High Kanamycin Concentration as Another Stress Factor Additional to Temperature to Increase pDNA Production in E. coli DH5α Batch and Fed-Batch Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Plasmid
2.2. Medium and Inoculum
2.2.1. Batch Cultures
2.2.2. Fed-Batch Cultures
2.2.3. Inoculum
2.3. pDNA Production at Different Temperatures and Kanamycin Concentrations in Batch Culture
Experimental Factorial Design 22 with Central Component to Evaluate the Effect of Temperature and Kanamycin Concentration on pDNA and NPTII Production
2.4. pDNA Production in High-Cell Density Culture (HCDC) with High Kanamycin Concentration
2.5. Analytical Methods
2.5.1. DCW and Glycerol Determination
2.5.2. Plasmid DNA Quantification
sc-pDNA Quantification
2.5.3. Organic Acids and NPTII Quantification
2.5.4. Scanning Electronic Microscopy (SEM)
3. Results and Discussion
3.1. Statistical Analysis of Experimental Design 22 with a Central Component Used in Batch Culture
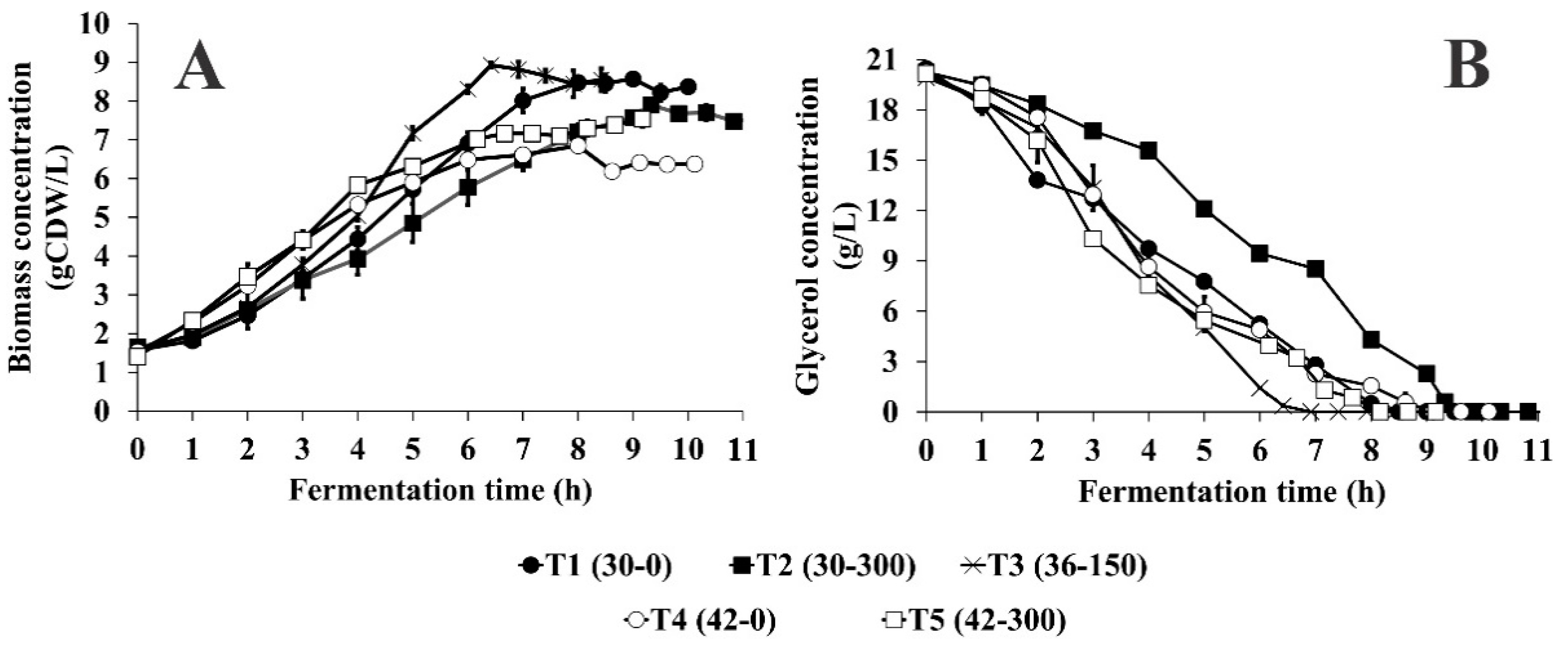
3.2. Effect of Temperature and Initial Kanamycin Concentration on Biomass Concentration Increment (∆X), Growth Rate (µmax), and Biomass Yield on Glycerol (YX/S) in Batch Culture
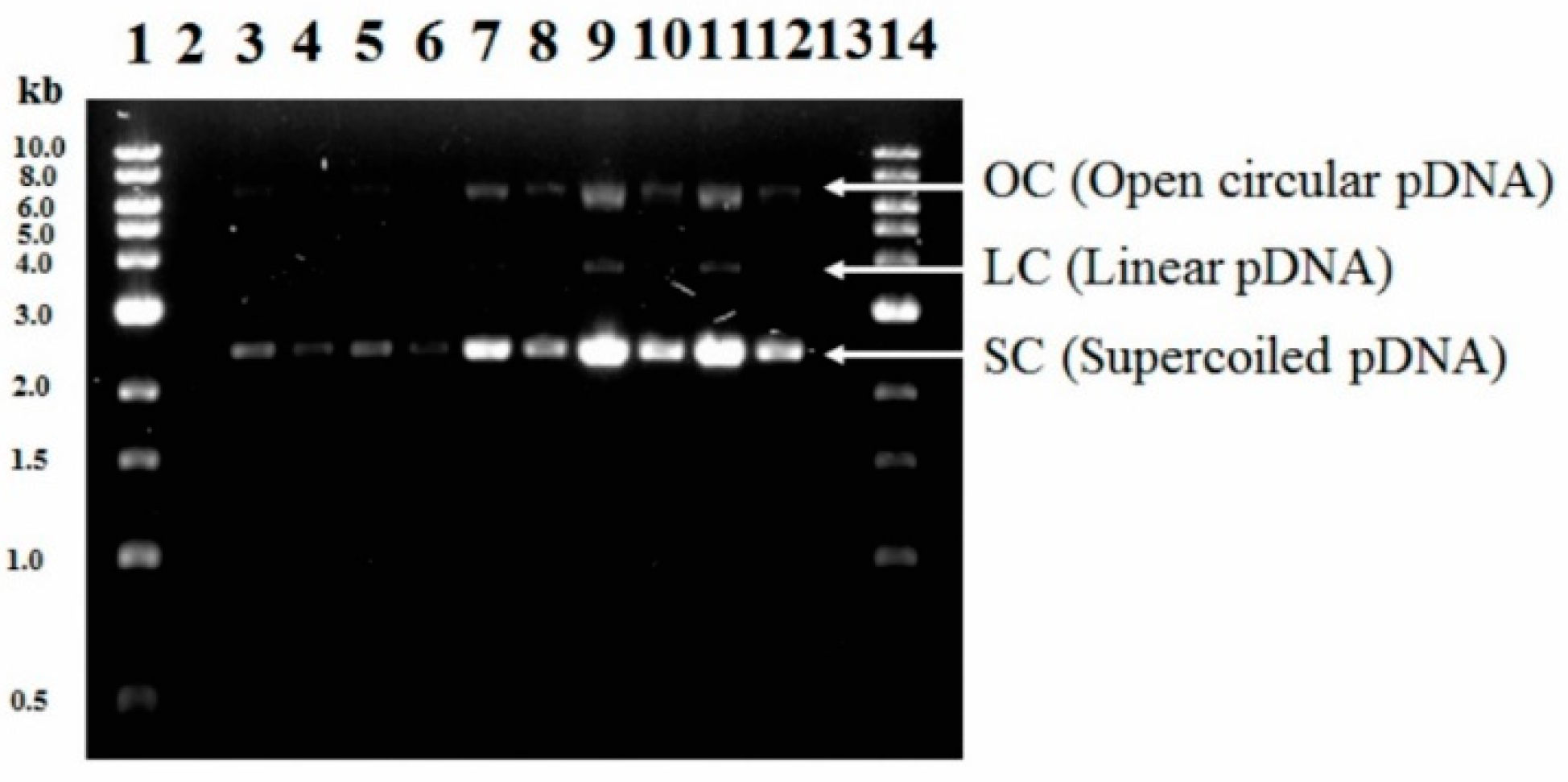
3.3. Effect of Temperature and Initial Kanamycin Concentration on Volumetric (pDNA) and Specific (YpDNA/X) Yields, Volumetric Productivity (pDNA-VP) of pDNA and % sc-pDNA in Batch Culture
3.4. Effect of Temperature and Initial Kanamycin Concentration on Metabolic Burden: Concentration (NPTII) and Specific Yield (YNPTII/X) of Neomycin Phosphotransferase II in Batch Culture
3.5. Effect of Temperature and Initial Kanamycin Concentration on Overflow Metabolism in Batch Culture
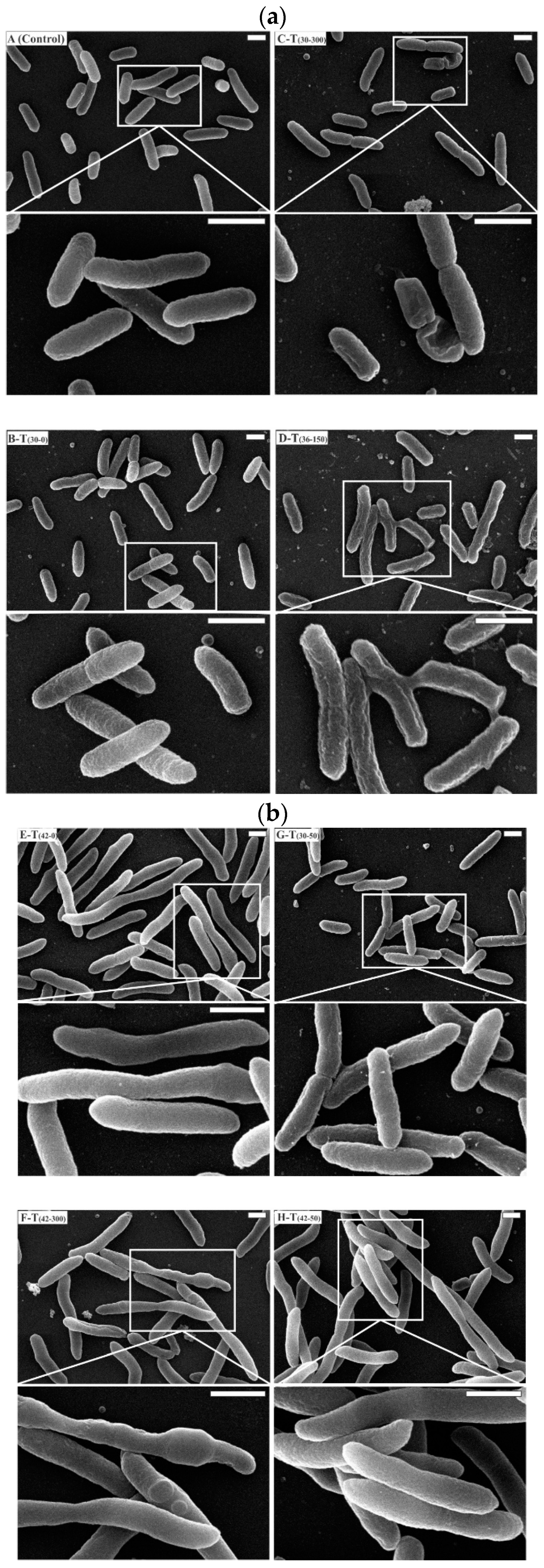
3.6. Cell Damage and Morphology Changes at Different Temperatures and Initial Kanamycin Concentration in Batch Culture
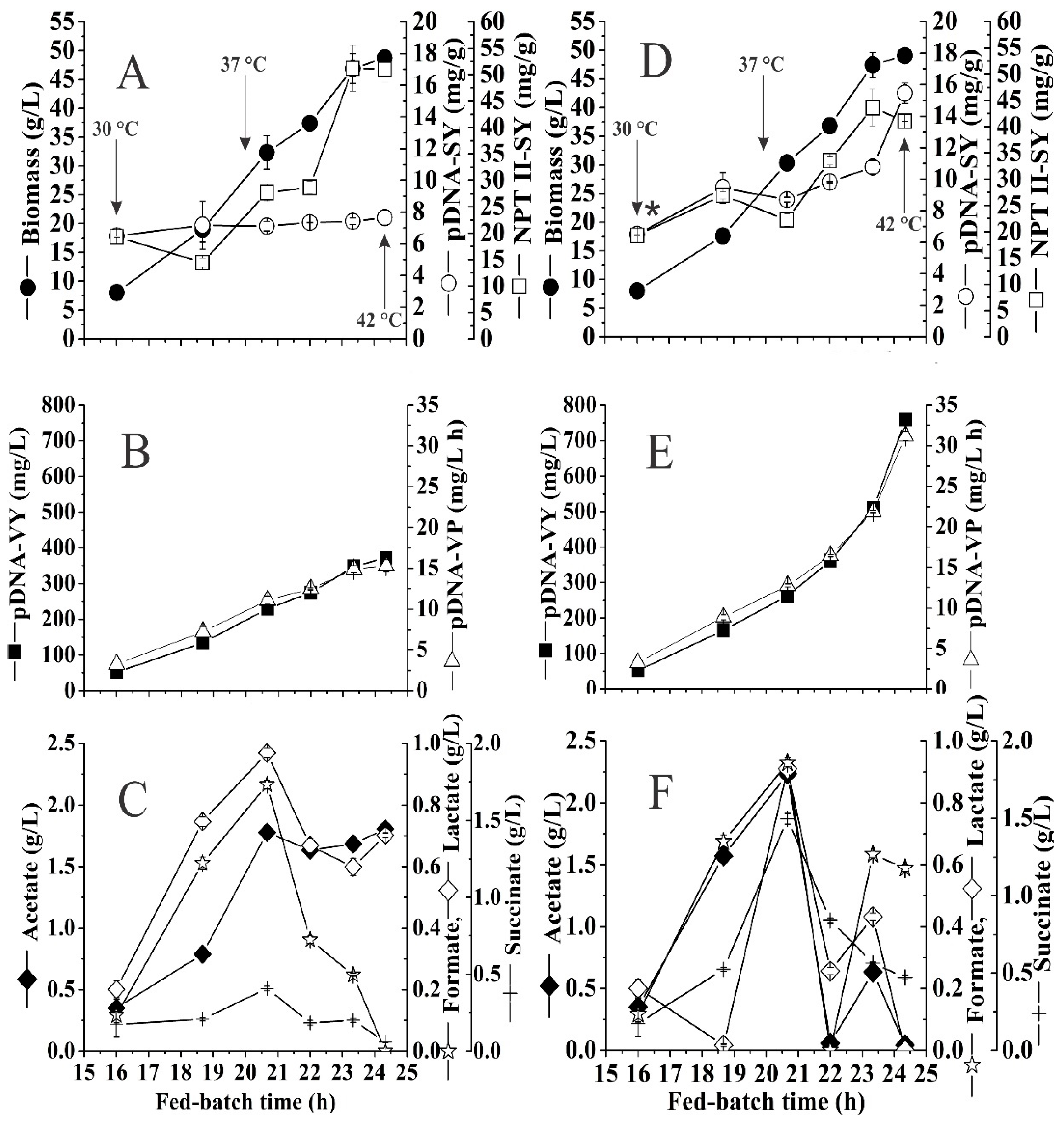
3.7. Effect of High Kanamycin Concentration on pDNA Production in HCDC
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Leitner, W.W.; Ying, H.; Restifo, N.P. DNA and RNA-based vaccines: Principles, progress and prospects. Vaccine 2000, 18, 765–777. [Google Scholar] [CrossRef]
