Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target?
Abstract
1. Introduction
2. Impact of agr and sarA
3. Impact on Biofilm-Associated Intrinsic Resistance
4. Impact on Oxacillin Resistance
5. Biofilm Inhibitors as Therapeutic Options
6. Diversity in S. aureus
7. Biofilm Formation as a Function of Methicillin Resistance
8. Why sarA Mutants Do Not Form a Biofilm
9. Why Increased Protease Production Limits Biofilm Formation
10. Impact of sarA on Other Critical Phenotypes
11. Protease-Mediated Post-Translational Regulation (Figure 1)
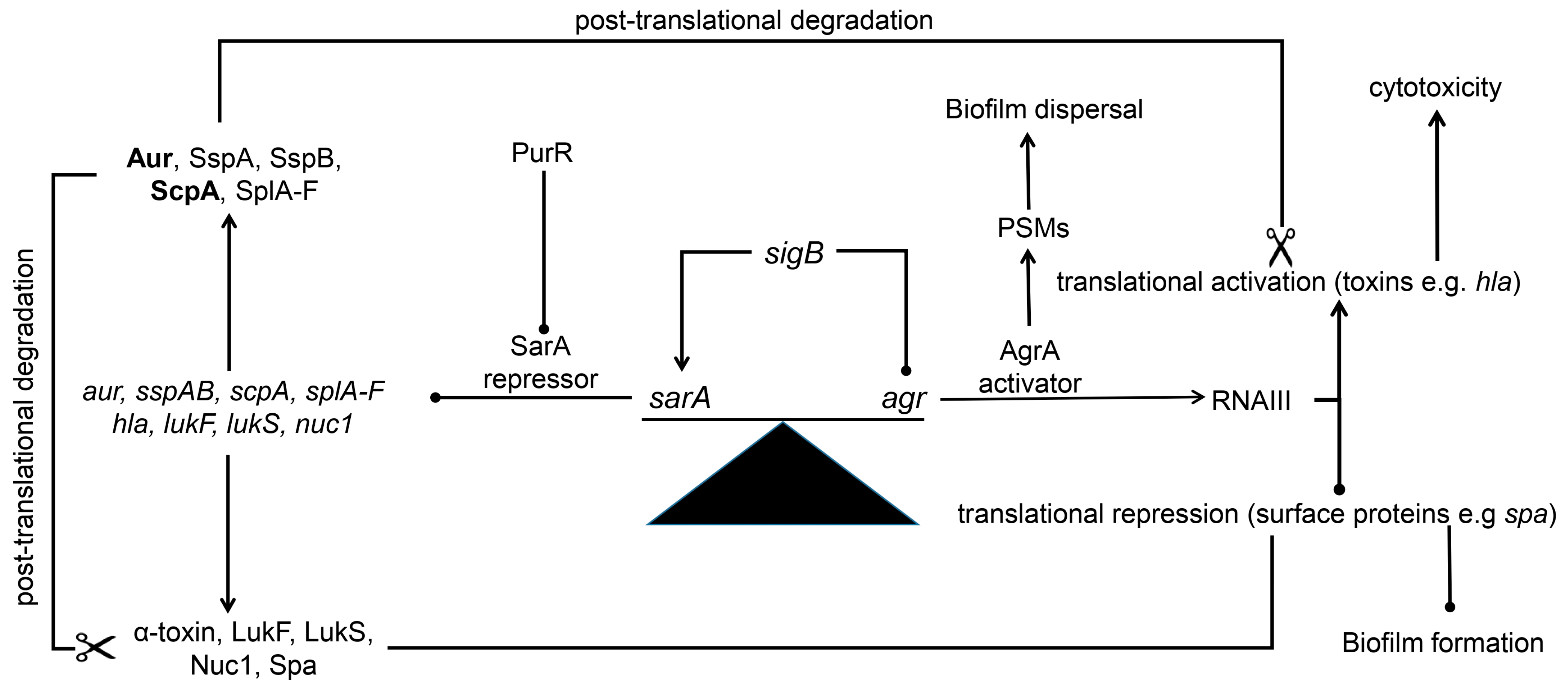
12. How sarA Limits Protease Production
13. Relative Importance of Individual Proteases
14. The Impact of Other S. aureus Regulatory Loci
15. Another Side of the sarA Protease Paradigm
16. Regulatory Loci Not Identified in the NTML Screen
17. What About S. epidermidis?
18. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdelhamid, A.B.; Yousef, A.E. Combating bacterial biofilms: Current and emerging antibiofilm strategies for treating persistent infections. Antibiotics 2022, 12, 1005. [Google Scholar] [CrossRef] [PubMed]
- Coifu, O.; Moser, C.; Jensen, P.Ø.; HØiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar]
- Gimza, B.D.; Cassat, J.E. Mechanisms of antibiotic failure during Staphylococcus Aureus Osteomyelitis. Front. Immunol. 2021, 12, 638085. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Otto, M. Approaches to combating methicillin-resistant Staphylococcus aureus (MRSA) biofilm infections. Expert. Opin. Investig. Drugs 2024, 33, 1–3. [Google Scholar] [CrossRef]
- Ricciardi, B.F.; Muthukrishnan, G.; Masters, E.; Ninomiya, M.; Lee, C.C.; Schwarz, E.M. Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: Biofilm and beyond. Curr. Rev. Musculoskelet. Med. 2018, 11, 389–400. [Google Scholar] [CrossRef]
- Tran, N.N.; Morrisette, T.; Jorgensen, S.C.J.; Orench-Benvenutti, J.M.; Kebriaei, R. Current therapies and challenges for the treatment of Staphylococcus aureus biofilm-related infections. Pharmacotherapy 2023, 43, 816–832. [Google Scholar] [CrossRef]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Kragh, K.N.; Tolker-Nielsen, T.; Lichtenberg, M. The non-attached biofilm aggregate. Commun. Biol. 2023, 6, 898. [Google Scholar] [CrossRef]
- Hofstee, M.I.; Riool, M.; Terjajevs, I.; Thompson, K.; Stoddart, M.J.; Richards, R.G.; Zaat, S.A.J.; Moriarty, T.F. Three-dimensional in vitro Staphylococcus aureus abscess communities display antibiotic tolerance and protection from neutrophil clearance. Infect. Immun. 2020, 88, e00293-20. [Google Scholar] [CrossRef]
- Atwood, D.N.; Beenken, K.E.; Lantz, T.L.; Meeker, D.G.; Lynn, W.B.; Mills, W.B.; Spencer, H.J.; Smeltzer, M.S. Regulatory mutations impacting antibiotic susceptibility in an established Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2016, 60, 1826–1829. [Google Scholar] [CrossRef]
- Johansen, M.I.; Petersen, M.E.; Faddy, E.; Seefeldt, A.M.; Mitkin, A.A.; Østergaard, L.; Meyer, R.L.; Jørgensen, N.P. Efficacy of rifampicin combination therapy against MRSA prosthetic vascular graft infections in a rat model. Biofilm 2024, 7, 100189. [Google Scholar] [CrossRef] [PubMed]
- Meeker, D.G.; Beenken, K.E.; Mills, W.B.; Loughran, A.J.; Spencer, H.J.; Lynn, W.B.; Smeltzer, M.S. Evaluation of antibiotics active against methicillin-resistant Staphylococcus aureus based on activity in an established biofilm. Antimicrob. Agents Chemother. 2016, 60, 5688–5694. [Google Scholar] [CrossRef] [PubMed]
- Tello-Díaz, C.; Muñoz, E.; Palau, M.; Gomis, X.; Gavaldà, J.; Gil-Sala, D.; Fernández-Hidalgo, N.; Bellmunt-Montoya, S. Antibiotic efficacy against methicillin-susceptible Staphylococcus aureus biofilms on synthetic and biological vascular grafts. Ann. Vasc. Surg. 2024, 108, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 2011, 14, 593–598. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Gerdes, K.; Lewis, K.; McKinney, J.D. A problem of persistence: Still more questions than answers. Nat. Rev. Microbiol. 2013, 11, 587–591. [Google Scholar] [CrossRef]
- Mirani, Z.A.; Aziz, M.; Khan, S.I. Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J. Antibiot. 2015, 68, 98–105. [Google Scholar] [CrossRef]
- Vasudevan, S.; David, H.; Chanemougam, L.; Ramani, J.; Ramesh Sangeetha, M.; Solomon, A.P. Emergence of persister cells in Staphylococcus aureus: Calculated or fortuitous move? Crit. Rev. Microbiol. 2024, 50, 64–75. [Google Scholar] [CrossRef]
- Waters, E.M.; Rowe, S.E.; O’Gara, J.P.; Conlon, B.P. 2016. Convergence of Staphylococcus aureus persister and biofilm research: Can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 2016, 12, e1006012. [Google Scholar] [CrossRef]
- Atwood, D.N.; Loughran, A.J.; Courtney, A.P.; Anthony, A.C.; Meeker, D.G.; Spencer, H.J.; Gupta, R.K.; Lee, C.Y.; Beenken, K.E.; Smeltzer, M.S. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 2015, 4, 436–451. [Google Scholar] [CrossRef]
