The Role of Drug Resistance in Candida Inflammation and Fitness
Abstract
1. Introduction
2. Material and Methods
2.1. Candida Species Isolates
2.2. Measure of Drug Susceptibility Phenotype of Candida Strains
2.3. Cell Culture
2.4. DLD-1 Stimulation with Candida Strains
2.5. Measurement of Cytokines Following DLD-1 Stimulation with Candida Strains
2.6. Statistical Analyses
3. Results
3.1. Candida Species, Drug Susceptibility Phenotypes, and Definitions
3.2. Quantity of Cytokines Induced over 6 and 24 h by Candida Strains According to the Global Fluconazole Susceptibility Phenotypes
3.3. Comparison Between Cytokines Levels Induced by Different Candida Species Within Homogenous Susceptibility Phenotype
3.4. Comparison Between Cytokines Levels Induced by Different Susceptibility Phenotype of Candida Within Homogenous Species
3.5. Comparisons Between Cytokine Levels Induced by C. auris and Individual Candida Species, Fluconazole Susceptible and Resistant
3.6. Profiles of Cytokine Inductions According to Fluconazole Susceptibility Phenotype and Species of Stimulating Candida Strains
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levtz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef]
- Hohmann, F.B.; Chaves, R.C.F.; Olivato, G.B.; Souza, G.M.; Galindo, V.B.; Silva, M., Jr.; Martino, M.D.V.; Menezes, F.G.; Corrêa, T.D. Characteristics, risk factors, and outcomes of bloodstream Candida infections in the intensive care unit: A retrospective cohort study. J. Int. Med. Res. 2023, 51, 3000605221131122. [Google Scholar] [CrossRef]
- Basmaciyan, L.; Bon, F.; Paradis, T.; Lapaquette, P.; Dalle, F. Candida albicans interactions with the host: Crossing the intestinal epithelial barrier. Tissue Barriers. 2019, 7, 1612661. [Google Scholar] [CrossRef]