- Singer, A.; Eiteman, M.A.; Altman, E. DNA plasmid production in different host strains of Escherichia coli. J. Ind. Microbiol. Biotechnol. 2009, 36, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Okonkowski, J.; Kizer-Bentley, L.; Listner, K.; Robinson, D.; Chartrain, M. Development of a robust, versatile, and scalable inoculum train for the production of a DNA vaccine. Biotechnol. Prog. 2005, 21, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Krishna Rao, D.V.; Ramu, C.T.; Rao, J.V.; Narasu, M.L.; Bhujanga Rao, A.K.S. Impact of dissolved oxygen concentration on some key parameters and production of rhG-CSF in batch fermentation. J. Ind. Microbiol. Biotechnol. 2008, 35, 991–1000. [Google Scholar] [CrossRef]
- Borja, G.M.; Meza Mora, E.; Barrón, B.; Gosset, G.; Ramírez, O.T.; Lara, A.R. Engineering Escherichia coli to increase plasmid DNA production in high cell-density cultivations in batch mode. Microb. Cell Fact. 2012, 11, 132. [Google Scholar] [CrossRef]
- Trivedi, R.N.; Akhtar, P.; Meade, J.; Bartlow, P.; Ataai, M.M.; Khan, S.A.; Domach, M.M. High-Level Production of Plasmid DNA by E. coli DH5α/sacB by Introducing inc Mutations. Appl. Environ. Microbiol. 2014, 80, 7154–7160. [Google Scholar] [CrossRef]
- Islas-Lugo, F.; Vega-Estrada, J.; Alvis, C.A.; Ortega-López, J.; del Carmen Montes-Horcasitas, M. Developing strategies to increase plasmid DNA production in Escherichia coli DH5α using batch culture. J. Biotechnol. 2016, 233, 66–73. [Google Scholar] [CrossRef]
- Lopes, M.B.; Martins, G.; Calado, C.R.C. Kinetic modeling of plasmid bioproduction in Escherichia coli DH5α cultures over different carbon-source compositions. J. Biotechnol. 2014, 186, 38–48. [Google Scholar] [CrossRef]
- Xu, Z.-N.; Shen, W.-H.; Chen, H.; Cen, P.-L. Effects of medium composition on the production of plasmid DNA vector potentially for human gene therapy. J. Zhejiang Univ. Sci. B 2005, 6, 396–400. [Google Scholar] [CrossRef]
- Jaén, K.E.; Lara, A.R.; Ramírez, O.T. Effect of heating rate on pDNA production by E. coli. Biochem. Eng. J. 2013, 79, 230–238. [Google Scholar]
- Silva, F.; Passarinha, L.; Sousa, F.; Queiroz, J.A.; Domingues, F.C. Influence of growth conditions on plasmid DNA production. J. Microbiol. Biotechnol. 2009, 19, 1408–1414. [Google Scholar] [PubMed]
- Rodrigues, J.L.; Rodrigues, L.R. Potential Applications of the Escherichia coli Heat Shock Response in Synthetic Biology. Trends Biotechnol. 2018, 36, 186–198. [Google Scholar] [CrossRef] [PubMed]
- García-Rendón, A.; Munguía-Soto, R.; Montesinos-Cisneros, R.M.; Guzman, R.; Tejeda-Mansir, A. Performance analysis of exponential-fed perfusion cultures for pDNA vaccines production. J. Chem. Technol. Biotechnol. 2017, 92, 342–349. [Google Scholar] [CrossRef]