- Campbell, M.J.; Beenken, K.E.; Ramirez, A.M.; Smeltzer, M.S. The major role of sarA in limiting Staphylococcus aureus extracellular protease production in vitro is correlated with decreased virulence in diverse clinical isolates in osteomyelitis. Virulence 2023, 14, 2175496. [Google Scholar] [CrossRef]
- Rom, J.S.; Ramirez, A.M.; Beenken, K.E.; Sahukhal, G.S.; Elasri, M.O.; Smeltzer, M.S. The impact of msaABCR on sarA-associated phenotypes are different in divergent clinical isolates of Staphylococcus aureus. Infect. Immun. 2020, 88, e00530-19. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Urish, K.L.; Cassat, J.E. Staphylococcus aureus osteomyelitis: Bone, bugs, and surgery. Infect. Immun. 2020, 88, e00932-19. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.A.; Cassat, J.E. Advances in the local and targeted delivery of anti-infective agents for management of osteomyelitis. Expert Rev. Anti Infect. Ther. 2017, 15, 851–860. [Google Scholar] [CrossRef]
- Antony, S.; Farran, Y. Prosthetic joint and orthopedic device related infections. The role of biofilm in the pathogenesis and treatment. Infect. Disord. Drug Targets. 2016, 16, 22–27. [Google Scholar] [CrossRef]
- Cierny, G. Surgical treatment of osteomyelitis. Plast. Reconstr. Surg. 2011, 127 (Suppl. S1), 190S–204S. [Google Scholar] [CrossRef]
- Conterno, L.O.; Turchi, M.D. Antibiotics for treating chronic osteomyelitis in adults. Cochrane Database Syst. Rev. 2013, 6, CD004439. [Google Scholar] [CrossRef]
- Hackett, D.J.; Rothenberg, A.C.; Chen, A.F.; Gutowski, C.; Jaekel, D.; Tomek, I.M.; Parsley, B.S.; Ducheyne, P.; Manner, P.A. The economic significance of orthopaedic infections. J. Am. Acad. Orthop. Surg. 2015, 23 (Suppl. S23), S1–S7. [Google Scholar] [CrossRef]
- Premkumar, A.; Kolin, D.A.; Farley, K.X.; Wilson, J.M.; McLawhorn, A.S.; Cross, M.B.; Sculco, P.K. Projected economic burden of periprosthetic joint infection of the hip and knee in the United States. J. Arthroplas. 2021, 36, 1484–1489. [Google Scholar] [CrossRef]
- Kremers, H.M.; Nwojo, M.E.; Ransom, J.E.; Wood-Wentz, C.M.; Melton, L.J.; Huddleston, P.M. Trends in the epidemiology of osteomyelitis: A population-based study, 1969–2009. J. Bone Joint Surg. Am. 2015, 97, 837–845. [Google Scholar] [CrossRef]
- Masters, E.A.; Ricciardi, B.F.; Bentley, K.L.M.; Moriarty, T.F.; Schwarz, E.M.; Muthukrishnan, G. Skeletal infections: Microbial pathogenesis, immunity and clinical management. Nat. Rev. Microbiol. 2022, 20, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, G.; Masters, E.A.; Daiss, J.L.; Schwarz, E.M. Mechanisms of immune evasion and bone tissue colonization that make Staphylococcus aureus the primary pathogen in osteomyelitis. Curr. Osteoporos. Rep. 2019, 17, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.S.; Thomas, J.R.; Hickmon, S.G.; Skinner, R.A.; Nelson, C.L.; Griffith, D.; Parr TRJr Evans, R.P. Characterization of a rabbit model of staphylococcal osteomyelitis. J. Orthop. Res. 1997, 15, 414–421. [Google Scholar] [CrossRef]
- Masters, E.A.; Muthukrishnan, G.; Ho, L.; Gill, A.L.; de Mesy Bentley, K.L.; Galloway, C.A.; McGrath, J.L.; Awad, H.A.; Gill, S.R.; Schwarz, E.M. Staphylococcus aureus cell wall biosynthesis modulates bone invasion and osteomyelitis pathogenesis. Front. Microbiol. 2021, 12, 723498. [Google Scholar] [CrossRef]
- Josse, J.; Velard, F.; Gangloff, S.C. Staphylococcus aureus vs. osteoblast: Relationship and consequences in osteomyelitis. Front. Cell Infect. Microbiol. 2015, 5, 85. [Google Scholar] [CrossRef]
- Krauss, J.L.; Roper, P.M.; Ballard, A.; Shih, C.C.; Fitzpatrick, J.A.J.; Cassat, J.E.; Ng, P.Y.; Pavlos, N.J.; Veis, D.J. Staphylococcus aureus infects osteoclasts and replicates intracellularly. mBio 2019, 10, e02447-19. [Google Scholar] [CrossRef]
- Tricou, L.P.; Mouton, W.; Cara, A.; Trouillet-Assant, S.; Bouvard, D.; Laurent, F.; Diot, A.; Josse, J. Staphylococcus aureus can use an alternative pathway to be internalized by osteoblasts in absence of β1 integrins. Sci. Rep. 2024, 14, 28643. [Google Scholar] [CrossRef]
- Richter, K.; Van den Driessche, F.; Coenye, T. Innovative approaches to treat Staphylococcus aureus biofilm-related infections. Essay Biochem. 2017, 61, 61–70. [Google Scholar]
- Kiedrowski, M.R.; Horswill, A.R. New approaches for treating staphylococcal biofilm infections. Ann. N.Y. Acad. Sci. 2011, 1241, 104–121. [Google Scholar] [CrossRef]
- Schlander, M.; Hernandez-Villafuerte, K.; Cheng, C.Y.; Mestre-Ferrandiz, J.; Baumann, M. 2021. How much does it cost to research and develop a new drug? A systematic review and assessment. Pharmacoeconomics 2021, 39, 1243–1269. [Google Scholar] [CrossRef]