- Sheppard, D.C.; Filler, S.G. Host cell invasion by medically important fungi. Cold Spring Harb. Perspect. Med. 2014, 5, a019687. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, L.; Xu, Z.; Zhang, J.; Jiang, Y.Y.; Cao, Y.; Yan, T. Innate immune cell response upon Candida albicans infection. Virulence 2016, 7, 512–526. [Google Scholar] [CrossRef]
- Jensen, O.; Trujillo, E.; Hanson, L.; Ost, K.S. Controlling Candida: Immune regulation of commensal fungi in the gut. Infect. Immun. 2024, 92, e0051623. [Google Scholar] [CrossRef]
- Plato, A.; Hardison, S.E.; Brown, G.D. Pattern recognition receptors in antifungal immunity. Semin. Immunopathol. 2015, 37, 97–106. [Google Scholar] [CrossRef]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A.R. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef]
- Klemptner, R.L.; Sherwood, J.S.; Tugizimana, F.; Dubery, I.A.; Piater, L.A. Ergosterol, an orphan fungal microbe-associated molecular pattern (MAMP). Mol. Plant Pathol. 2014, 15, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Rossard, S.; Roblin, G.; Atanassova, R. Ergosterol triggers characteristic elicitation steps in Beta vulgaris leaf tissues. J. Exper. Botany. 2010, 61, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Targeting pathogen sterols: Defence and counterdefence? PLoS Pathog. 2017, 13, e1006297. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautema-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistence: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 12, e383–e392. [Google Scholar] [CrossRef] [PubMed]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef]
- Clancy, C.J.; Nguyen, M.H. At what cost echinocandin resistance? J. Infect. Dis. 2011, 204, 499–501. [Google Scholar] [CrossRef]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular evolution of antifungal drug resistance. Microbiol. Spectr. 2017, 71, 753–775. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.A.; Coste, A.T.; Ischer, F.; Parker, J.E.; Kelly, S.L.; Pinto, E.; Sanglard, D. Azole resistance by loss of function of the sterol Δ5,6-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob. Agents Chemother. 2012, 56, 1960–1968. [Google Scholar] [CrossRef]