- Gomes, L.C.; Mergulh, F.J. Effects of antibiotic concentration and nutrient medium composition on Escherichia coli biofilm formation and green fluorescent protein expression. FEMS Microbiol. Lett. 2017, 364, 1–8. [Google Scholar] [CrossRef]
- Lara, A.R.; Jaén, K.E.; Folarin, O.; Keshavarz-Moore, E.; Büchs, J. Effect of the oxygen transfer rate on oxygen-limited production of plasmid DNA by Escherichia coli. Biochem. Eng. J. 2019, 150, 107303. [Google Scholar] [CrossRef]
- Cranenburgh, R.M.; Lewis, K.S.; Hanak, J.A.J. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2004, 7, 197–203. [Google Scholar] [CrossRef]
- Nelson, J.; Rodriguez, S.; Finlayson, N.; Williams, J.; Carnes, A. Antibiotic-free production of a herpes simplex virus 2 DNA vaccine in a high yield cGMP process. Hum. Vaccin. Immunother. 2013, 2211–2215. [Google Scholar] [CrossRef]
- Vandermeulen, G.; Marie, C.; Scherman, D.; Préat, V. New Generation of Plasmid Backbones Devoid of Antibiotic Resistance Marker for Gene Therapy Trials. Mol. Ther. 2009, 19, 1942–1949. [Google Scholar] [CrossRef]
- Mairhofer, J.; Lara, A.R. Chapter 38 Advances in Host and Vector Development for the Production of Plasmid DNA Vaccines; Humana Press: New York, NY, USA, 2014; Volume 1139, ISBN 9781493903450. [Google Scholar]
- Hassan, S.; Keshavarz-Moore, E.; Ward, J. A cell engineering strategy to enhance supercoiled plasmid DNA production for gene therapy. Biotechnol. Bioeng. 2016, 113, 2064–2071. [Google Scholar] [CrossRef]
- Rozkov, A.; Avignone-Rossa, C.A.; Ertl, P.F.; Jones, P.; O’Kennedy, R.D.; Smith, J.J.; Dale, J.W.; Bushell, M.E. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol. Bioeng. 2004, 88, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.S.; Koepsel, R.R.; Ataai, M.M.; Domach, M.M. Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb. Cell Fact. 2009, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Panayotatos, N. Recombinant protein production with minimal-antibiotic-resistance vectors. Gene 1988, 74, 357–363. [Google Scholar] [CrossRef]
- Rahn, A.; Drummelsmith, J.; Whitfield, C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: Relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1999, 181, 2307–2313. [Google Scholar] [PubMed]
- Ebel, W.; Trempy, J.E. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 1999, 181, 577–584. [Google Scholar] [PubMed]
