGCGATTTGTTC | forward | 8.88E-08 | Glutathione amide reductase | Ribonuclease J 2 | 1.661 | 0.026 | −0.411 | 0.591 |
| LL195 | GAGTCACTCACCCGCTTAAAAACAGATCACTGGAATGCG | reverse | 1.65E-07 | putative cyclic di-GMP phosphodiesterase PdeG | Malolactic enzyme | −0.527 | 0.348 | −0.221 | 0.707 |
| LL195 | GCACCATTCGCACTCTTCAACATCGATATCGTTAGAGCG | reverse | 2.93E-16 | Ribonucleoside-diphosphate reductase 2 subunit alpha | hypothetical protein | 1.620 | 0.013 | 0.141 | 0.842 |
| LL195 | CACTAATTCCCCCATTTATAAAATAAAATCGGTAAAGCG | reverse | 2.78E-14 | High-affinity heme uptake system protein IsdE | Iron-regulated surface determinant protein A | 1.012 | 0.143 | 1.706 | 0.001 |
| LL195 | CGCTTGCTCTTTCTCATTGATTAATTGTTCATATTGCGC | forward | 2.58E-07 | putative protein YhaN | putative metallophosphoesterase YhaO | −0.614 | 0.282 | −0.139 | 0.825 |
| LL195 | CCATAAGCGACATTATCATAAATCGAAAACGGGAATGGG | reverse | 9.55E-07 | Phosphate import ATP-binding protein PstB 3 | Phosphate import ATP-binding protein PstB 3 | 0.767 | 0.360 | 0.468 | 0.447 |
| LL195 | CAGGCATTCAAACTTGCCAACAAATATACGAATAAAGCG | reverse | 1.93E-07 | Heptaprenyl diphosphate synthase component 2 | 3′,5′-cyclic adenosine monophosphate phosphodiesterase CpdA | −0.772 | 0.187 | −1.617 | 0.005 |
| N12-0605 | GAACAAATCGCGCTTGCAAAAATCATTGAAGATAGAGAG | reverse | 8.88E-08 | Glutathione amide reductase | hypothetical protein | 0.248 | 0.864 | 0.324 | 0.739 |
| N12-0605 | CTCTGTCATTGTTTCGCTAATTAAGTGGCTGATTTCTGT | forward | 7.55E-07 | Putative TrmH family tRNA/rRNA methyltransferase | Putative aminopeptidase YsdC | 0.152 | 0.879 | 0.157 | 0.881 |
| N12-0605 | CCCCCAGTAATCATCTTCTTCGTTAATATCATTAAAGTG | reverse | 3.67E-07 | hypothetical protein | hypothetical protein | 0.998 | 0.121 | 0.093 | 0.921 |
| N12-0605 | CTCATCCATTGCTACGTCGTTTACATTTGCTAATTCTTG | forward | 6.03E-07 | hypothetical protein | hypothetical protein | 0.104 | 0.916 | −0.116 | 0.940 |
| N12-0605 | AGCATTCACTTTATCTCCTACTAAATAGGTGTATTCTGC | forward | 1.48E-07 | Internalin-A | hypothetical protein | −0.204 | 0.855 | −0.950 | 0.264 |
| N12-0605 | CGCTGTCCCTTTATCGTCGTTAACAAATGTTAATGCCTC | forward | 1.81E-15 | Listeriolysin O | 1-phosphatidylinositol phosphodiesterase | 2.764 | 0.012 | 0.729 | 0.267 |
| N12-0605 | GCCTCATACTGACATTCAATCGCCGAAGCAGGTAAAGCG | reverse | 2.19E-16 | hypothetical protein | hypothetical protein | 0.639 | 0.420 | 0.753 | 0.289 |
| N12-0605 | CGGTGTAACTATATTTTTAATCACATTTGGATATTCTTC | forward | 9.16E-07 | Cyclic-di-AMP phosphodiesterase GdpP | Accessory gene regulator A | −0.356 | 0.721 | −0.100 | 0.934 |
| N12-0605 | CGCTTTATTCGTATATTTGTTGGCAAGTTTGAATGCCTG | forward | 1.93E-07 | Heptaprenyl diphosphate synthase component 2 | hypothetical protein | −0.704 | 0.298 | −0.287 | 0.717 |