- Flowers, S.A.; Barker, K.S.; Berkow, E.l.; Toner, G.; Chadwick, S.D.; Gygax, S.E.; Morschhäuser, J.; Rogers, P.D. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 2012, 11, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Dykes, C.; Demeter, L.M. Clinical significance of human immunodeficiency virus type 1 replication fitness. Clin. Microbiol. Rev. 2007, 20, 550–578. [Google Scholar] [CrossRef][Green Version]
- Goig, G.A.; Windels, E.M.; Loiseau, C.; Stritt, C.; Biru, L.; Borrell, S.; Brites, D.; Gagneux, S. Ecology, global diversity and evolutionary mechanisms in the Mycobacterium tuberculosis complex. Nat. Rev. Microbiol. 2025. epub ahead of print. [Google Scholar] [CrossRef]
- Borrell, S.; Gagneux, S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung. Dis. 2009, 13, 1456–1466. [Google Scholar][Green Version]
- Cowen, L.E.; Kohn, L.M.; Anderson, J.B. Divergence in fitness and evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 2001, 183, 2971–2978. [Google Scholar] [CrossRef]
- Anderson, J.B. Evolution of antifungal-drug resistance: Mechanisms and pathogen fitness. Nat. Rev. Microbiol. 2005, 3, 547–556. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Garcia-Effron, G.; Lewis, R.E.; Gamarra, S.; Leventakos, K.; Perlin, D.S.; Kontoyiannis, D.P. Fitness and virulence costs of Candida albicans FKS1 Hot Spot Mutations associated with echinocandin resistance. J. Infect. Dis. 2011, 204, 626–635. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antifungal Susceptibility Testing of Yeasts, 3rd ed.; CLSI supplement M27M44S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- NCCLS. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 2nd ed.; Approved standard, NCCLS document M27-A2; NCCLS: Wayne, PA, USA, 2002. [Google Scholar]
