Role of Saccharomyces cerevisiae Fcy Proteins and Their Homologs in the Catabolism of Modified Heterocyclic Pyrimidine Bases
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of Modified Pyrimidine Heterocyclic Bases
- General Information
2.1.1. Synthesis of 2-Methylthiouracil
2.1.2. Synthesis of 2-Methylthiocytosine
2.1.3. Synthesis of 3-Methyluracil
- (i)
- Protection of N-1 with BOC-group: To a solution of uracil (0.224 g, 2 mmol) and (t-BOC)2O (0.436 g, 2 mmol) in acetonitrile (5 mL), DMAP (2.2 mg, 0.02 mmol) was added, and the mixture was stirred at room temperature for 3 h. The solvent was removed under vacuum, and the residue was used in the next step without purification.
- (ii)
- N-3 methylation: The crude material from the previous (i) step was diluted with DMF (10 mL), and NaH (120 mg, 3.1 mmol, 60% in mineral oil) was added. The suspension was stirred at room temperature for 30 min, and then CH3I (194 µL, 3.1 mmol) was added. The reaction mixture was stirred for 2 h and then poured into cold water (20 mL), and the product was extracted with ethyl acetate (3 × 20 mL). The extracts were dried (Na2SO4), evaporated to dryness, and used in the next step.
- (iii)
- N-1 BOC deprotection: The crude material was dissolved in chloroform (10 mL), and Trifluoroacetic acid (1.5 mL, 19.6 mmol) was added. The reaction mixture was stirred at room temperature for 24 h. After the reaction was completed (TLC), the solvent was evaporated under reduced pressure, and the residue was purified through flash column chromatography (silica gel, chloroform/methanol mixture, 10/0→10/0.5). Overall yield 100 mg (40%), white solid, Rf = 0.48 (CHCl3/MeOH–9/1). HPLC purity 97%. 1H NMR (DMSO-d6, 400 MHz): Δ = 3.11 (s, 3H, CH3), 5.59 (dd, J = 7.6, 1.1 Hz, 1H, CH=CH), 7.44 (dd, J = 7.5, 5.9 Hz, 1H, CH=CH), 11.13 (s, 1H, NH). 13C NMR (DMSO-d6, 101 MHz): Δ = 26.86, 100.02, 140.84, 152.07, 163.77.
2.2. Yeast Strains and Growth Media
2.3. Genome-Wide Screening Procedure
2.4. Growth of Selected YKO Mutants and Retesting on MD–Agar Medium
2.5. Determination of the Growth Rate of Selected YKO Mutants