- Arya, R.; Princy, S.A. An insight into pleiotropic regulators Agr and Sar: Molecular probes paving the new way for antivirulent therapy. Future Microbiol. 2013, 8, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Ravikumar, R.; Santhosh, R.S.; Princy, S.A. SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front. Microbiol. 2015, 6, 416. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zheng, W.; Li, Z. High-throughput screening strategies for the development of anti-virulence inhibitors against Staphylococcus aureus. Curr. Med. Chem. 2019, 26, 2297–2312. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.P.; Williams, P.; Chan, W.C. Attenuating Staphylococcus aureus virulence gene regulation: A medicinal chemistry perspective. J. Med. Chem. 2013, 56, 1389–1404. [Google Scholar] [CrossRef]
- Hsieh, R.C.; Liu, R.; Burgin, D.J.; Otto, M. Understanding mechanisms of virulence in MRSA: Implications for antivirulence treatment strategies. Expert Rev. Anti Infect. Ther. 2023, 21, 911–928. [Google Scholar] [CrossRef]
- Horswill, A.R.; Gordon, C.P. Structure-activity relationship studies of small molecule modulators of the staphylococcal accessory gene regulator. J Med. Chem. 2020, 63, 2705–2730. [Google Scholar] [CrossRef]
- Otto, M. Critical assessment of the prospects of quorum-quenching therapy for Staphylococcus aureus infection. Int. J. Mol. Sci. 2023, 24, 4025. [Google Scholar] [CrossRef]
- Piewngam, P.; Chiou, J.; Chatterjee, P.; Otto, M. Alternative approaches to treat bacterial infections: Targeting quorum-sensing. Expert Rev. Anti Infect. Ther. 2020, 18, 499–510. [Google Scholar] [CrossRef]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, J10, e1004174. [Google Scholar] [CrossRef]
- Tan, L.; Li, S.R.; Jiang, B.; Hu, X.M.; Li, S. Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Vinodhini, V.; Kavitha, M. Deciphering agr quorum sensing in Staphylococcus aureus: Insights and therapeutic prospects. Mol. Biol. Rep. 2024, 51, 155. [Google Scholar] [CrossRef] [PubMed]
- Beenken, K.E.; Campbell, M.J.; Smeltzer, M.S. The ability of sarA to limit protease production plays a key role in the pathogenesis of Staphylococcus aureus osteomyelitis irrespective of the functional status of agr. Infect. Immun. 2025, 93, e0047324. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J.; Beenken, K.E.; Ramirez, A.M.; Smeltzer, M.S. Increased production of aureolysin and staphopain A is a primary determinant of the reduced virulence of Staphylococcus aureus sarA mutants in osteomyelitis. mBio 2024, 15, e0338323. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, A.F.; Hickmon, S.G.; Skinner, R.A.; Thomas, J.R.; Nelson, C.L.; Smeltzer, M.S. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 1995, 63, 3373–3380. [Google Scholar] [CrossRef]
- Loughran, A.J.; Gaddy, D.; Beenken, K.E.; Meeker, D.G.; Morello, R.; Zhao, H.; Byrum, S.D.; Tackett, A.J.; Cassat, J.E.; Smeltzer, M.S. Impact of sarA and phenol-soluble modulins on the pathogenesis of osteomyelitis in diverse clinical isolates of Staphylococcus aureus. Infect. Immun. 2016, 84, 2586–2894. [Google Scholar] [CrossRef]
- Rom, J.S.; Atwood, D.N.; Beenken, K.E.; Meeker, D.G.; Loughran, A.J.; Spencer, H.J.; Lantz, T.L.; Smeltzer, M.S. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 2017, 8, 1776–1790. [Google Scholar] [CrossRef]
- Beenken, K.E.; Campbell, M.J.; Byrum, S.D.; Edmondson, R.D.; Mackintosh, S.G.; Tackett, A.J.; Smeltzer, M.S. Staphylococcus aureus proteins implicated in the reduced virulence of sarA and sarA/agr mutants in osteomyelitis. Microorganisms. 2025, 13, 181. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Chabelskaya, S.; Bordeau, V.; Felden, B. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res. 2014, 42, 4847–4858. [Google Scholar] [CrossRef]
- Chevalier, C.; Boisset, S.; Romilly, C.; Masquida, B.; Fechter, P.; Geissmann, T.; Vandenesch, F.; Romby, P. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010, 6, e1000809. [Google Scholar] [CrossRef]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995, 14, 4569–4577. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Chen, L.; Joo, H.S.; Cheung, G.Y.; Kreiswirth, B.N.; Otto, M. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e28781. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.; Manna, A.C.; Projan, S.J.; Cheung, A.L. SarA a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Bi. Chem. 1999, 274, 37169–37176. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.; Manna, A.C.; Cheung, A.L. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 1998, 30, 991–1001. [Google Scholar] [CrossRef]
- Cheung, A.L.; Bayer, M.G.; Heinrichs, J.H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 1997, 179, 3963–3971. [Google Scholar] [CrossRef]
- Cheung, A.L.; Projan, S.J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 1994, 176, 4168–4172. [Google Scholar] [CrossRef]
- Beenken, K.E.; Blevins, J.S.; Smeltzer, M.S. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 2003, 71, 4206–4211. [Google Scholar] [CrossRef]
- Beenken, K.E.; Mrak, L.N.; Griffin, L.M.; Zielinska, A.K.; Shaw, L.N.; Rice, K.C.; Horswill, A.R.; Bayles, K.W.; Smeltzer, M.S. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5, e10790. [Google Scholar] [CrossRef]