- Navarro-Arias, M.J.; Hernández-Chávez, M.J.; García-Carnero, L.C.; Amezcua-Hernández, D.G.; Lozoya-Pérez, N.E.; Estrada-Mata, E.; Martínez-Duncker, I.; Franco, B.; Mora-Montes, H.M. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect. Drug Resist. 2019, 12, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Mata, E.; Navarro-Arias, M.J.; Pérez-García, L.A.; Mellado-Mojica, E.; López, M.G.; Csonka, K.; Gacser, A.; Mora-Montes, H.M. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 2016, 6, 1527. [Google Scholar] [CrossRef]
- Wachtler, B.; Citiulo, F.; Jablonowski, N.; Förster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-epithelial interactions: Dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE 2012, 7, e36952. [Google Scholar] [CrossRef] [PubMed]
- Prieto, D.; Carpena, N.; Maneu, V.; Gil, M.L.; Pla, J.; Gozalbo, D. TLR2 modulates gut colonization and dissemination of Candida albicans in a murine model. Microbes Infect. 2016, 18, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Singh-Babak, S.D.; Babak, T.; Diezmann, S.; Hill, J.A.; Xie, J.L.; Chen, Y.L.; Poutanen, S.M.; Rennie, R.P.; Heitman, J.; Cowen, L.E. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012, 8, e1002718. [Google Scholar] [CrossRef]
- Cowen, L.E.; Sanglard, D.; Calabrese, D.; Sirjusingh, C.; Anderson, J.B.; Kohn, L.M. Evolution of drug resistance in experimental population of Candida albicans. J. Bacteriol. 2000, 182, 1515–1522. [Google Scholar] [CrossRef]
- Vincent, B.M.; Lancaster, A.K.; Scherz-Shouval, R.; Whitesell, L.; Lindquist, S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013, 11, e1001692. [Google Scholar] [CrossRef]
- Andersson, D.I. The biological cost of mutational antibiotoc resistance: Any practical conclusion? Curr. Opin. Microbiol. 2006, 9, 461–465. [Google Scholar] [CrossRef]