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A Database of RNA Modifications and Related Information. 2023 Update. Nucleic Acids Res. 2024, 52, D239–D244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, Z. tRNA Modifications: Greasing the Wheels of Translation and Beyond. RNA Biol. 2025, 22, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. RNA Nucleotide Methylation: 2021 Update. WIREs RNA 2022, 13, e1691. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The Expanding World of tRNA Modifications and Their Disease Relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef]
- Arevalo, C.P.; Bolton, M.J.; Le Sage, V.; Ye, N.; Furey, C.; Muramatsu, H.; Alameh, M.G.; Pardi, N.; Drapeau, E.M.; Parkhouse, K.; et al. A Multivalent Nucleoside-Modified mRNA Vaccine against All Known Influenza Virus Subtypes. Science 2022, 378, 899–904. [Google Scholar] [CrossRef]
- Ren, D.; Mo, Y.; Yang, M.; Wang, D.; Wang, Y.; Yan, Q.; Guo, C.; Xiong, W.; Wang, F.; Zeng, Z. Emerging Roles of tRNA in Cancer. Cancer Lett. 2023, 563, 216170. [Google Scholar] [CrossRef]
- Echaide, M.; Chocarro de Erauso, L.; Bocanegra, A.; Blanco, E.; Kochan, G.; Escors, D. mRNA Vaccines against SARS-CoV-2: Advantages and Caveats. Int. J. Mol. Sci. 2023, 24, 5944. [Google Scholar] [CrossRef]
- Delaunay, S.; Helm, M.; Frye, M. RNA Modifications in Physiology and Disease: Towards Clinical Applications. Nat. Rev. Genet. 2024, 25, 104–122. [Google Scholar] [CrossRef]
- Traube, F.R.; Carell, T. The Chemistries and Consequences of DNA and RNA Methylation and Demethylation. RNA Biol. 2017, 14, 1099–1107. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes 2023, 14, 873. [Google Scholar] [CrossRef]
- Chen, Y.; Hong, T.; Wang, S.; Mo, J.; Tian, T.; Zhou, X. Epigenetic Modification of Nucleic Acids: From Basic Studies to Medical Applications. Chem. Soc. Rev. 2017, 46, 2844–2872. [Google Scholar] [CrossRef] [PubMed]
- Duschinsky, R. The Synthesis of 5-Fluoropyrimidines. J. Am. Chem. Soc. 1957, 79, 4559–4560. [Google Scholar] [CrossRef]
- Chang, Y.C.; Lamichhane, A.K.; Cai, H.; Walter, P.J.; Bennett, J.E.; Kwon-Chung, K.J. Moderate Levels of 5-Fluorocytosine Cause the Emergence of High Frequency Resistance in Cryptococci. Nat. Commun. 2021, 12, 3418. [Google Scholar] [CrossRef]