| N12-0605 | CGCTTTACCGATTTTATTTTATAAATGGGGGAATTAGTG | forward | 2.78E-14 | Iron-regulated surface determinant protein A | High-affinity heme uptake system protein IsdE | 2.261 | 0.003 | 1.593 | 0.089 |
| N12-0605 | CGCTCTAACGATATCGATGTTGAAGAGTGCGAATGGTGC | forward | 2.93E-16 | Ribonucleoside-diphosphate reductase 2 subunit alpha | Ribonucleoside-diphosphate reductase subunit beta | 1.063 | 0.211 | 0.957 | 0.204 |
| N12-0605 | CAAGAAATGGCGCTTTTCTTCAACAAAAAAGAGAAAGCA | reverse | 1.18E-07 | RNA polymerase sigma-H factor | Protein translocase subunit SecE | −1.214 | 0.027 | 0.495 | 0.603 |
| N12-0605 | GCTGAAGTAGCACTTGAAAAAGACGATATCGACTCTGCG | reverse | 8.24E-07 | Beta-barrel assembly-enhancing protease | hypothetical protein | 0.029 | 0.977 | 1.196 | 0.081 |
| N12-0605 | CGCATTCCAGTGATCTGTTTTTAAGCGGGTGAGTGACTC | forward | 1.65E-07 | putative cyclic di-GMP phosphodiesterase PdeG | hypothetical protein | −0.688 | 0.296 | −0.810 | 0.205 |
| N12-0605 | CCCATTCCCGTTTTCGATTTATGATAATGTCGCTTATGG | forward | 9.55E-07 | Phosphate import ATP-binding protein PstB 3 | Phosphate-specific transport system accessory protein PhoU | −0.116 | 0.928 | 0.295 | 0.801 |
| N13-1507 | GAACAAATTGCGCTCGCAAAAATCATTGAAGATAGAGAG | reverse | 8.60E-07 | Glutathione amide reductase | hypothetical protein | 4.418 | 0.000 | 2.365 | 0.000 |
| N13-1507 | CTCTGTCATTGTTTCGCTAATTAAGTGGCTGATTTCTGT | forward | 7.55E-07 | Putative TrmH family tRNA/rRNA methyltransferase | Putative aminopeptidase YsdC | 0.314 | 0.290 | 0.011 | 0.981 |
| N13-1507 | CCCCCAGTAATCATCTTCTTCGTTAATATCATTAAAGTG | reverse | 3.67E-07 | hypothetical protein | hypothetical protein | 1.047 | 0.005 | 1.084 | 0.001 |
| N13-1507 | CTCATCCATTGCTACGTCGTTTACATTTGCTAATTCTTG | forward | 6.03E-07 | hypothetical protein | hypothetical protein | −0.024 | 0.963 | 2.358 | 0.000 |
| N13-1507 | AGCATTCACTTTATCTCCTACTAAATAGGTGTATTCTGC | forward | 1.48E-07 | Internalin-A | hypothetical protein | 0.482 | 0.353 | NA | NA |
| N13-1507 | CGCTGTCCCTTTATCGTCGTTAACAAATGTTAATGCCTC | forward | 1.81E-15 | Listeriolysin O | 1-phosphatidylinositol phosphodiesterase | 4.286 | 0.000 | 1.503 | 0.000 |
| N13-1507 | GCCTCATACTGACATTCAATCGCCGAAGCAGGTAAAGCG | reverse | 2.19E-16 | hypothetical protein | hypothetical protein | 1.287 | 0.005 | 2.146 | 0.000 |
| N13-1507 | CGGTGTAACTATATTTTTAATCACATTTGGATATTCTTC | forward | 9.16E-07 | Cyclic-di-AMP phosphodiesterase GdpP | Accessory gene regulator A | −0.285 | 0.406 | 0.945 | 0.005 |
| N13-1507 | CGCTTTATTCGTATATTTGTTGGCAAGTTTGAATGCCTG | forward | 1.93E-07 | Heptaprenyl diphosphate synthase component 2 | hypothetical protein | −1.672 | 0.000 | −1.265 | 0.000 |