- Stevenson, G.; Andrianopoulos, K.; Hobbs, M.; Reeves, P.R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1996, 178, 4885–4893. [Google Scholar] [CrossRef] [PubMed]
- Taber, H.W.; Mueller, J.P.; Miller, P.F.; Arrow, A.M.Y.S. Bacterial Uptake of Aminoglycoside Antibiotics. Microbiol. Rev. 1987, 51, 439–457. [Google Scholar]
- Goltermann, L.; Good, L.; Bentin, T. Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in Escherichia coli. J. Biol. Chem. 2013, 288, 10483–10489. [Google Scholar] [CrossRef]
- Guisbert, E.; Herman, C.; Lu, C.Z.; Gross, C.A. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004, 18, 2812–2821. [Google Scholar] [CrossRef]
- Vanbogelen, R.A.; Neidhardt, F.C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 5589–5593. [Google Scholar] [CrossRef]
- An, T.; Sun, H.; Li, G.; Zhao, H.; Wong, P.K. Differences in photoelectrocatalytic inactivation processes between E. coli and its isogenic single gene knockoff mutants: Destruction of membrane framework or associated proteins? Appl. Catal. B Environ. 2016, 188, 360–366. [Google Scholar] [CrossRef]
- Myriam, S.C.; Eric, D.; Jaime, O.L. Production optimisation of a DNA vaccine candidate against leishmaniasis in flask culture. Afr. J. Biotechnol. 2013, 12, 4874–4880. [Google Scholar] [CrossRef]
- Feizollahzadeh, S.; Kouhpayeh, S.; Rahimmansh, I.; Khanahmad, H.; Sabzehei, F.; Ganjalikhani-hakemi, M.; Andalib, A.; Hejazi, Z.; Rezaei, A. The increase in protein and plasmid yields of E. coli with optimized concentration of ampicillin as selection marker. Iran. J. Biotechnol. 2017, 15, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Bondioli, P.; Della Bella, L. An alternative spectrophotometric method for the determination of free glycerol in biodiesel. Eur. J. Lipid Sci. Technol. 2005, 107, 153–157. [Google Scholar] [CrossRef]
- Soto, R.; Caspeta, L.; Barrón, B.; Gosset, G.; Ramírez, O.T.; Lara, A.R. High cell-density cultivation in batch mode for plasmid DNA production by a metabolically engineered E. coli strain with minimized overflow metabolism. Biochem. Eng. J. 2011, 56, 165–171. [Google Scholar] [CrossRef]
- Munguía-Soto, R.; García-Rendõn, A.; Garibay-Escobar, A.; Guerrero-Germán, P.; Tejeda-Mansir, A. Segregated growth kinetics of Escherichia coli DH5α-NH36 in exponential-fed perfusion culture for pDNA vaccine production. Biotechnol. Appl. Biochem. 2015, 62, 795–805. [Google Scholar] [CrossRef]