- Shelton, J.; Lu, X.; Hollenbaugh, J.A.; Cho, J.H.; Amblard, F.; Schinazi, R.F. Metabolism, Biochemical Actions, and Chemical Synthesis of Anticancer Nucleosides, Nucleotides, and Base Analogs. Chem. Rev. 2016, 116, 14379–14455. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, X.; Xi, Z.; Chattopadhyaya, J. Nucleic Acids in Medicinal Chemistry and Chemical Biology: Drug Development and Clinical Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2023. [Google Scholar] [CrossRef]
- Bartoletti, M.; Mozaffari, E.; Amin, A.N.; Doi, Y.; Loubet, P.; Rivera, C.G.; Roshon, M.; Rawal, A.; Kaiser, E.; Nicolae, M.V.; et al. A Systematic Review and Meta-Analysis of the Effectiveness of Remdesivir to Treat SARS-CoV-2 Infection in Hospitalized Patients: Have the Guidelines Evolved with the Evidence? Clin. Infect. Dis. 2025, ciaf111. [Google Scholar] [CrossRef]
- Hollenstein, M. Enzymatic Synthesis of Base-Modified Nucleic Acids. In Handbook of Chemical Biology of Nucleic Acids; Sugimoto, N., Ed.; Springer Nature: Singapore, 2023; pp. 1–39. [Google Scholar] [CrossRef]
- Van Giesen, K.J.D.; Thompson, M.J.; Meng, Q.; Lovelock, S.L. Biocatalytic Synthesis of Antiviral Nucleosides, Cyclic Dinucleotides, and Oligonucleotide Therapies. J. Am. Chem. Soc. Au 2023, 3, 13–24. [Google Scholar] [CrossRef]
- Chevallier, M.-R. Cloning and Transcriptional Control of a Eucaryotic Permease Gene. Mol. Cell Biol. 1982, 2, 977–984. [Google Scholar] [CrossRef]
- Kurtz, J.E.; Exinger, F.; Erbs, P.; Jund, R. New Insights into the Pyrimidine Salvage Pathway of Saccharomyces cerevisiae: Requirement of Six Genes for Cytidine Metabolism. Curr. Genet. 1999, 36, 130–136. [Google Scholar] [CrossRef]
- Weber, E.; Rodriguez, C.; Chevallier, M.R.; Jund, R. The Purine-cytosine Permease Gene of Saccharomyces cerevisiae: Primary Structure and Deduced Protein Sequence of the FCY2 Gene Product. Mol. Microbiol. 1990, 4, 585–596. [Google Scholar] [CrossRef]