| N13-1507 | CGCTTTACCGATTTTATTTTATAAATGGGGGAATTAGTG | forward | 2.78E-14 | Iron-regulated surface determinant protein A | High-affinity heme uptake system protein IsdE | 1.624 | 0.001 | 2.351 | 0.001 |
| N13-1507 | CGCTCTAACGATATCGATGTTGAAGAGTGCGAATGGTGC | forward | 2.93E-16 | Ribonucleoside-diphosphate reductase 2 subunit alpha | Ribonucleoside-diphosphate reductase subunit beta (thioredoxin two genes downstream) | 1.508 | 0.000 | 1.653 | 0.000 |
| N13-1507 | TGCTTTCTCTTTTTTGTTGAAGAAAAGCGCCATTTCTTG | forward | 1.18E-07 | RNA polymerase sigma-H factor | putative protein YacP | −0.867 | 0.004 | −0.786 | 0.026 |
| N13-1507 | GCTGAAGTAGCACTTGAAAAAGACGATATCGACTCTGCG | reverse | 8.24E-07 | Beta-barrel assembly-enhancing protease | hypothetical protein | −1.407 | 0.000 | 0.359 | 0.312 |
| N13-1507 | CGCATTCCAGTGATCTGTTTTTAAGCGGGTGAGTGACTC | forward | 1.65E-07 | putative cyclic di-GMP phosphodiesterase PdeG | hypothetical protein | −0.353 | 0.309 | −0.288 | 0.421 |
| N13-1507 | CCATAAGCGACATTATCATAAATCGAAAACGGGAATGGG | reverse | 9.55E-07 | Phosphate import ATP-binding protein PstB 3 | Phosphate import ATP-binding protein PstB 3 | 3.181 | 0.000 | 1.230 | 0.024 |
| Motif 14_0047 | |||||||||
| Strain | Motif sequence | Strand 1 | Score | Product up 2 | Product down 2 | FC up 3 | FDR up 4 | FC down 3 | FDR down 4 |
| LL195 | TGATAATAATTCTCA | reverse | 4.17E-08 | Iron(3+)-hydroxamate-binding protein YxeB | putative siderophore transport system ATP-binding protein YusV | 0.222 | 0.820 | 0.531 | 0.341 |
| LL195 | AGAAAATCATTTTCA | forward | 9.75E-07 | Fe(2+) transporter FeoB | hypothetical protein | 1.829 | 0.008 | 1.534 | 0.002 |
| LL195 | TGAGAATGATTTTCA | reverse | 1.99E-08 | Fe-S protein maturation auxiliary factor SufT | hypothetical protein | 0.740 | 0.210 | 0.927 | 0.045 |
| LL195 | TGGTAGCCATTTTCA | forward | 5.38E-07 | hypothetical protein | putative protein YwqG | −1.006 | 0.048 | −0.165 | 0.774 |
| LL195 | TGAAAACAATTTTCA | forward | 3.78E-07 | Phosphoglucomutase | Aldose 1-epimerase | −0.761 | 0.216 | −0.624 | 0.289 |
| LL195 | TGGAAACAATTTTCA | forward | 6.42E-07 | hypothetical protein | hypothetical protein | 0.893 | 0.209 | 0.812 | 0.239 |
| LL195 | TGAAAGTGATTTCCA | reverse | 1.09E-07 | O-acetyl-ADP-ribose deacetylase | Energy-dependent translational throttle protein EttA | 2.519 | 0.000 | −1.055 | 0.176 |
| LL195 | TGAAAATGATTTTCA | reverse | 2.07E-09 | Putative N-acetylmannosamine-6-phosphate 2-epimerase | Ribosomal RNA small subunit methyltransferase G | 0.106 | 0.923 | −0.749 | 0.137 |
| N12-0605 | TGAAAATCATTTTCA | forward | 2.07E-09 | PTS system glucitol/sorbitol-specific EIIA component | Fe(3+)-citrate-binding protein YfmC | 0.103 | 0.927 | 1.426 | 0.012 |