- Carnes, A.E.; Hodgson, C.P.; Williams, J.A. Inducible Escherichia coli fermentation for increased plasmid DNA production. Biotechnol. Appl. Biochem. 2006, 45, 155. [Google Scholar]
- Lin-Chao, S.; Chen, W.-T.; Wong, T.-T. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol. Microbiol. 1992, 6, 3385–3393. [Google Scholar] [CrossRef]
- Miller, C.; Thomsen, L.E.; Gaggero, C.; Mosseri, R.; Ingmer, H.; Cohen, S.N. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 2004, 305, 1629–1631. [Google Scholar] [CrossRef]
- Sousa, Â.; Sousa, F.; Queiroz, J.A. Advances in chromatographic supports for pharmaceutical-grade plasmid DNA purification. J. Sep. Sci. 2012, 35, 3046–3058. [Google Scholar] [CrossRef]
- Prazeres, D.M.F.; Ferreira, G.N.M.; Monteiro, G.A.; Cooney, C.L.; Cabral, J.M.S. Large-scale production of pharmaceutical-grade plasmid DNA for gene therapy: Problems and bottlenecks. Trends Biotechnol. 1999, 17, 169–174. [Google Scholar] [CrossRef]
- Morgan, G.J.; Hatfull, G.F.; Casjens, S.; Hendrix, R.W. Bacteriophage Mu genome sequence: Analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002, 317, 337–359. [Google Scholar] [CrossRef] [PubMed]
- Axenov, A.; Jang, S.; Jones, M. Pretreatment with Sub-minimal Inhibitory Concentration of Kanamycin Increases RpoH Levels in Escherichia coli SC122. J. Exp. Microbiol. Immunol. 2013, 17, 29–33. [Google Scholar]
- Narberhaus, F.; Balsiger, S. Structure-function studies of Escherichia coli RpoH (σ32) by in vitro linker insertion mutagenesis. J. Bacteriol. 2003, 185, 2731–2738. [Google Scholar] [CrossRef]
- Phue, J.N.; Sang, J.L.; Trinh, L.; Shiloach, J. Modified Escherichia coli B (BL21), a superior producer of plasmid DNA compared with Escherichia coli K (DH5α). Biotechnol. Bioeng. 2008, 101, 831–836. [Google Scholar] [CrossRef]
- Cavallero, A.; Eftimiadi, C.; Radin, L.; Schito, G.C. Suppression of tricarboxylic acid cycle in Escherichia coli exposed to sub-MICs of aminoglycosides. Antimicrob. Agents Chemother. 1990, 34, 295–301. [Google Scholar] [CrossRef]
- Bryan, L.E.; Kwan, S. Roles of ribosomal binding, membrane potential, and electron transport in bacterial upake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 1983, 23, 835–845. [Google Scholar] [CrossRef]
- Faraji, R.; Parsa, A.; Torabi, B.; Withrow, T. Effects of kanamycin on the Macromolecular Composition of kanamycin Sensitive Escherichia coli DH5 Strain. J. Exp. Microbiol. Immunol. 2006, 9, 31–38. [Google Scholar]