- Altman, D.R.; Sullivan, M.J.; Chacko, K.I.; Balasubramanian, D.; Pak, T.R.; Sause, W.E.; Kumar, K.; Sebra, R.; Deikus, G.; Attie, O.; et al. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect. Immun. 2018, 86, e00331-18. [Google Scholar] [CrossRef]
- Gor, V.; Takemura, A.J.; Nishitani, M.; Higashide, M.; Medrano Romero, V.; Ohniwa, R.L.; Morikawa, K. Finding of agr phase variants in Staphylococcus aureus. mBio 2019, 10, e00796-19. [Google Scholar] [CrossRef]
- Suligoy, C.M.; Lattar, S.M.; Noto Llana, M.; González, C.D.; Alvarez, L.P.; Robinson, D.A.; Gómez, M.I.; Buzzola, F.R.; Sordelli, D.O. Mutation of agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front. Cell Infect. Microbiol. 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology (Reading) 2008, 154 Pt 8, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.S.; Kafer, J.M.; Wasserman, G.A.; Velickovic, L.; Mathema, B.; Holzman, R.S.; Knipe, T.A.; Becker, K.; von Eiff, C.; Peters, G.; et al. Nasal carriage as a source of agr-defective Staphylococcus aureus bacteremia. J. Infect. Dis. 2012, 206, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; Paquette, K.M.; Tikh, I.B.; Volper, E.M.; Greenberg, E.P. Generation of virulence factor variants in Staphylococcus aureus biofilms. J. Bacteriol. 2007, 189, 7961–7967. [Google Scholar] [CrossRef]
- Weiss, E.C.; Zielinska, A.; Beenken, K.E.; Spencer, H.J.; Daily, S.J.; Smeltzer, M.S. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrob. Agents Chemother. 2009, 53, 4096–4102. [Google Scholar] [CrossRef]
- Weiss, E.C.; Spencer, H.J.; Daily, S.J.; Weiss, B.D.; Smeltzer, M.S. Impact of sarA on antibiotic susceptibility of Staphylococcus aureus in a catheter-associated in vitro model of biofilm formation. Antimicrob. Agents Chemother. 2009, 53, 2475–2482. [Google Scholar] [CrossRef]
- Lamichhane-Khadka, R.; Cantore, S.A.; Riordan, J.T.; Delgado, A.; Norman, A.E.; Dueñas, S.; Zaman, S.; Horan, S.; Wilkinson, B.J.; Gustafson, J.E. sarA inactivation reduces vancomycin-intermediate and ciprofloxacin resistance expression by Staphylococcus aureus. Int. J. Antimicrob. Agents. 2009, 34, 136–141. [Google Scholar] [CrossRef]
- Bischoff, M.; Entenza, J.M.; Giachino, P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 2001, 183, 5171–5179. [Google Scholar] [CrossRef]
- Karlsson, A.; Arvidson, S. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 2002, 70, 4239–4246. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Bischoff, M.; Lattar, S.M.; Noto Llana, M.; Pförtner, H.; Niemann, S.; Geraci, J.; Van de Vyver, H.; Fraunholz, M.J.; Cheung, A.L.; et al. Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 2015, 11, e1004870. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Geraci, J.; Löffler, B. Staphylococcus aureus regulator sigma B is important to develop chronic infections in hematogenous murine osteomyelitis model. Pathogens. 2017, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.T.; O’Leary, J.O.; Gustafson, J.E. Contributions of sigB and sarA to distinct multiple antimicrobial resistance mechanisms of Staphylococcus aureus. Int. J. Antimicrob. Agents. 2006, 28, 54–61. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Löffler, B. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 2016, 62, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Tuchscherr, L.; Löffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple metabolic pathways impact the expression of virulence factors in small-colony variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. ChemMedChem. 2021, 16, 65–80. [Google Scholar] [CrossRef]
- Molina, K.C.; Morrisette, T.; Miller, M.A.; Huang, V.; Fish, D.N. The emerging role of β-Lactams in the treatment of methicillin-resistant Staphylococcus aureus bloodstream infections. Antimicrob. Agents Chemother. 2020, 64, e00468-20. [Google Scholar] [CrossRef]
- Li, L.; Cheung, A.; Bayer, A.S.; Chen, L.; Abdelhady, W.; Kreiswirth, B.N.; Yeaman, M.R.; Xiong, Y.Q. The global regulon sarA regulates β-lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J. Infect. Dis. 2016, 214, 1421–1429. [Google Scholar] [CrossRef]
- Alekshun, M.N. New advances in antibiotic development and discovery. Expert Opin. Investig. Drugs. 2005, 14, 117–134. [Google Scholar]
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2017, 133, 4–19. [Google Scholar] [CrossRef]
- Årdal, C.; Balasegaram, M.; Laxminarayan, R.; McAdams, D.; Outterson, K.; Rex, J.H.; Sumpradit, N. Antibiotic development—economic, regulatory and societal challenges. Nat. Rev. Microbiol. 2020, 18, 267–274. [Google Scholar] [CrossRef]
- Wang, C.H.; Hsieh, Y.H.; Powers, Z.M.; Kao, C.Y. Defeating antibiotic-resistant bacteria: Exploring alternative therapies for a post-antibiotic era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Carrel, M.; Perencevich, E.N.; David, M.Z. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg. Infect. Dis. 2015, 21, 1973–1980. [Google Scholar] [CrossRef]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2013, 4, e00537-12. [Google Scholar] [CrossRef]