- Selmecki, A.M.; Dulmage, K.; Cowen, L.E.; Anderson, J.B.; Berman, J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal resistance. PLoS Genet. 2009, 5, e1000705. [Google Scholar] [CrossRef]
- Borghi, E.; Andreoni, S.; Cirasola, D.; Ricucci, V.; Sciota, R.; Morace, G. Antifungal resistance does not necessarily affect Candida glabrata fitness. J. Chemother. 2014, 26, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; van de Veerdonk, F.L.; Brown, A.J.P.; Netea, M.G. Candida albicans morphogenesis and host defence: Discriminating invasion from colonization. Nat. Rev. Microbiol. 2013, 10, 112–122. [Google Scholar] [CrossRef]
- Tong, Y.; Tang, J. Candida albicans infection and intestinal immunity. Microbiol. Res. 2017, 198, 27–35. [Google Scholar] [CrossRef]
- Imbert, S.; Castain, L.; Pons, A.; Jacob, S.; Meyer, I.; Palous, M.; Vezinet, C.; Langeron, O.; Hennequin, C.; Monse, A.; et al. Discontinuation of echinocandin and azole treatments led to the disappearance of an FKS alteration but not azole resistance during clonal Candida glabrata persistent candidaemia. Clin. Microbiol. Infect. 2016, 22, 891.e5–891.e8. [Google Scholar] [CrossRef] [PubMed]
- Zang, Q.; Sahin, O.; McDermott, P.F.; Payot, S. Fitness of antimicrobial-resistant Campylobacter and Salmonella. Microbes Infect. 2006, 8, 1972–1978. [Google Scholar] [CrossRef]
- Alame Emane, A.K.; Guo, X.; Takiff, H.E.; Liu, S. Drug resistance, fitness and compensatory mutations in Mycobacterium tuberculosis. Tuberculosis 2021, 129, 102091. [Google Scholar] [CrossRef]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Kumar, N.; Pandey, R.; Meis, J.F.; Chowdhary, A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016, 13, 77–82. [Google Scholar] [CrossRef]
- Chatterjee, S.; Alampalli, S.V.; Nageshan, R.K.; Chettiar, S.T.; Joshi, S.; Tatu, U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom. 2015, 16, 686. [Google Scholar] [CrossRef]