- Engel, S.R.; Aleksander, S.; Nash, R.S.; Wong, E.D.; Weng, S.; Miyasato, S.R.; Sherlock, G.; Cherry, J.M. Saccharomyces Genome Database: Advances in Genome Annotation, Expanded Biochemical Pathways, and Other Key Enhancements. Genetics 2025, 229, iyae185. [Google Scholar] [CrossRef]
- Huang, R.Y.; Eddy, M.; Vujcic, M.; Kowalski, D. Genome-Wide Screen Identifies Genes Whose Inactivation Confer Resistance to Cisplatin in Saccharomyces cerevisiae. Cancer Res. 2005, 65, 5890–5897. [Google Scholar] [CrossRef] [PubMed]
- Guetsova, M.L.; Lecoq, K.; Daignan-Fornier, B. The Isolation and Characterization of Saccharomyces cerevisiae Mutants That Constitutively Express Purine Biosynthetic Genes. Genetics 1997, 147, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Paluszynski, J.P.; Klassen, R.; Rohe, M.; Meinhardt, F. Various Cytosine/Adenine Permease Homologues Are Involved in the Toxicity of 5-Flourocytosine in Saccharomyces cerevisiae. Yeast 2006, 23, 707–715. [Google Scholar] [CrossRef]
- Yang, D.H.; Lamichhane, A.K.; Kwon-Chung, K.J.; Chang, Y.C. Factors Influencing the Nitrogen-Source Dependent Flucytosine Resistance in Cryptococcus Species. mBio 2023, 14, e0345122. [Google Scholar] [CrossRef]
- Urbonavičius, J.; Čekytė, A.; Tauraitė, D. N4-Methylcytosine Supports the Growth of Escherichia Coli Uracil Auxotrophs. Int. J. Mol. Sci. 2025, 26, 1812. [Google Scholar] [CrossRef]
- Borek, C.; Reichle, V.F.; Kellner, S. Synthesis and Metabolic Fate of 4-Methylthiouridine in Bacterial tRNA. ChemBioChem 2020, 21, 2768–2771. [Google Scholar] [CrossRef]
- Zhang, Q.; Xia, Z.; Mitten, M.J.; Lasko, L.M.; Klinghofer, V.; Bouska, J.; Johnson, E.F.; Penning, T.D.; Luo, Y.; Giranda, V.L.; et al. Hit to Lead Optimization of a Novel Class of Squarate-Containing Polo-like Kinases Inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 7615–7622. [Google Scholar] [CrossRef]
- Jaime-Figueroa, S.; Zamilpa, A.; Guzmán, A.; Morgans, J. N-3-Alkylation of Uracil and Derivatives via N-1-BOC Protection. Synth. Commun. 2001, 31, 3739–3746. [Google Scholar] [CrossRef]