- Cassat, J.E.; Dunman, P.M.; McAleese, F.; Murphy, E.; Projan, S.J.; Smeltzer, M.S. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 2005, 187, 576–592. [Google Scholar] [CrossRef]
- Elasri, M.O.; Thomas, J.R.; Skinner, R.A.; Blevins, J.S.; Beenken, K.E.; Nelson, C.L.; Smeltzer, M.S. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 2002, 30, 275–280. [Google Scholar] [CrossRef]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 2011, 79, 1153–1165. [Google Scholar] [CrossRef]
- Kaushik, A.; Kest, H.; Sood, M.; Steussy, B.W.; Thieman, C.; Gupta, S. Biofilm producing methicillin-rsistant Staphylococcus aureus (MRSA) infections in humans: Clinical implications and management. Pathogens 2024, 13, 76. [Google Scholar] [CrossRef]
- O’Reilly, M.; Kreiswirth, B.; Foster, T.J. Cryptic alpha-toxin gene in toxic shock syndrome and septicaemia strains of Staphylococcus aureus. Mol. Microbiol. 1990, 4, 1947–1955. [Google Scholar] [CrossRef]
- Caiazza, N.C.; O’Toole, G.A. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 2003, 185, 3214–3217. [Google Scholar] [CrossRef] [PubMed]
- Rom, J.S.; Beenken, K.E.; Ramirez, A.M.; Walker, C.M.; Echols, E.J.; Smeltzer, M.S. Limiting protease production plays a key role in the pathogenesis of the divergent clinical isolates of Staphylococcus aureus LAC and UAMS-1. Virulence 2021, 12, 584–600. [Google Scholar] [CrossRef]
- Baba, T.; Bae, T.; Schneewind, O.; Takeuchi, F.; Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008, 190, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Carpaij, N.; Willems, R.J.; Rice, T.W.; Weinstein, R.A.; Hinds, J.; Witney, A.A.; Lindsay, J.A.; Bonten, M.J.; Fluit, A.C. Genetic variation in spatio-temporal confined USA300 community-associated MRSA isolates: A shift from clonal dispersion to genetic evolution? PLoS ONE 2011, 6, e16419. [Google Scholar] [CrossRef] [PubMed]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Porcella, S.F.; Martens, C.; Whitney, A.R.; Braughton, K.R.; Chen, L.; Craig, C.T.; Tenover, F.C.; Kreiswirth, B.N.; Musser, J.M.; et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J. Clin. Microbiol. 2010, 48, 4504–4511. [Google Scholar] [CrossRef]
- Tewhey, R.; Cannavino, C.R.; Leake, J.A.; Bansal, V.; Topol, E.J.; Torkamani, A.; Bradley, J.S.; Schork, N.J. Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics. 2012, 13, 508. [Google Scholar] [CrossRef]
- Sassi, M.; Sharma, D.; Brinsmade, S.R.; Felden, B.; Augagneur, Y. Genome sequence of the clinical isolate Staphylococcus aureus subsp. aureus strain UAMS-1. Genome Announc. 2015, 3, e01584-14. [Google Scholar]
- Hernández-Cuellar, E.; Tsuchiya, K.; Valle-Ríos, R.; Medina-Contreras, O. Differences in biofilm formation by methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. Diseases 2023, 11, 160. [Google Scholar] [CrossRef]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Loughran, A.J.; Atwood, D.N.; Anthony, A.C.; Harik, N.S.; Spencer, H.J.; Beenken, K.E.; Smeltzer, M.S. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen 2014, 3, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Beenken, K.E.; Mrak, L.N.; Zielinska, A.K.; Atwood, D.N.; Loughran, A.J.; Griffin, L.M.; Matthews, K.A.; Anthony, A.M.; Spencer, H.J.; Skinner, R.A.; et al. Impact of the functional status of saeRS on in vivo phenotypes of Staphylococcus aureus sarA mutants. Mol. Microbiol. 2014, 92, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Mrak, L.N.; Zielinska, A.K.; Beenken, K.E.; Mrak, I.N.; Atwood, D.N.; Griffin, L.M.; Lee, C.Y.; Smeltzer, M.S. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS ONE 2012, 7, e38453. [Google Scholar] [CrossRef]
- Ramirez, A.M.; Byrum, S.D.; Beenken, K.E.; Washam, C.; Edmondson, R.D.; Mackintosh, S.G.; Spencer, H.J.; Tackett, A.J.; Smeltzer, M.S. Exploiting correlations between protein abundance and the functional status of saeRS and sarA to identify virulence factors of potential importance in the pathogenesis of Staphylococcus aureus osteomyelitis. ACS Infect. Dis. 2020, 6, 237–249. [Google Scholar] [CrossRef]
- Karlsson-Kanth, A.; Tegmark-Wisell, K.; Arvidson, S.; Oscarsson, J. Natural human isolates of Staphylococcus aureus selected for high production of proteases and alpha-hemolysin are sigmaB deficient. Int. J. Med. Microbiol. 2006, 296, 229–236. [Google Scholar] [CrossRef]
- Herbert, S.; Ziebandt, A.K.; Ohlsen, K.; Schäfer, T.; Hecker, M.; Albrecht, D.; Novick, R.; Götz, F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010, 78, 2877–2889. [Google Scholar] [CrossRef]
- Le, K.Y.; Dastgheyb, S.; Ho, T.V.; Otto, M. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front. Cell Infect. Microbiol. 2014, 4, 167. [Google Scholar] [CrossRef]