| N12-0605 | TGAAAATTGTTTCCA | reverse | 6.42E-07 | hypothetical protein | Protein SapB | 0.037 | 0.977 | 0.099 | 0.940 |
| N12-0605 | TGAAAATCATTCTCA | forward | 1.99E-08 | hypothetical protein | Fe-S protein maturation auxiliary factor SufT | 0.710 | 0.281 | 1.343 | 0.037 |
| N12-0605 | TGAAAATTGTTTTCA | reverse | 3.78E-07 | Phosphoglucomutase | Nucleotide-binding protein YvcJ | −0.854 | 0.223 | 0.424 | 0.569 |
| N12-0605 | TGAAAATGGCTACCA | reverse | 5.38E-07 | hypothetical protein | Putative ion-transport protein YfeO | −0.542 | 0.500 | −0.562 | 0.668 |
| N13-1507 | TGAAAATCATTTTCA | forward | 2.07E-09 | PTS system glucitol/sorbitol-specific EIIA component | Fe(3+)-citrate-binding protein YfmC | 1.276 | 0.019 | 1.784 | 0.000 |
| N13-1507 | TGAAAATTGTTTCCA | reverse | 6.42E-07 | hypothetical protein | Protein SapB | 3.061 | 0.000 | 3.743 | 0.000 |
| N13-1507 | TGAAAATCATTCTCA | forward | 1.99E-08 | hypothetical protein | Fe-S protein maturation auxiliary factor SufT | 0.376 | 0.294 | 1.456 | 0.000 |
| N13-1507 | TGGTAGCCATTTTCA | forward | 5.38E-07 | hypothetical protein | putative protein YwqG | −0.605 | 0.178 | 0.357 | 0.364 |
| N13-1507 | TGAAAACAATTTTCA | forward | 3.78E-07 | Phosphoglucomutase | Aldose 1-epimerase | −1.563 | 0.000 | −1.892 | 0.000 |
| Strain ID | Clonal Complex | Serotype | Isolation Source | Acid Phenotype | RNAseq Replicates |
|---|---|---|---|---|---|
| LL195 | CC1 | 4b | Outbreak strain | Resistant | 3 |
| N12-0605 | CC1 | 4b | Meat product | Resistant | 3 |
| N13-1507 | CC6 | 4b | Blood | Resistant | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horlbog, J.A.; Stevens, M.J.A.; Stephan, R.; Guldimann, C. Global Transcriptional Response of Three Highly Acid-Tolerant Field Strains of Listeria monocytogenes to HCl Stress. Microorganisms 2019, 7, 455. https://doi.org/10.3390/microorganisms7100455
Horlbog JA, Stevens MJA, Stephan R, Guldimann C. Global Transcriptional Response of Three Highly Acid-Tolerant Field Strains of Listeria monocytogenes to HCl Stress. Microorganisms. 2019; 7(10):455. https://doi.org/10.3390/microorganisms7100455
Chicago/Turabian StyleHorlbog, Jule Anna, Marc J. A. Stevens, Roger Stephan, and Claudia Guldimann. 2019. "Global Transcriptional Response of Three Highly Acid-Tolerant Field Strains of Listeria monocytogenes to HCl Stress" Microorganisms 7, no. 10: 455. https://doi.org/10.3390/microorganisms7100455
APA StyleHorlbog, J. A., Stevens, M. J. A., Stephan, R., & Guldimann, C. (2019). Global Transcriptional Response of Three Highly Acid-Tolerant Field Strains of Listeria monocytogenes to HCl Stress. Microorganisms, 7(10), 455. https://doi.org/10.3390/microorganisms7100455