- Sherman, F.; Fink, G.; Hicks, J. Methods in Yeast Genetics: A Laboratory Course Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1987. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Stanislauskienė, R.; Laurynėnas, A.; Rutkienė, R.; Aučynaitė, A.; Tauraitė, D.; Meškienė, R.; Urbelienė, N.; Kaupinis, A.; Valius, M.; Kaliniene, L.; et al. YqfB Protein from Escherichia coli: An Atypical Amidohydrolase Active towards N4-Acylcytosine Derivatives. Sci. Rep. 2020, 10, 788. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Maitra, P.K. Induction of Specific Enzymes of the Oxidative Pentose Phosphate Pathway by Glucono-δ-Lactone in Saccharomyces cerevisiae. J. Gen. Microbiol. 1992, 138, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Kwolek-Mirek, M.; Maslanka, R.; Molon, M. Disorders in NADPH Generation via Pentose Phosphate Pathway Influence the Reproductive Potential of the Saccharomyces cerevisiae Yeast Due to Changes in Redox Status. J. Cell. Biochem. 2019, 120, 8521–8533. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Straub, M.L.; Souciet, J.L.; Potier, S.; De Montigny, J. New Plasmid System to Select for Saccharomyces Cerevisiae Purine-Cytosine Permease Affinity Mutants. J. Bacteriol. 2001, 183, 4386–4388. [Google Scholar] [CrossRef]
- Hope, W.W.; Tabernero, L.; Denning, D.W.; Anderson, M.J. Molecular Mechanisms of Primary Resistance to Flucytosine in Candida Albicans. Antimicrob. Agents Chemother. 2004, 48, 4377–4386. [Google Scholar] [CrossRef]
- Yoo, H.S.; Cunningham, T.S.; Cooper, T.G. The Allantoin and Uracil Permease Gene Sequences of Saccharomyces cerevisiae Are Nearly Identical. Yeast 1992, 8, 997–1006. [Google Scholar] [CrossRef]
- Vickers, M.F.; Yao, S.Y.M.; Baldwin, S.A.; Young, J.D.; Cass, C.E. Nucleoside Transporter Proteins of Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 25931–25938. [Google Scholar] [CrossRef]
- Pinson, B.; Chevallier, J.; Urban-Grimal, D. Only One of the Charged Amino Acids Located in Membrane-Spanning Regions Is Important for the Function of the Saccharomyces cerevisiae Uracil Permease. Biochem. J. 1999, 339, 37–42. [Google Scholar] [CrossRef]
- Jund, R.; Lacroute, F. Genetic and Physiological Aspects of Resistance to 5-Fluoropyrimidines in Saccharomyces cerevisiae. J. Bacteriol. 1970, 102, 607–615. [Google Scholar] [CrossRef]