- DelMain, E.A.; Moormeier, D.E.; Endres, J.L.; Hodges, R.E.; Sadykov, M.R.; Horswill, A.R.; Bayles, K.W. Stochastic expression of sae-dependent virulence genes during Staphylococcus aureus biofilm development is dependent on SaeS. mBio 2020, 11, e03081-19. [Google Scholar] [CrossRef]
- Grundmeier, M.; Hussain, M.; Becker, P.; Heilmann, C.; Peters, G.; Sinha, B. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 2004, 72, 7155–7163. [Google Scholar] [CrossRef]
- Cue, D.; Junecko, J.M.; Lei, M.G.; Blevins, J.S.; Smeltzer, M.S.; Lee, C.Y. SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS ONE 2015, 10, e0123027. [Google Scholar] [CrossRef]
- Ramirez, A.M.; Beenken, K.E.; Bryum, S.D.; Tackett, A.J.; Shaw, L.N.; Gimza, B.D.; Smeltzer, M.S. 2020. SarA plays a predominant role in controlling the production of extracellular proteases in diverse clinical isolates of Staphylococcus aureus. Virulence 2020, 11, 1738–1762. [Google Scholar] [CrossRef]
- Tsang, L.H.; Cassat, J.E.; Shaw, L.N.; Beenken, K.; Smeltzer, M.S. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 2008, 3, e3361. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef]
- Beenken, K.E.; Spencer, H.; Griffin, L.M.; Smeltzer, M.S. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 2012, 80, 1634–1638. [Google Scholar] [CrossRef]
- Sugimoto, S.; Sato, F.; Miyakawa, R.; Chiba, A.; Onodera, S.; Hori, S.; Mizunoe, Y. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci. Rep. 2018, 8, 2254. [Google Scholar] [CrossRef]
- Gries, C.M.; Biddle, T.; Bose, J.L.; Kielian, T.; Lo, D.D. Staphylococcus aureus fibronectin binding protein A mediates biofilm development and infection. Infect. Immun. 2020, 88, e00859-19. [Google Scholar] [CrossRef]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penadés, J.R.; Lasa, I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef]
- Fitzgerald, J.R.; Sturdevant, D.E.; Mackie, S.M.; Gill, S.R.; Musser, J.M. Evolutionary genomics of Staphylococcus aureus: Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci USA 2001, 98, 8821–8826. [Google Scholar] [CrossRef]
- Byrum, S.D.; Loughran, A.J.; Beenken, K.E.; Orr, L.M.; Mackintosh, S.G.; Edmondson, R.D.; Tackett, A.J.; Smeltzer, M.S. Label-free proteomic approach to characterize protease-dependent and independent effects of sarA inactivation on the Staphylococcus aureus exoproteome. ACS J. Proteome Res. 2018, 17, 3384–3395. [Google Scholar] [CrossRef]
- Seebach, E.; Kraus, F.V.; Elschner, T.; Kubatzky, K.F. Staphylococci planktonic and biofilm environments differentially affect osteoclast formation. Inflamm. Res. 2023, 72, 1465–1484. [Google Scholar] [CrossRef] [PubMed]
- Mouton, W.; Josse, J.; Jacqueline, C.; Abad, L.; Trouillet-Assant, S.; Caillon, J.; Bouvard, D.; Bouchet, M.; Laurent, F.; Diot, A. Staphylococcus aureus internalization impairs osteoblastic activity and early differentiation process. Sci. Rep. 2021, 11, 17685. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, A.E.; Mendoza-Bertelli, A.; Ledo, C.; Gonzalez, C.D.; Noto Llana, M.; Blanco, C.; Sordelli, D.O.; Putman, N.E.; Cassat, J.E.; Delpino, M.V.; et al. Neutralization of Staphylococcus aureus protein A prevents exacerbated osteoclast activity and bone loss during osteomyelitis. Antimicrob. Agents Chemother. 2023, 67, e0114022. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J.; Bustamante-Gomez, C.; Fu, Q.; Beenken, K.E.; Reyes-Pardo, H.; Smeltzer, M.S.; O’Brien, C.A. RANKL-mediated osteoclast formation is required for bone loss in a murine model of Staphylococcus aureus osteomyelitis. Bone 2024, 187, 117181. [Google Scholar] [CrossRef]
- Gresham, H.D.; Lowrance, J.H.; Caver, T.E.; Wilson, B.S.; Cheung, A.L.; Lindberg, F.P. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J. Immunol. 2000, 164, 3713–3722. [Google Scholar] [CrossRef]
- Kubica, M.; Guzik, K.; Koziel, J.; Zarebski, M.; Richter, W.; Gajkowska, B.; Golda, A.; Maciag-Gudowska, A.; Brix, K.; Shaw, L.; et al. A potential new pathway for Staphylococcus aureus dissemination: The silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 2008, 3, e1409. [Google Scholar] [CrossRef]
- Rasigade, J.P.; Trouillet-Assant, S.; Ferry, T.; Diep, B.A.; Sapin, A.; Lhoste, Y.; Ranfaing, J.; Badiou, C.; Benito, Y.; Bes, M.; et al. PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS ONE 2013, 8, e63176. [Google Scholar] [CrossRef]
- Kalinka, J.; Hachmeister, M.; Geraci, J.; Sordelli, D.; Hansen, U.; Niemann, S.; Oetermann, S.; Peters, G.; Löffler, B.; Tuchscherr, L. Staphylococcus aureus isolates from chronic osteomyelitis are characterized by high host cell invasion and intracellular adaptation, but still induce inflammation. Int. J. Med. Microbiol. 2014, 304, 1038–1049. [Google Scholar] [CrossRef]
- Massey, R.C.; Kantzanou, M.N.; Fowler, T.; Day, N.P.; Schofield, K.; Wann, E.R.; Berendt, A.R.; Höök, M.; Peacock, S.J. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell Microbiol. 2001, 3, 839–851. [Google Scholar] [CrossRef]