- Lockhart, E.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Roger, P.D.; Perlin, D.S. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 2015, 5, a019752. [Google Scholar] [CrossRef] [PubMed]
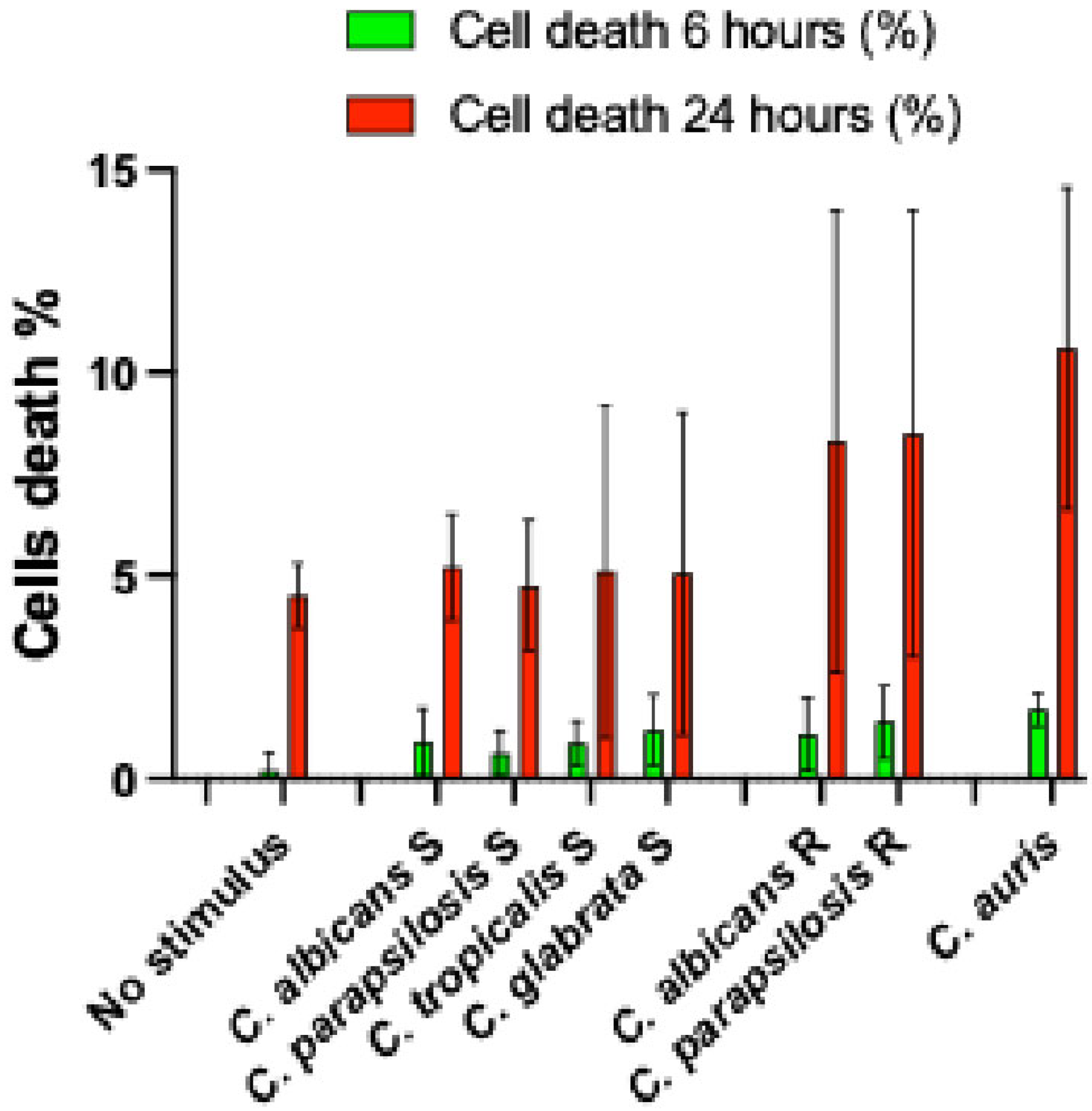
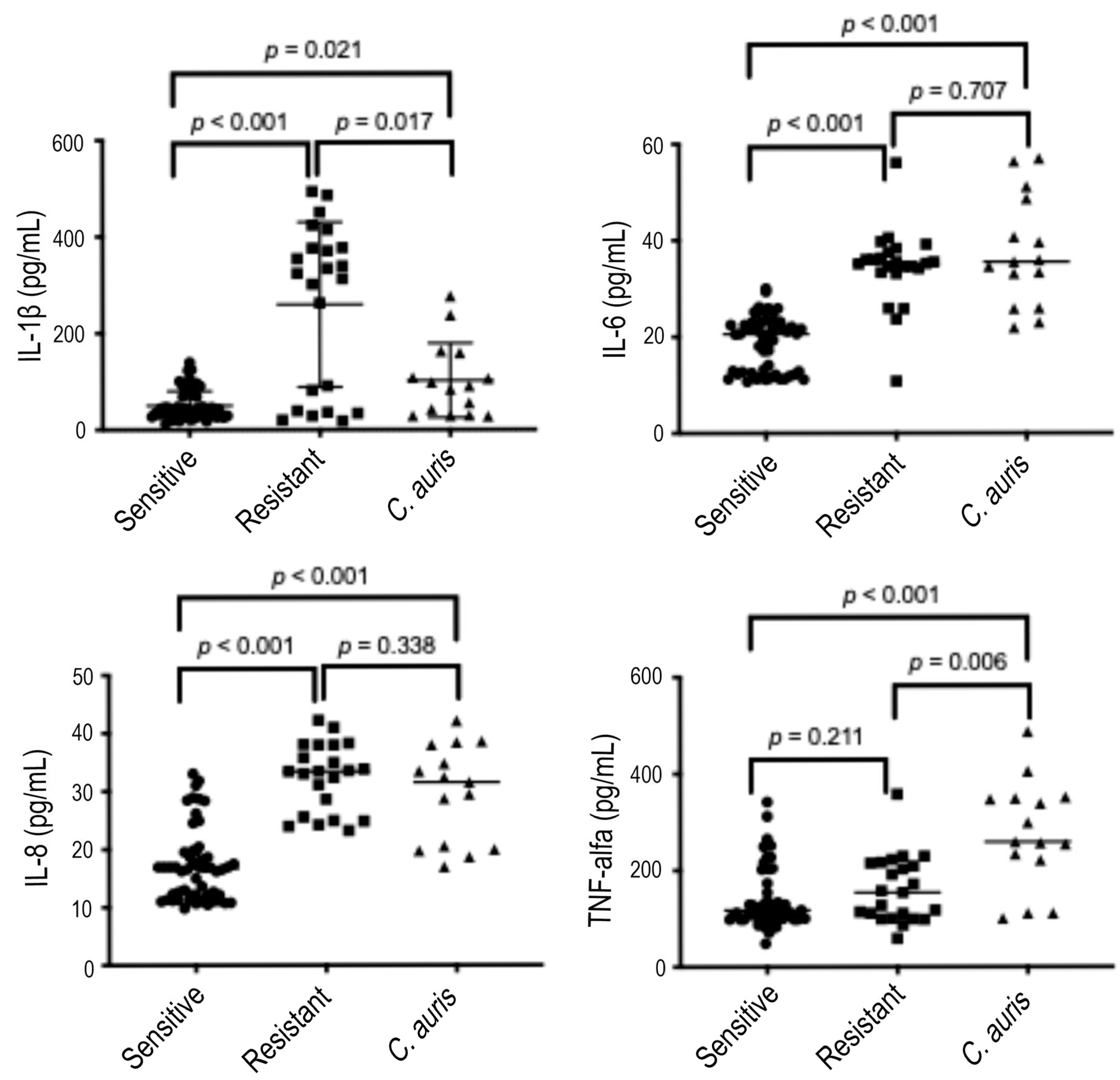
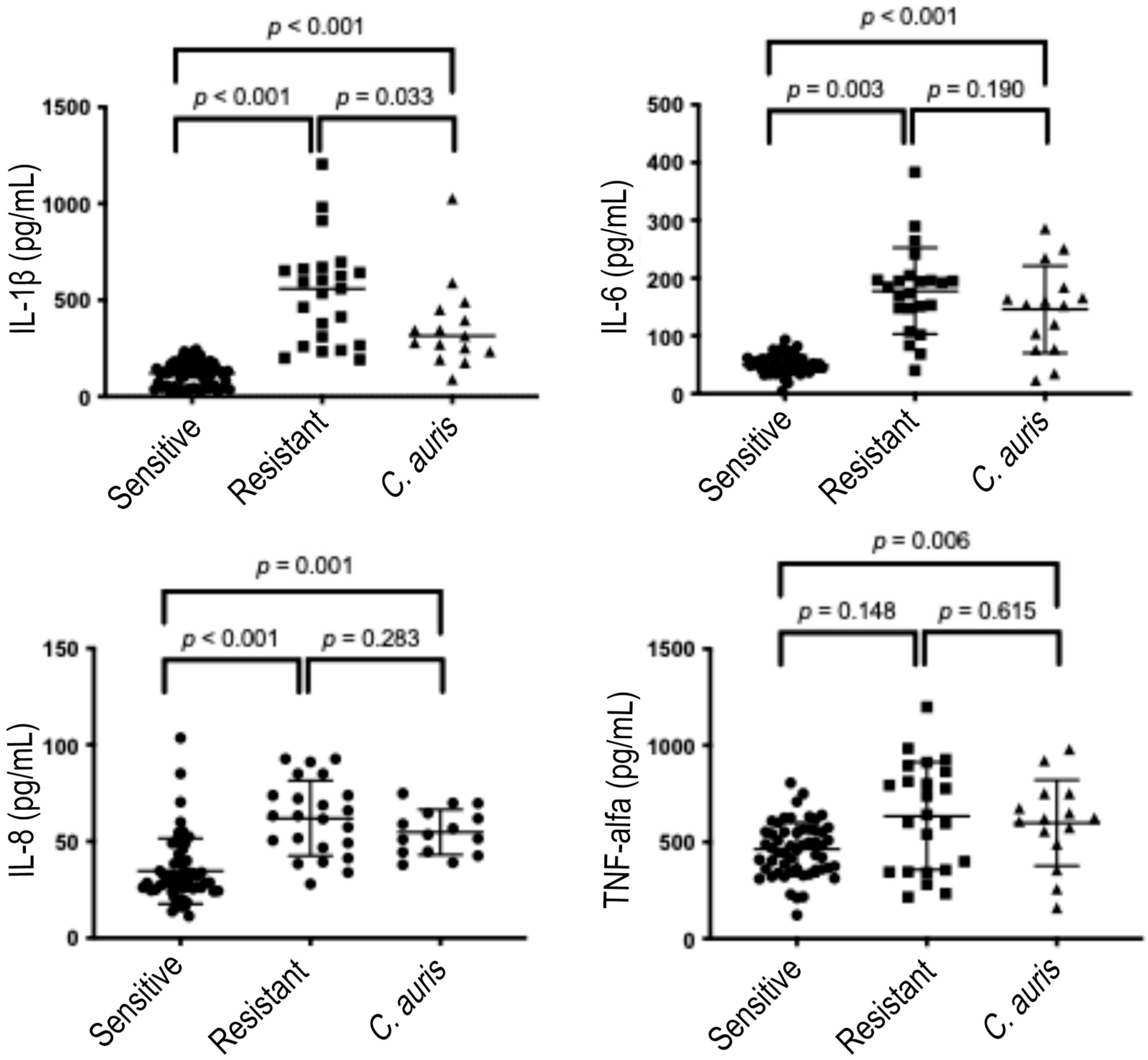
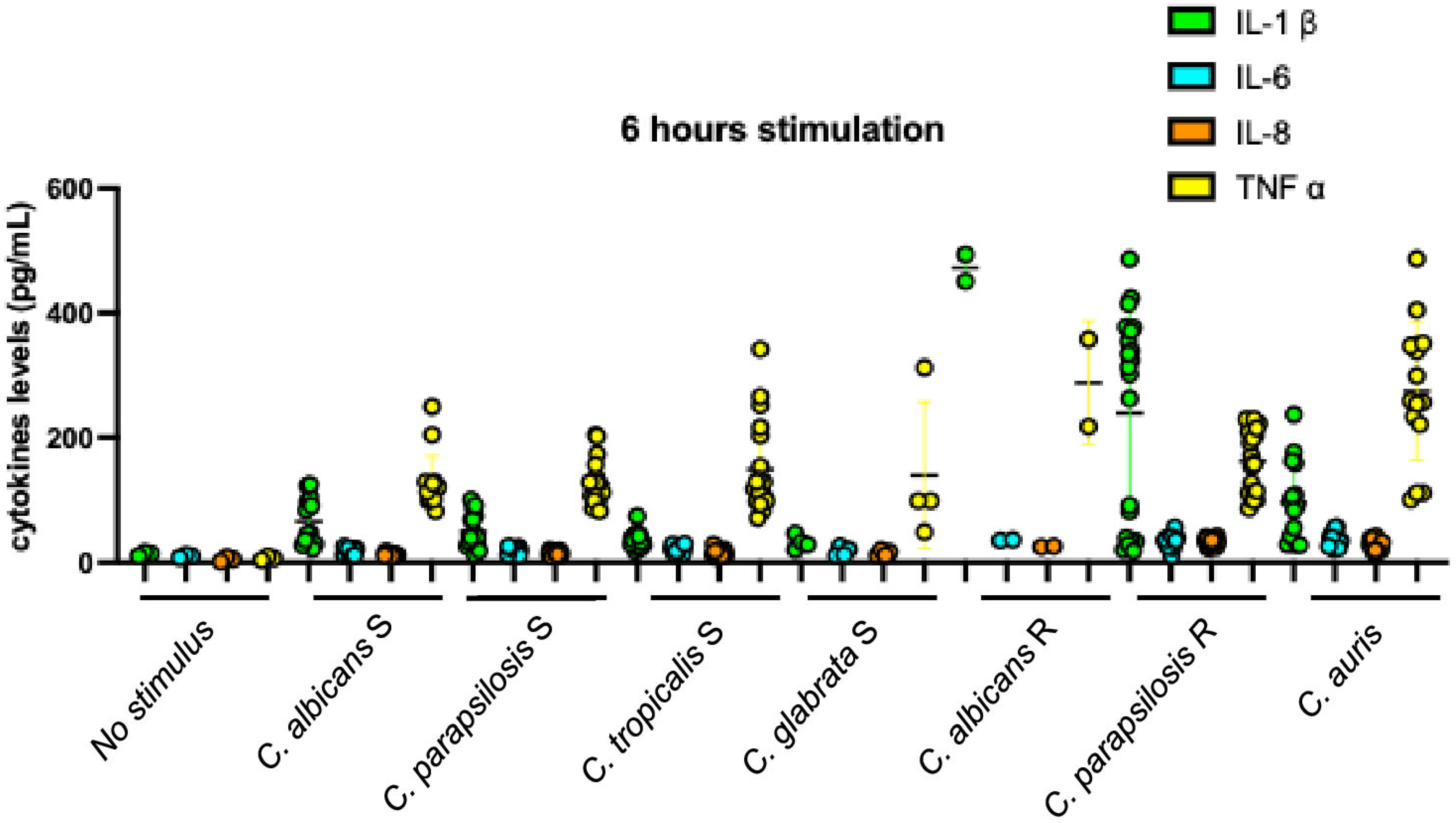
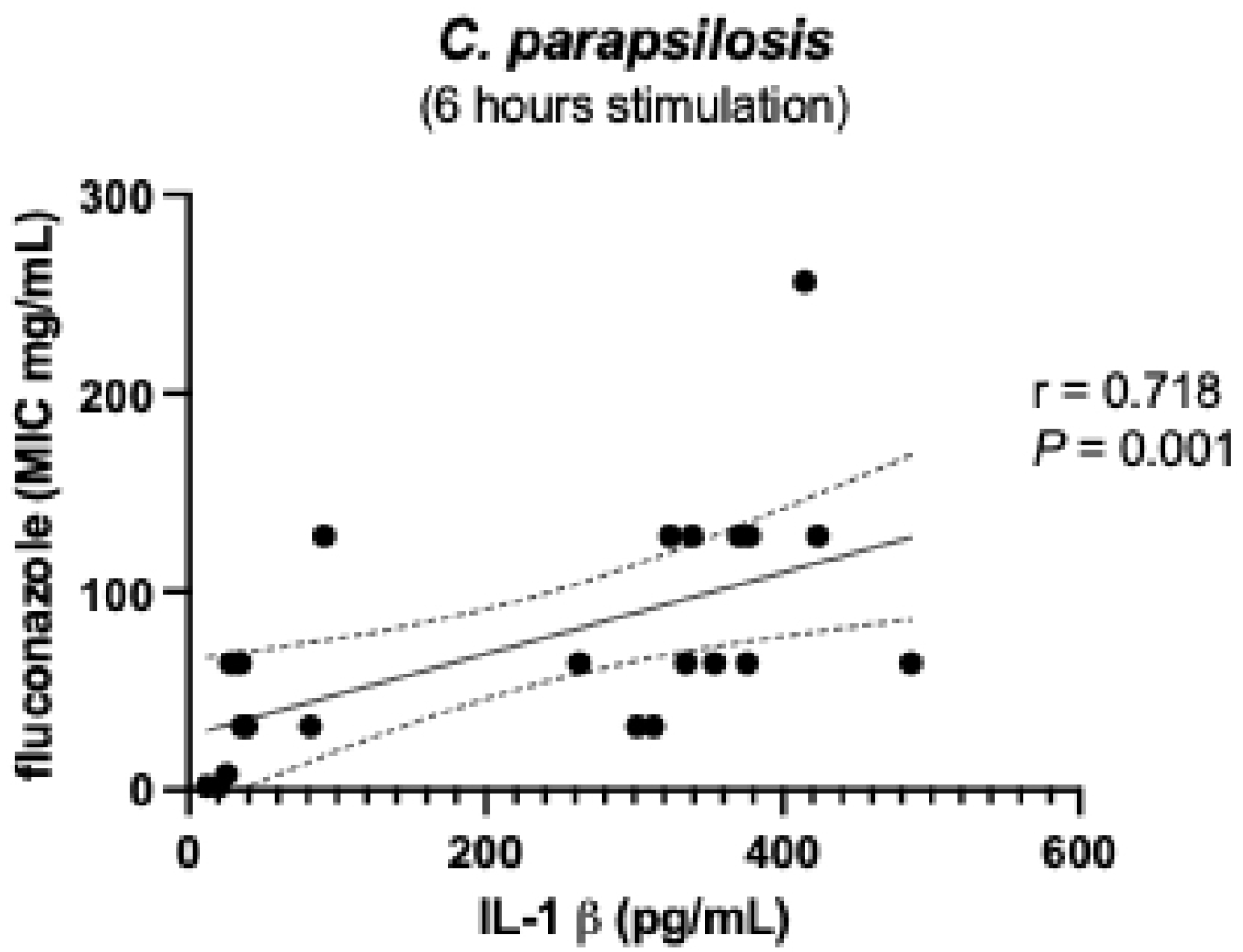
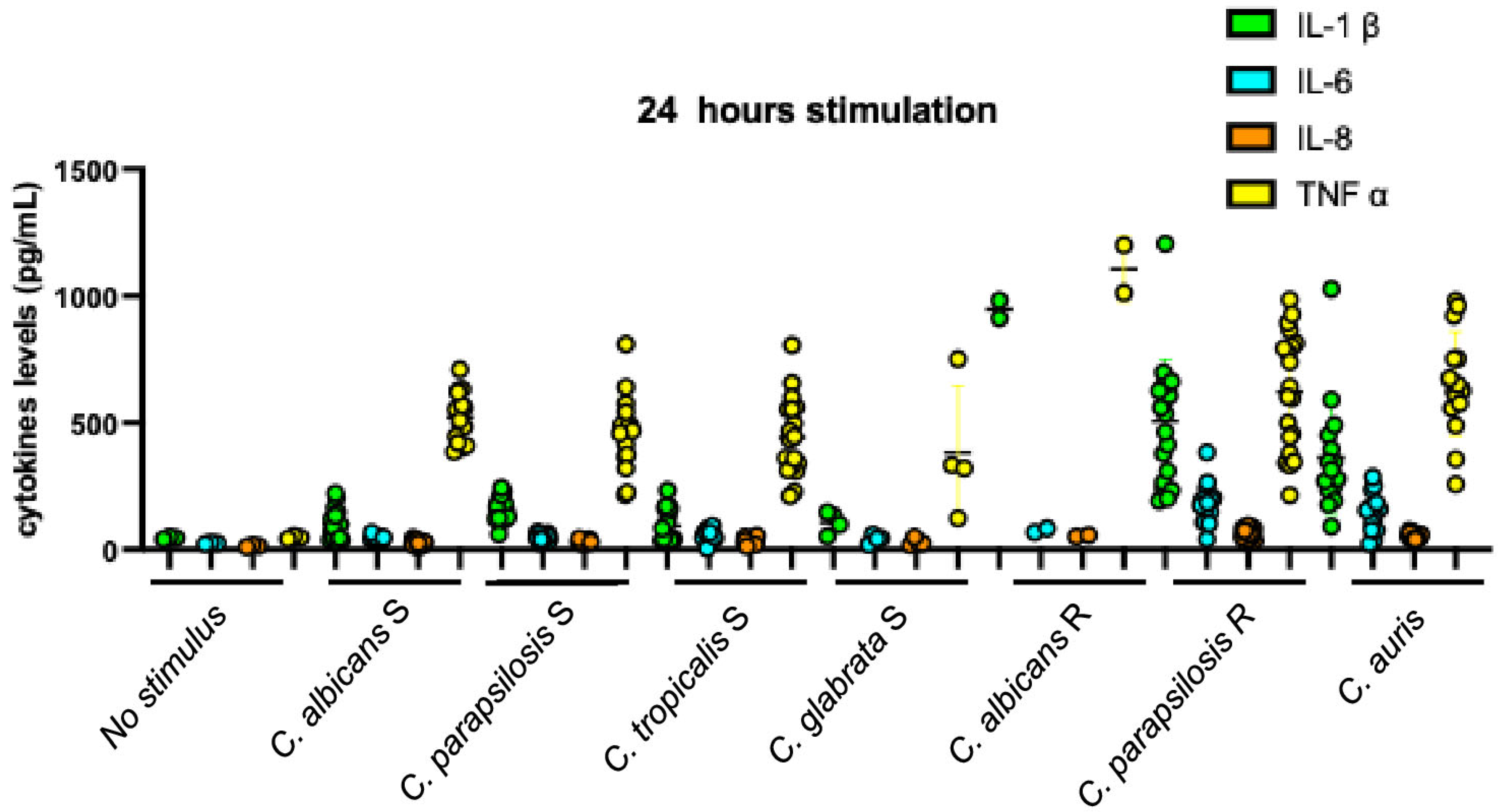
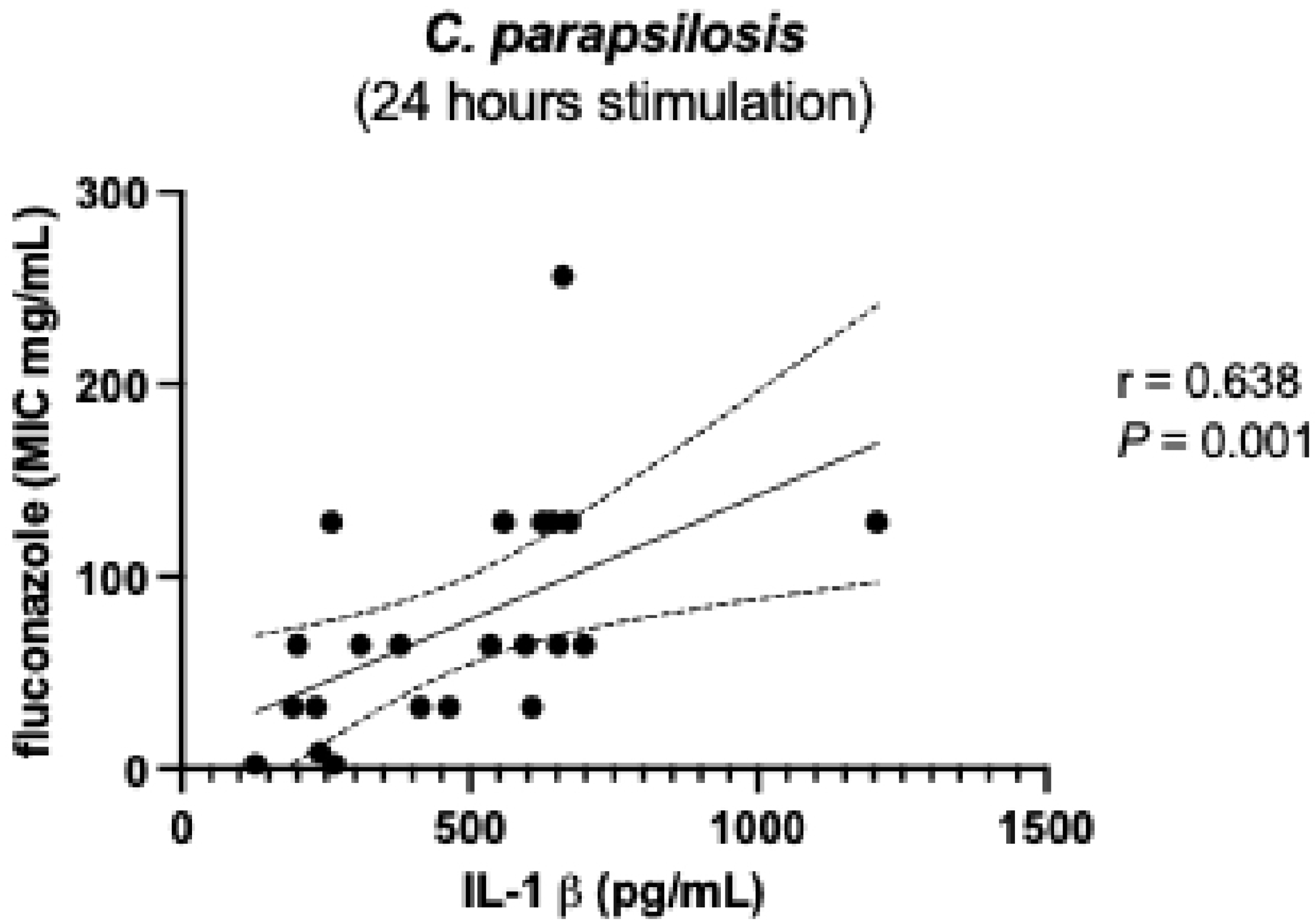
| C. albicans | C. parapsilosis | C. tropicalis | C. glabrata | C. auris | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N strain | MIC mg/L | N strain | MIC mg/L | N strain | MIC mg/L | N strain | MIC mg/L | N Strain | MIC mg/L | N strain | MIC mg/L | ||||||||
| FZ | AB | FZ | AB | FZ | AB | FZ | AB | FZ | AB | FZ | AB | VOR | CAS | ||||||
| A1 | 1 | 1 | P1 | 8 | 1 | P21 | 32 | 1 | T1 | 2 | 1 | G1 | 2 | 0.25 | Au1 | >256 | 2 | 2 | 0.12 |
| A2 | 0.5 | 1 | P2 | 0.25 | 0.5 | P22 | 32 | 2 | T2 | 4 | 1 | G2 | 2 | 1 | Au2 | >256 | 2 | 2 | 0.12 |
| A3 | 2 | 0.5 | P3 | 0.12 | 1 | P23 | 32 | 2 | T3 | 1 | 1 | G3 | 0.5 | 0.25 | Au3 | >256 | 1 | 2 | 0.25 |
| A4 | 0.25 | 0.5 | P4 | 2 | 1 | P24 | 2 | 4 | T4 | 1 | 1 | G4 | 2 | 0.5 | Au4 | >256 | 2 | 4 | 0.12 |
| A5 | 0.25 | 1 | P5 | 0.5 | 1 | P25 | 64 | 0.5 | T5 | 0.5 | 1 | Au5 | >256 | 2 | 2 | 0.12 | |||
| A6 | 0.25 | 1 | P6 | 0.5 | 0.25 | P26 | 128 | 1 | T6 | 0.5 | 1 | Au6 | >256 | 2 | 2 | 0.12 | |||
| A7 | 0.5 | 0.5 | P7 | 0.5 | 1 | P27 | 64 | 0.5 | T7 | 1 | 1 | Au7 | >256 | 2 | 2 | 0.12 | |||
| A8 | 0.25 | 1 | P8 | 0.5 | 1 | P28 | 64 | 2 | T8 | 1 | 0.5 | Au8 | >256 | 4 | 4 | 0.06 | |||
| A9 | 0.25 | 1 | P9 | 1 | 1 | P29 | 128 | 2 | T9 | 1 | 1 | Au9 | >256 | 2 | 2 | >4 | |||
| A10 | 0.5 | 0.5 | P10 | 0.5 | 0.5 | P30 | 128 | 1 | T10 | 1 | 1 | Au10 | >256 | 2 | 4 | 0.12 | |||