- Belenky, P.A.; Moga, T.G.; Brenner, C. Saccharomyces cerevisiae YOR071C Encodes the High Affinity Nicotinamide Riboside Transporter Nrt1. J. Biol. Chem. 2008, 283, 8075–8079. [Google Scholar] [CrossRef]
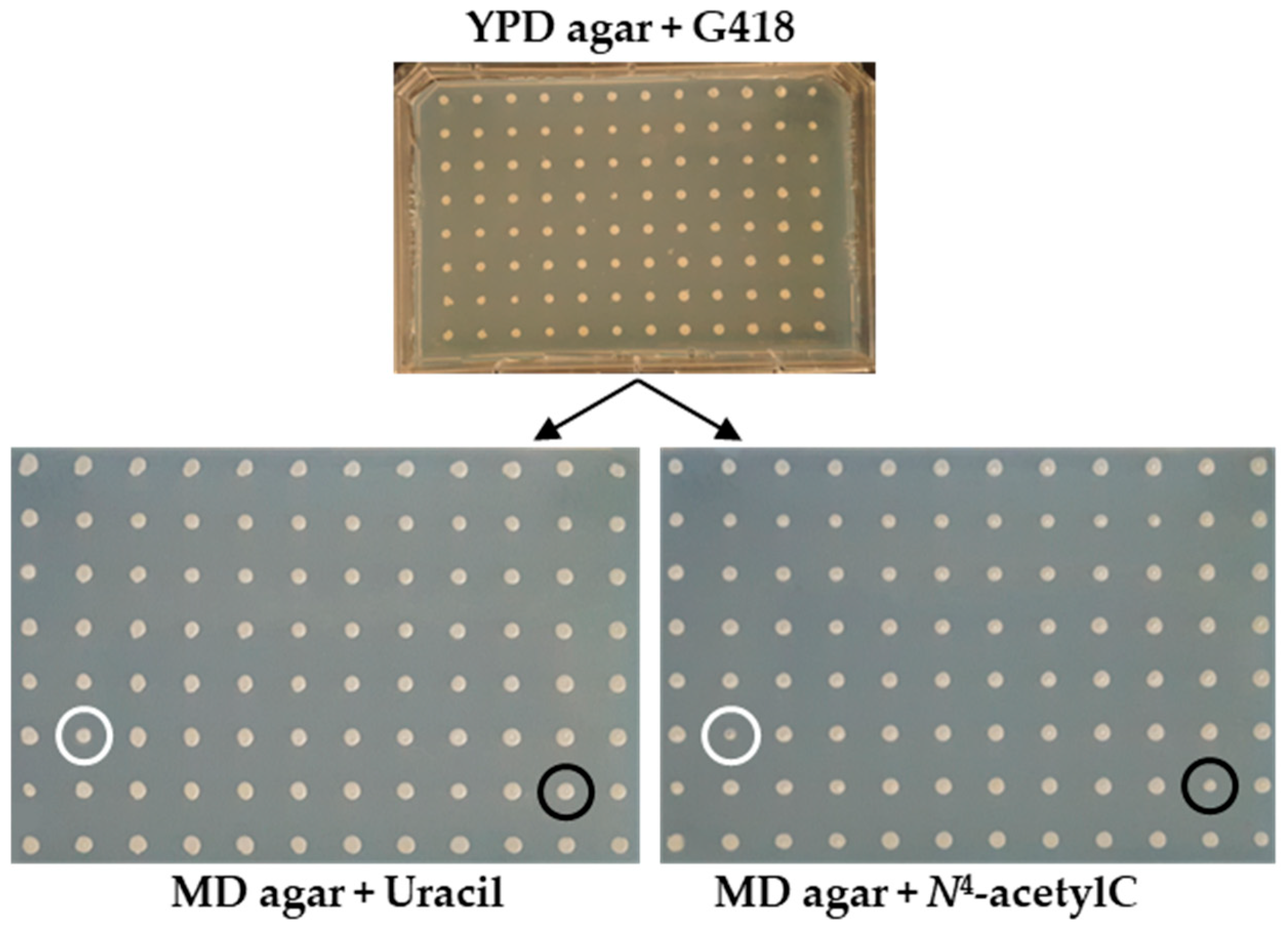

| Primary Screen | Retesting | |||||
|---|---|---|---|---|---|---|
| ORF ID | Gene Name | MD + U | MD + N4-AcetylC | MD + U | MD + N4-AcetylC | Description |
| YPR062W | FCY1 | G++ | G+/− | +++ | +/− | Cytosine deaminase |
| YER060W | FCY21 | G++ | G+ | +++/− | ++/− | Putative purine–cytosine permease |
| YEL029C | BUD16 | G++ | G+/− | +++ | +/− | Putative pyridoxal kinase |
| YHR183W | GND1 | G++ | G+ | +++/− | ++ | 6-phosphogluconate dehydrogenase (decarboxylating) |
| YBR021W | FUR4 | G++ | G+ | +++ | ++ | Plasma membrane localized uracil permease |
| YER056C | FCY2 | G+ | G+/− | +/− | +/− | Purine–cytosine permease |
| YBL067C | UBP13 | G++ | G+ | +++/− | +++/− | Ubiquitin-specific protease that cleaves Ub–protein fusions |
| YBR159W | IFA38 | G++ | G+ | +++/− | +++/− | Microsomal beta-keto-reductase |
| YCR023C | G++ | G+ | +++/− | +++/− | Vacuolar membrane protein of unknown function | |
| YDL201W | TRM8 | G++ | G+ | +++/− | +++/− | Catalytic subunit of a tRNA methyltransferase complex |
| YDR370C | DXO1 | G++ | G+ | +++ | +++ | Cytoplasmic 5′ exoribonuclease |
| YDR369C | XRS2 | G++ | G+ | +++ | +++ | FHA domain-containing component of the Mre11 complex |
| YDR227W | SIR4 | G++ | G+ | +++/− | +++/− | SIR protein involved in the assembly of silent chromatin domains |
| YER005W | YND1 | G++ | G+/− | +++ | +++ | Apyrase with wide substrate specificity |
| YDR452W | PPN1 | G++ | G+/− | +++ | +++ | Dual endo- and exopolyphosphatase with a role in phosphate metabolism |
| YDR482C | CWC21 | G++ | G+ | +++/− | +++/− | Protein involved in RNA splicing by the spliceosome |
| YIR032C | DAL3 | G++ | G+ | +++/− | +++/− | Ureidoglycolate lyase |
| YJL133W | MRS3 | G++ | G+ | +++/− | +++/− | Iron transporter; mediates Fe2+ transport across the inner mito membrane |
| YLR363C | NMD4 | G++ | G+ | +++ | +++ | Protein that may be involved in nonsense-mediated mRNA decay |
| YLR376C | PSY3 | G++ | G+ | +++ | +++ | Component of the Shu complex (aka PCSS complex) |