- Martins, L.M.; Pedro, A.Q.; Oppolzer, D.; Sousa, F.; Queiroz, J.A.; Passarinha, L.A. Enhanced biosynthesis of plasmid DNA from Escherichia coli VH33 using Box-Behnken design associated to aromatic amino acids pathway. Biochem. Eng. J. 2015, 98, 117–126. [Google Scholar] [CrossRef]
- Gonçalves, G.A.L.; Prazeres, D.M.F.; Monteiro, G.A.; Prather, K.L.J. De novo creation of MG1655-derived E. coli strains specifically designed for plasmid DNA production. Appl. Microbiol. Biotechnol. 2013, 97, 611–620. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Papo, N.; Barra, D.; Simmaco, M.; Bozzi, A.; Giulio, A.D.I.; Rinaldi, A.C. Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochem. J. 2004, 865, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Ackerley, D.F.; Barak, Y.; Lynch, S.V.; Curtin, J.; Matin, A. Effect of chromate stress on Escherichia coli K-12. J. Bacteriol. 2006, 188, 3371–3381. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, M.; Canham, L.T.; Reeves, C.L. Morphological changes (including filamentation) in Escherichia coli grown under starvation conditions on silicon wafers and other surfaces. Lett. Appl. Microbiol. 1999, 29, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Ow, D.S.W.; Nissom, P.M.; Philp, R.; Oh, S.K.W.; Yap, M.G.S. Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme Microb. Technol. 2006, 39, 391–398. [Google Scholar] [CrossRef]
- Mañas, P.; Mackey, B.M. Morphological and Physiological Changes Induced by High Hydrostatic Pressure in Exponential- and Stationary-Phase Cells of Escherichia coli: Relationship with Cell Death. Appl. Environ. Microbiol. 2004, 70, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Carnes, A.E.; Hodgson, C.P. Plasmid DNA vaccine vector design: Impact on efficacy, safety and upstream production. Biotechnol. Adv. 2009, 27, 353–370. [Google Scholar] [CrossRef]
- Jaén, K.E.; Velázquez, D.; Sigala, J.; Lara, A.R. Design of a microaerobically inducible replicon for high-yield plasmid DNA production. Biotechnol. Bioeng. 2019, 116, 514–2525. [Google Scholar] [CrossRef]
- Carnes, A.E.; Luke, J.M.; Vincent, J.M.; Schukar, A.; Anderson, S.; Hodgson, C.P.; Williams, J.A. Plasmid DNA fermentation strain and process-specific effects on vector yield, quality, and transgene expression. Biotechnol. Bioeng. 2011, 108, 354–363. [Google Scholar] [CrossRef]
| Nomenclature | Coded Levels | Real Input Variables Levels | ||
|---|---|---|---|---|
| X1 | X2 | Temperature (°C) | Kanamycin (mg/L) | |