| A11 | 0.5 | 0.5 | P11 | 0.5 | 0.5 | P31 | 32 | 0.5 | T11 | 0.5 | 0.5 | Au11 | >256 | 2 | 4 | 0.03 | |||
| A12 | 0.5 | 1 | P12 | 0.5 | 0.25 | P32 | 32 | 0.5 | T12 | 1 | 1 | Au12 | >256 | 2 | 4 | 0.25 | |||
| A13 | 1 | 0.5 | P13 | 0.5 | 0.5 | P33 | 64 | 1 | T13 | 2 | 1 | ||||||||
| A14 | <0.12 | 0.5 | P14 | 0.5 | 0.5 | P34 | 128 | 1 | T14 | 1 | 2 | ||||||||
| A15 | 0.25 | 1 | P15 | 0.25 | 0.5 | P35 | 256 | 0.5 | T15 | 4 | 1 | ||||||||
| A16 | 0.25 | 1 | P16 | 0.5 | 0.5 | P36 | 128 | 0.5 | T16 | 2 | 1 | ||||||||
| A17 | >256 | 1 | P17 | 1 | 1 | P37 | 64 | 1 | T17 | 1 | 1 | ||||||||
| A18 | >256 | 1 | P18 | 1 | 1 | P38 | 128 | 1 | T18 | 1 | 1 | ||||||||
| P19 | 64 | 0.5 | P39 | 64 | 1 | T19 | 4 | 1 | |||||||||||
| P20 | 16 | 0.5 | |||||||||||||||||
| Stimulation By | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cytokine and incubation time (hours) | Sensitive C. albicans vs. | Sensitive C. parapsilosis vs. | Sensitive C. tropicalis vs. | Sensitive C. albicans vs. | Sensitive C. parapsilosis vs. | Resistant C. albicans | |||
| Sensitive C. parapsilosis | Sensitive C. tropicalis | Sensitive C. glabrata | Sensitive C. tropicalis | Sensitive C. glabrata | Sensitive C. glabrata | Resistant C. albicans | Resistant C. parapsilosis | Resistant C. parapsilosis | |
| P | P | P | P | P | P | P | P | P | |
| IL-1β (6) | 0.058 | 0.057 | 0.097 | 0.186 | 0.253 | 0456 | 0.001 | 0.013 | 0.012 * |
| IL-1β (24) | 0.018 * | 0.554 | 0.307 | 0.086 | 0.166 | 0.493 | 0.015 | <0.001 | 0.032 * |
| IL-6 (6) | 0.426 | 0.017 * | 0.871 | 0.048 * | 0.950 | 0.259 | 0.003 | <0.001 | 0.707 |
| IL-6 (24) | 0.357 | 0.008 * | 0.867 | 0.096 | 0.300 | 0.144 | 0.015 | <0.001 | 0.032 * |
| IL-8 (6) | 0.046 * | 0.041 * | 0.133 | 0.800 | 0.853 | 0.709 | 0.001 | <0.001 | 0.216 |
| IL-8 (24) | 0.075 | 0.130 | 0.904 | 0.889 | 0.187 | 0.318 | 0.003 | <0.001 | 0.557 |
| TNF-α (6) | 0.979 | 0.601 | 0.253 | 0.573 | 0.525 | 0.399 | 0.029 | 0.023 | 0.431 |