| YLR375W | STP3 | G++ | G+ | +++/− | +++/− | Zinc-finger protein of unknown function |
| YMR068W | AVO2 | G++ | G+ | +++/− | +++/− | Subunit of TORC2, a regulator of plasma membrane (PM) homeostasis |
| YMR105C | PGM2 | G++ | G− | +++/− | +++/− | Phosphoglucomutase |
| YMR121C | RPL15B | G++ | G+ | +++/− | +++/− | Ribosomal 60S subunit protein L15B |
| YMR120C | ADE17 | G++ | G+ | +++/− | +++/− | Enzyme of ‘de novo’ purine biosynthesis |
| YNL307C | MCK1 | G++ | G+ | +++/− | +++/− | Dual-specificity ser/thr and tyrosine protein kinase |
| YNL219C | ALG9 | G++ | G+ | +++ | +++ | Mannosyltransferase, involved in N-linked glycosylation |
| YNR032W | PPG1 | G++ | G+ | ++ | ++ | Serine protein phosphatase involved in the formation of the FAR complex |
| YOL009C | MDM12 | G++ | G+ | +++/− | + | Mitochondrial outer membrane protein, ERMES complex subunit |
| YBL042C | FUI1 | G++ | G+ | +++/− | +++/− | High-affinity uridine permease; localizes to the plasma membrane |
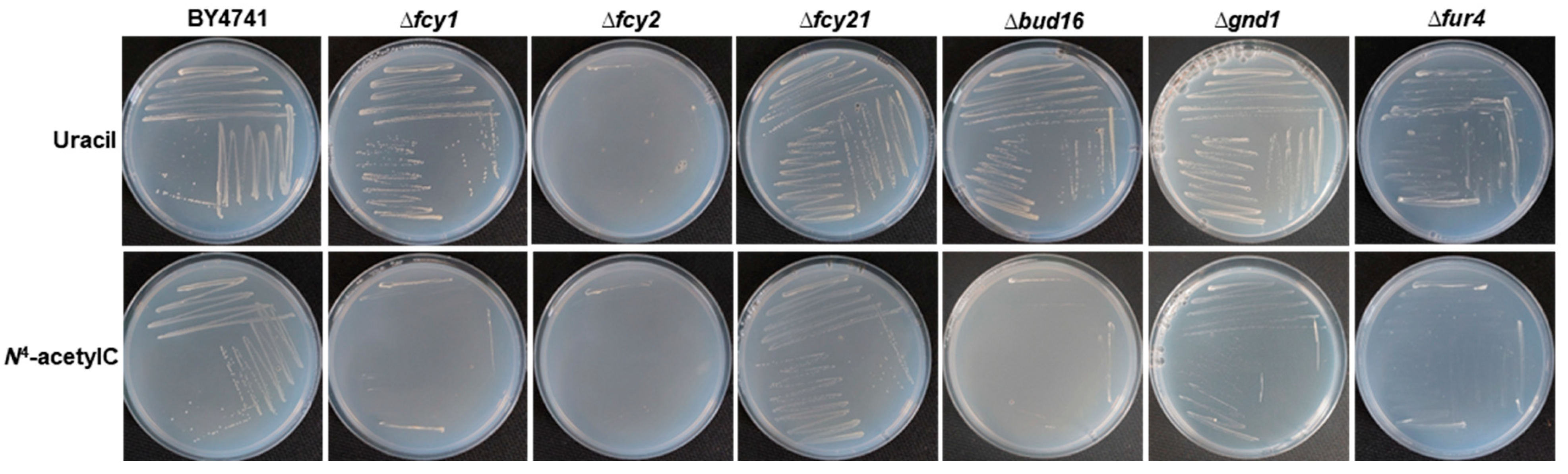
| Sample | Growth Rate Constant, h−1 | Generation Time, h |
|---|---|---|
| BY4741 + uracil | 0.103 ± 0.001 | 6.756 ± 0.080 |
| BY4741 + N4-acetylC | 0.077 ± 0.017 | 9.794 ± 1.868 |
| Δfcy1 + uracil | 0.133 ± 0.006 * | 5.237 ± 0.258 |
| Δfcy1 + N4-acetylC | 0.025 ± 0.002 | 27.965 ± 1.631 |
| Δfcy2 + uracil | 0.029 ± 0.002 * | 24.275 ± 1.336 |
| Δfcy2 + N4-acetylC | 0.018 ± 0.005 | 42.172 ± 8.479 |
| Δfcy21 + uracil | 0.066 ± 0.001 * | 10.594 ± 0.167 |
| Δfcy21 + N4-acetylC | 0.023 ± 0.002 | 30.103 ± 2.252 |
| Δbud16 + uracil | 0.052 ± 0.016 * | 16.929 ± 6.105 |
| Δbud16 + N4-acetylC | 0.030 ± 0.003 | 23.688 ± 1.910 |
| Δgnd1 + uracil | 0.053 ± 0.014 * | 16.441 ± 6.157 |
| Δgnd1 + N4-acetylC | 0.020 ± 0.001 | 34.540 ± 1.601 |
| Δfur4 + uracil | 0.066 ± 0.003 * | 10.494 ± 0.533 |
| Δfur4 + N4-acetylC | 0.020 ± 0.002 | 34.492 ± 3.310 |
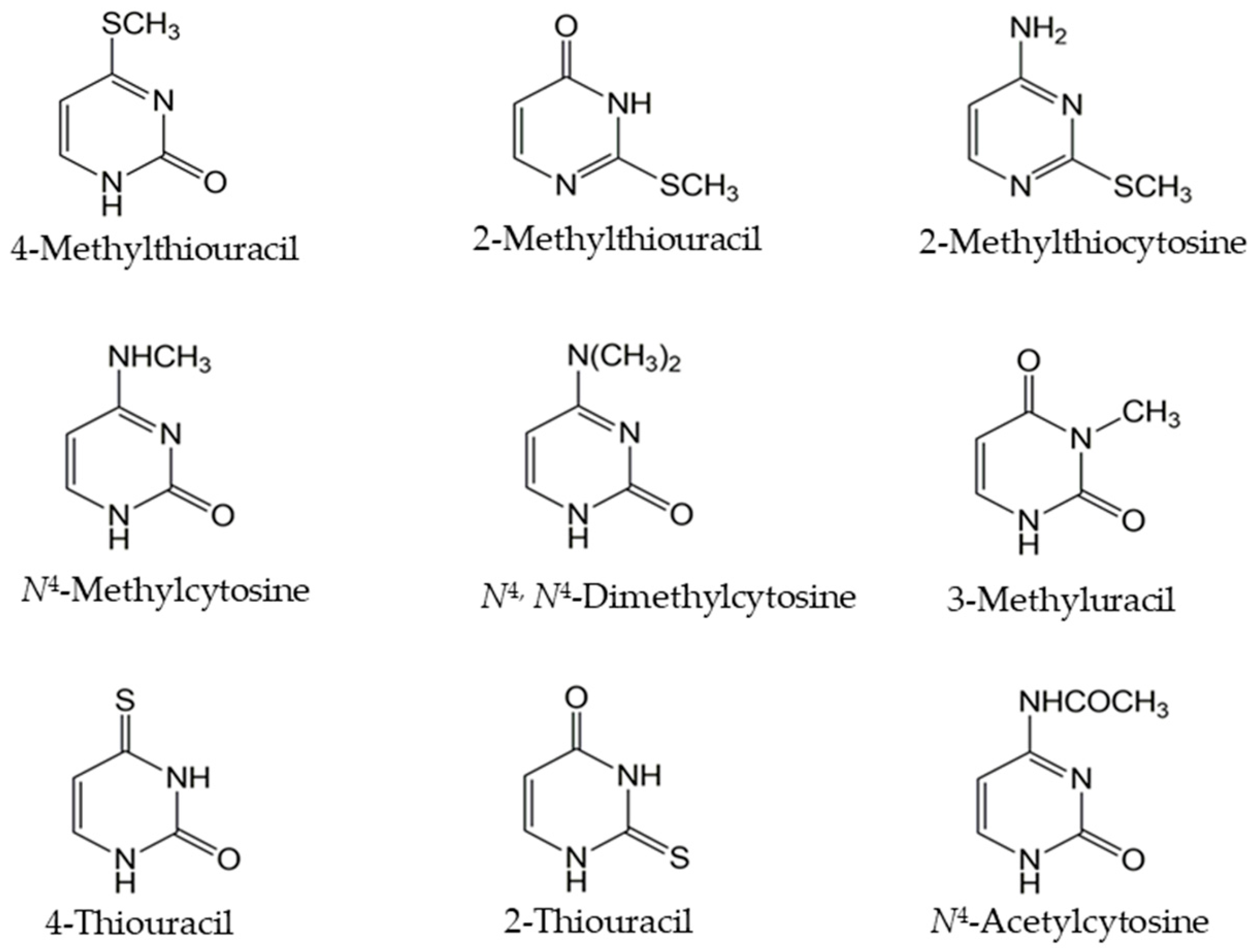
| ORF ID | WT | YPR062W | YER060W | YER056C | YER060W-A | YIR028W | YBL042C | YBR021W | YOR071C |
|---|---|---|---|---|---|---|---|---|---|
| Compound/gene | ∆fcy1 | ∆fcy21 | ∆fcy2 | ∆fcy22 | ∆dal4 | ∆fui1 | ∆fur4 | ∆nrt1 | |