| T(30-0) | − | − | 30 | 0 |
| T(30-300) | − | + | 30 | 300 |
| T(36-150) | 0 | 0 | 36 | 150 |
| T(42-0) | + | − | 42 | 0 |
| T(42-300) | + | + | 42 | 300 |
| Output Variable | Model | Term | F-Value | T-Value | p-Value | ||
|---|---|---|---|---|---|---|---|
| T-Value | Predicted R-Square % | Adjusted R-Square % | |||||
| pDNA Volumetric Yield | 1278.05 | 99.61 | 99.86 | A | 5441.28 | 73.77 | 0.000 |
| B | 462.92 | 21.52 | 0.000 | ||||
| AB | 271.15 | 16.47 | 0.000 | ||||
| Central point | −14.62 | 0.000 | |||||
| pDNA Specific Yield | 1465.21 | 99.66 | 99.88 | A | 6366.57 | 79.79 | 0.000 |
| B | 335.05 | 18.3 | 0.000 | ||||
| AB | 139.32 | 11.8 | 0.000 | ||||
| Central point | −22.03 | 0.000 | |||||
| pDNA Volumetric Productivity | 1501.93 | 99.66 | 99.88 | A | 6250.64 | 79.06 | 0.000 |
| B | 694.52 | 26.35 | 0.000 | ||||
| AB | 551.25 | 23.48 | 0.000 | ||||
| Central point | −3.5 | 0.025 | |||||
| NPTII Volumetric Yield | 647.58 | 99.23 | 99.72 | A | 96.01 | 9.8 | 0.001 |
| B | 65.69 | 8.1 | 0.001 | ||||
| AB | 91.34 | 9.56 | 0.001 | ||||
| Central point | 54.52 | 0.000 | |||||
| NPTII Specific Yield | 988.45 | 99.49 | 99.82 | A | 360.41 | 18.98 | 0.000 |
| B | 148.69 | 12.19 | 0.000 | ||||
| AB | 143.83 | 11.99 | 0.000 | ||||
| Central point | 65.37 | 0.000 | |||||
| Treatment | T(30-0) | * T(30-50) | T(30-300) | T(36-150) | T(42-0) | * T(42-50) | T(42-300) |
|---|---|---|---|---|---|---|---|
| Temperature (°C) | 30 °C | 36 °C | 42 °C | ||||
| Kanamycin (mg/L) | 0 | 50 | 300 | 150 | 0 | 50 | 300 |
| ∆X (g/L) | 6.90 ± 0.00 | 6.43 ± 0.05 | 6.21 ± 0.20 | 7.40 ± 0.06 | 5.32 ± 0.13 | 5.51 ± 0.26 | 5.95 ± 0.16 |
| YX/S (g/g) | 0.34 ± 0.01 | 0.32 ± 0.01 | 0.31 ± 0.00 | 0.37 ± 0.00 | 0.26 ± 0.01 | 0.27 ± 0.01 | 0.30 ± 0.01 |
| µ (h−1) | 0.288 ± 0.02 | 0.247 ± 0.01 | 0.189 ± 0.01 | 0.322 ± 0.01 | 0.353 ± 0.01 | 0.292 ± 0.03 | 0.349 ± 0.00 |
| pDNA (mg/L) | 27.2 ± 0.76 | 26.3 ± 0.51 | 33.9 ± 2.47 | 57.4 ± 1.71 | 102.2 ± 1.36 | 97.6 ± 4.22 | 151.8 ± 2.55 |
| YpDNA/X (mg/g) | 3.2 ± 0.09 | 3.1 ± 0.16 | 4.3 ± 0.34 | 6.4 ± 0.11 | 14.9 ± 0.21 | 12.9 ± 1.93 | 20.1 ± 0.23 |
| pDNA-VP (mg/L h) | 3.2 ± 0.09 | 3.3 ± 0.22 | 3.6 ± 0.27 | 9.0 ± 0.27 | 11.9 ± 0.16 | 13.9 ± 1.04 | 19.7 ± 0.33 |
| NPT II (mg/L) | 125 ± 10 | 152 ± 10 | 116 ± 11 | 558 ± 33 | 127 ± 11 | 186 ± 10 | 244 ± 10 |
| YNPTII/X (mg/g) | 14.5 ± 3.27 | 18.05 ± 2.12 | 14.6 ± 1.46 | 61.9 ± 2.67 | 18.5 ± 1.54 | 24.6 ± 1.14 | 32.4 ± 0.41 |