| TNF-α (24) | 0.929 | 0.054 | 0.152 | 0.090 | 0.195 | 0.403 | 0.041 | 0.043 | 0.008 * |
| Stimulation By | ||||||
|---|---|---|---|---|---|---|
| Cytokine and incubation time (hours) | C. auris versus | |||||
| Sensitive C. albicans | Sensitive C. parapsilosis | Sensitive C. tropicalis | Sensitive C. glabrata | Resistant C. albicans | Resistant C. parapsilosis | |
| P | P | P | P | P | P | |
| IL-1β (6) | 0.262 * | 0.151 * | 0.007 | 0.062 * | 0.057 | 0.066 |
| IL-1β (24) | <0.001 | <0.001 | <0.001 | 0.004 | 0.059 | 0.063 |
| IL-6 (6) | <0.001 | <0.001 | 0.004 | 0.002 | 0.941 | 0.663 |
| IL-6 (24) | 0.002 | <0.001 | 0.005 | 0.014 | 0.235 | 0.088 |
| IL-8 (6) | <0.001 | <0.001 | <0.001 | 0.040 | 0.529 | 0.230 |
| IL-8 (24) | <0.001 | <0.001 | <0.001 | 0.049 | 0.941 | 0.235 |
| TNF-α (6) | 0.005 | 0.001 | 0.001 | 0.036 | 0.926 | 0.003 * |
| TNF-α (24) | 0.030 | 0.002 | 0.001 | 0.077 | 0.015 * | 0.981 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piatti, G.; Vitale, A.; Schito, A.M.; Penco, S.; Saverino, D. The Role of Drug Resistance in Candida Inflammation and Fitness. Microorganisms 2025, 13, 1777. https://doi.org/10.3390/microorganisms13081777
Piatti G, Vitale A, Schito AM, Penco S, Saverino D. The Role of Drug Resistance in Candida Inflammation and Fitness. Microorganisms. 2025; 13(8):1777. https://doi.org/10.3390/microorganisms13081777
Chicago/Turabian StylePiatti, Gabriella, Alberto Vitale, Anna Maria Schito, Susanna Penco, and Daniele Saverino. 2025. "The Role of Drug Resistance in Candida Inflammation and Fitness" Microorganisms 13, no. 8: 1777. https://doi.org/10.3390/microorganisms13081777
APA StylePiatti, G., Vitale, A., Schito, A. M., Penco, S., & Saverino, D. (2025). The Role of Drug Resistance in Candida Inflammation and Fitness. Microorganisms, 13(8), 1777. https://doi.org/10.3390/microorganisms13081777