| Uracil | +++ | +++ | +++/− | +/− | +/− | +++/− | +++/− | +++ | +++/− |
| Cytosine | +++ | +++/− | ++ | +/− | +/− | +++/− | +++/− | +++/− | +++/− |
| N4-Acetylcytosine | +++ | +/− | ++/− | +/− | +/− | +++/− | +++/− | ++ | +++ |
| 4-Methylthiouracil | +++/− | + | + | +/− | +/− | ++ | +++/− | +/− | +++/− |
| N4-Methylcytosine | +++/− | +++/− | + | +/− | +/− | +++/− | +++/− | +/− | +++/− |
| N4,N4-Dimethylcytosine | +++/− | ++ | ++/− | +/− | +/− | + | ++/− | +/− | ++/− |
| 2-Thiouracil | +++/− | +++/− | ++ | +/− | +/− | +++/− | +++/− | +/− | ++ |
| 4-Thiouracil | +++/− | ++ | ++ | +/− | +/− | +++/− | +++/− | +/− | +++/− |
| 2-Methylthiouracil | +/− | +/− | +/− | +/− | +/− | +/− | + | +/− | + |
| 2-Methylthiocytosine | +/− | +/− | +/− | +/− | +/− | +/− | +/− | +/− | +/− |
| 3-Methyluracil | +/− | + | +/− | +/− | +/− | +/− | +/− | +/− | +/− |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbonavičius, J.; Vepštaitė-Monstavičė, I.; Lukša-Žebelovič, J.; Servienė, E.; Tauraitė, D. Role of Saccharomyces cerevisiae Fcy Proteins and Their Homologs in the Catabolism of Modified Heterocyclic Pyrimidine Bases. Microorganisms 2025, 13, 1506. https://doi.org/10.3390/microorganisms13071506
Urbonavičius J, Vepštaitė-Monstavičė I, Lukša-Žebelovič J, Servienė E, Tauraitė D. Role of Saccharomyces cerevisiae Fcy Proteins and Their Homologs in the Catabolism of Modified Heterocyclic Pyrimidine Bases. Microorganisms. 2025; 13(7):1506. https://doi.org/10.3390/microorganisms13071506
Chicago/Turabian StyleUrbonavičius, Jaunius, Iglė Vepštaitė-Monstavičė, Juliana Lukša-Žebelovič, Elena Servienė, and Daiva Tauraitė. 2025. "Role of Saccharomyces cerevisiae Fcy Proteins and Their Homologs in the Catabolism of Modified Heterocyclic Pyrimidine Bases" Microorganisms 13, no. 7: 1506. https://doi.org/10.3390/microorganisms13071506
APA StyleUrbonavičius, J., Vepštaitė-Monstavičė, I., Lukša-Žebelovič, J., Servienė, E., & Tauraitė, D. (2025). Role of Saccharomyces cerevisiae Fcy Proteins and Their Homologs in the Catabolism of Modified Heterocyclic Pyrimidine Bases. Microorganisms, 13(7), 1506. https://doi.org/10.3390/microorganisms13071506