- Niemann, S.; Nguyen, M.T.; Eble, J.A.; Chasan, A.I.; Mrakovcic, M.; Böttcher, R.T.; Preissner, K.T.; Roßlenbroich, S.; Peters, G.; Herrmann, M. More is not always better-the double-headed role of fibronectin in Staphylococcus aureus host cell invasion. mBio 2021, 12, e0106221. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front. Microbiol. 2020, 11, 2054. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.; Abdel Aziz, M.H. Crosstalk involving two-component systems in Staphylococcus aureus signaling networks. J.Bacteriol. 2024, 206, e0041823. [Google Scholar] [CrossRef] [PubMed]
- Girma, A. Staphylococcus aureus: Current perspectives on molecular pathogenesis and virulence. Cell Surf. 2024, 13, 100137. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Rawat, S. A genetic regulatory see-saw of biofilm and virulence in MRSA pathogenesis. Front. Microbiol. 2023, 14, 1204428. [Google Scholar] [CrossRef]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics 2022, 12, 12. [Google Scholar] [CrossRef]
- Cassat, J.E.; Hammer, N.D.; Campbell, J.P.; Benson, M.A.; Perrien, D.S.; Mrak, L.N.; Smeltzer, M.S.; Torres, V.J.; Skaar, E.P. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe. 2013, 13, 759–772. [Google Scholar] [CrossRef]
- Gimza, B.D.; Larias, M.I.; Budney, B.G.; Shaw, L.N. Mapping the global network of extracellular protease regulation in Staphylococcus aureus. mSphere 2019, 4, e00676-19. [Google Scholar] [CrossRef]
- Gimza, B.D.; Jackson, J.K.; Frey, A.M.; Budny, B.G.; Chaput, D.; Rizzo, D.N.; Shaw, L.N. Unraveling the impact of secreted proteases on hyprevirulence in Staphylococcus aureus. mBio 2021, 12, e03288-20. [Google Scholar] [CrossRef]
- Shaw, L.; Golonka, E.; Potempa, J.; Foster, S.J. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 2004, 150 Pt 1, 217–228. [Google Scholar] [CrossRef]
- Sterba, K.M.; Mackintosh, S.G.; Blevins, J.S.; Hurlburt, B.K.; Smeltzer, M.S. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 2003, 185, 4410–4417. [Google Scholar] [CrossRef]
- Atwood, D.N.; Beenken, K.E.; Loughran, A.J.; Meeker, D.G.; Lantz, T.L.; Graham, J.W.; Spencer, H.J.; Smeltzer, M.S. XerC contributes to diverse forms of Staphylococcus aureus infection via agr-dependent and agr-independent pathways. Infect. Immun. 2016, 84, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Ledger, E.V.K.; Lau, K.; Tate, E.W.; Edwards, A.M. XerC is required for the repair of antibiotic- and immune-mediated DNA damage in Staphylococcus aureus. Antimicrob. Agents Chemother. 2023, 67, e0120622. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, Y.; Ohniwa, R.L.; Morikawa, K. Identification of nucleoid associated proteins (NAPs) under oxidative stress in Staphylococcus aureus. BMC Microbiol. 2017, 17, 207. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, D.F.; Higginbotham, R.H.; Sterba, K.M.; Maleki, S.J.; Segall, A.M.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol. Microbiol. 2009, 74, 1445. [Google Scholar] [CrossRef]
- Lee, J.; Carda-Diéguez, M.; Žiemytė, M.; Vreugde, S.; Cooksley, C.; Crosby, H.A.; Horswill, A.R.; Mira, A.; Zilm, P.S.; Kidd, S.P. Functional mgrA influences genetic changes within a Staphylococcus aureus cell population over time. J. Bacteriol. 2022, 204, e0013822. [Google Scholar] [CrossRef]
- Goncheva, M.I.; Flannagan, R.S.; Heinrichs, D.E. De novo purine biosynthesis is required for intracellular growth of Staphylococcus aureus and for the hypervirulence phenotype of a purR mutant. Infect. Immun. 2020, 88, e00104-20. [Google Scholar] [CrossRef]
- Alkam, D.; Jenjaroenpunb, P.; Ramirez, A.M.; Spencer, H.J.; Smeltzer, M.S. The increased accumulation of Staphylococcus aureus virulence factors is maximized in a purR mutant by the increased production of SarA and decreased production of extracellular proteases. Infect. Immun. 2021, 89, e00718-20. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Li, Y.; Goncheva, M.I.; Elsayed, A.M.; Zhu, F.; Li, L.; Abdelhady, W.; Flannagan, R.S.; Yeaman, M.R.; Bayer, A.S.; et al. The purine biosynthesis repressor, PurR, contributes to vancomycin susceptibility of methicillin-resistant Staphylococcus aureus in experimental endocarditis. J. Infect. Dis. 2024, 229, 1648–1657. [Google Scholar] [CrossRef]
- Mootz, J.M.; Benson, M.A.; Heim, C.E.; Crosby, H.A.; Kavanaugh, J.S.; Dunman, P.M.; Kielian, T.; Torres, V.J.; Horswill, A.R. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 2015, 96, 388–404. [Google Scholar] [CrossRef]
- Guo, X.; Wang, L.; Zhang, J.; Liu, Q.; Wang, B.; Liu, D.; Gao, F.; Lanzi, G.; Zhao, Y.; Shi, Y. Thwarting resistance: MgrA inhibition with methylophiopogonanone a unveils a new battlefront against S. aureus. NPJ Biofilms Microbiomes 2024, 10, 15. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Smeltzer, M.S.; Elasri, M.O. Identification and characterization of msa (SA1233), a gene involved in expression of sarA and several virulence factors in Staphylococcus aureus. Microbiology 2006, 152 Pt 9, 2559–2572. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamoorthy, K.; Schwartz, A.; Nagarajan, V.; Elasri, M.O. The role of msa in Staphylococcus aureus biofilm formation. BMC Microbiol. 2008, 8, 221. [Google Scholar] [CrossRef]