| sc-pDNA (%) | 91.58 ± 1.25 | 92.76 ± 2.76 | 92.65 ± 1.46 | 89.98 ± 0.32 | 89.97 ± 3.6 | 90.6 ± 4.72 | 89.85 ± 2.33 |
| Treatment | T(30-0) | * T(30-50) | T(30-300) | T(36-150) | T(42-0) | * T(42-50) | T(42-300) |
|---|---|---|---|---|---|---|---|
| Temperature | 30 °C | 36 °C | 42 °C | ||||
| Kanamycin (mg/L) | 0 | 50 | 300 | 150 | 0 | 50 | 300 |
| Acetate (g/L) | 1.07 ± 0.12 | 0.67 ± 0.06 | 0.49 ± 0.08 | 0.59 ± 0.10 | 2.60 ± 0.11 | 4.62 ± 0.17 | 1.18 ± 0.21 |
| Lactate (g/L) | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.05 ± 0.01 | 0.10 ± 0.00 | 0.41 ± 0.04 | 0.29 ± 0.03 | 0.23 ± 0.06 |
| Succinate (g/L) | 0.17 ± 0.01 | 0.18 ± 0.02 | 0.19 ± 0.04 | 0.20 ± 0.05 | 0.63 ± 0.07 | 0.14 ± 0.02 | 0.39 ± 0.06 |
| Formate (g/L) | 0.07 ± 0.00 | 0.05 ± 0.01 | 0.11 ± 0.03 | 0.09 ± 0.00 | 0.11 ± 0.02 | 0.05 ± 0.00 | 0.09 ± 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grijalva-Hernández, F.; Vega-Estrada, J.; Escobar-Rosales, M.; Ortega-López, J.; Aguilar-López, R.; Lara, A.R.; Montes-Horcasitas, M.d.C. High Kanamycin Concentration as Another Stress Factor Additional to Temperature to Increase pDNA Production in E. coli DH5α Batch and Fed-Batch Cultures. Microorganisms 2019, 7, 711. https://doi.org/10.3390/microorganisms7120711
Grijalva-Hernández F, Vega-Estrada J, Escobar-Rosales M, Ortega-López J, Aguilar-López R, Lara AR, Montes-Horcasitas MdC. High Kanamycin Concentration as Another Stress Factor Additional to Temperature to Increase pDNA Production in E. coli DH5α Batch and Fed-Batch Cultures. Microorganisms. 2019; 7(12):711. https://doi.org/10.3390/microorganisms7120711
Chicago/Turabian StyleGrijalva-Hernández, Fernando, Jesús Vega-Estrada, Montserrat Escobar-Rosales, Jaime Ortega-López, Ricardo Aguilar-López, Alvaro R. Lara, and Ma. del Carmen Montes-Horcasitas. 2019. "High Kanamycin Concentration as Another Stress Factor Additional to Temperature to Increase pDNA Production in E. coli DH5α Batch and Fed-Batch Cultures" Microorganisms 7, no. 12: 711. https://doi.org/10.3390/microorganisms7120711
APA StyleGrijalva-Hernández, F., Vega-Estrada, J., Escobar-Rosales, M., Ortega-López, J., Aguilar-López, R., Lara, A. R., & Montes-Horcasitas, M. d. C. (2019). High Kanamycin Concentration as Another Stress Factor Additional to Temperature to Increase pDNA Production in E. coli DH5α Batch and Fed-Batch Cultures. Microorganisms, 7(12), 711. https://doi.org/10.3390/microorganisms7120711