- Batte, J.L.; Samanta, D.; Elasri, M.O. MsaB activates capsule production at the transcription level in Staphylococcus aureus. Microbiology 2016, 162, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Sahukhal, G.S.; Elasri, M.O. The msaABCR operon regulates persister formation by modulating energy metabolism in Staphylococcus aureus. Front. Microbiol. 2021, 12, 657753. [Google Scholar] [CrossRef]
- Sahukhal, G.S.; Elasri, M.O. Role of the msaABCR operon in cell wall biosynthesis, autolysis, integrity, and antibiotic resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e00680-19. [Google Scholar]
- Sahukhal, G.S.; Tucci, M.; Benghuzzi, H.; Wilson, G.; Elasri, M.O. The role of the msaABCR operon in implant-associated chronic osteomyelitis in Staphylococcus aureus USA300 LAC. BMC Microbiol. 2020, 20, 324. [Google Scholar] [CrossRef]
- Fluckiger, U.; Wolz, C.; Cheung, A.L. Characterization of a sar homolog of Staphylococcus epidermidis. Infect. Immun. 1998, 66, 2871–2878. [Google Scholar] [CrossRef]
- Tormo, M.A.; Martí, M.; Valle, J.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 2005, 187, 2348–2356. [Google Scholar] [CrossRef]
- Christner, M.; Heinze, C.; Busch, M.; Franke, G.; Hentschke, M.; Bayard Dühring, S.; Büttner, H.; Kotasinska, M.; Wischnewski, V.; Kroll, G.; et al. sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 2012, 86, 394–410. [Google Scholar] [CrossRef]
- Blevins, J.S.; Elasri, M.O.; Allmendinger, S.D.; Beenken, K.E.; Skinner, R.A.; Thomas, J.R.; Smeltzer, M.S. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 2003, 71, 516–523. [Google Scholar] [CrossRef]
- Booth, M.C.; Cheung, A.L.; Hatter, K.L.; Jett, B.D.; Callegan, M.C.; Gilmore, M.S. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect. Immun. 1997, 65, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Heyer, G.; Saba, S.; Adamo, R.; Rush, W.; Soong, G.; Cheung, A.; Prince, A. Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infect. Immun. 2002, 70, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Elmwall, J.; Kwiecinski, J.; Na, M.; Ali, A.A.; Osla, V.; Shaw, L.N.; Wang, W.; Sävman, K.; Josefsson, E.; Bylund, J.; et al. Galectin-3 is a target for proteases involved in the virulence of Staphylococcus aureus. Infect. Immun. 2017, J85, e00177-17. [Google Scholar] [CrossRef]
- Jordan, R.E.; Fernandez, J.; Brezski, R.J.; Greenplate, A.R.; Knight, D.M.; Raju, T.S.; Lynch, A.S. A peptide immunization approach to counteract a Staphylococcus aureus protease defense against host immunity. Immunol. Lett. 2016, 172, 29–39. [Google Scholar] [CrossRef]
- Jusko, M.; Potempa, J.; Kantyka, T.; Bielecka, E.; Miller, H.K.; Kalinska, M.; Dubin, G.; Garred, P.; Shaw, L.N.; Blom, A.M. Staphylococcal proteases aid in evasion of the human complement system. J. Innate Immun. 2014, 6, 31–46. [Google Scholar] [CrossRef]
- Kline, S.N.; Saito, Y.; Archer, N.K. Staphylococcus aureus proteases: Orchestrators of skin inflammation. DNA Cell Biol. 2024, 43, 483–491. [Google Scholar] [CrossRef]
- Lehman, M.K.; Nuxoll, A.S.; Yamada, K.J.; Kielian, T.; Carson, S.D.; Fey, P.D. Protease-mediated growth of Staphylococcus aureus on host proteins Is opp3 dependent. mBio 2019, 10, e02553-18. [Google Scholar] [CrossRef]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front. Cell Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef]
- Singh, V.; Phukan, U.J. Interaction of host and Staphylococcus aureus protease-system regulates virulence and pathogenicity. Med. Microbiol. Immunol. 2019, 208, 585–607. [Google Scholar] [CrossRef]
- Tyurin, Y.A.; Mustafin, I.G.; Faskhanov, R.S. Effects of Staphylococcus aureus supernatants on human lymphocyte surface receptors. Bull. Exp. Biol. Med. 2013, 154, 765–768. [Google Scholar] [CrossRef]
- Abbas, H.A.; Atallah, H.; El-Sayed, M.A.; El-Ganiny, A.M. Diclofenac mitigates virulence of multidrug-resistant Staphylococcus aureus. Arch. Microbiol. 2020, 202, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- El-Ganiny, A.M.; Gad, A.I.; El-Sayed, M.A.; Shaldam, M.A.; Abbas, H.A. The promising anti-virulence activity of candesartan, domperidone, and miconazole on Staphylococcus aureus. Braz. J. Microbiol. 2022, 53, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.M.; Yousef, N.; Shafik, S.M.; Abbas, H.A. Attenuating the virulence of the resistant superbug Staphylococcus aureus bacteria isolated from neonatal sepsis by ascorbic acid, dexamethasone, and sodium bicarbonate. BMC Microbiol. 2022, 22, 268. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.J.; Beenken, K.E.; Spencer, H.J.; Jayana, B.; Hester, H.; Sahukhal, G.S.; Elasri, M.O.; Smeltzer, M.S. Comparative evaluation of small molecules reported to be inhibitors of Staphylococcus aureus biofilm formation. mSpectrum 2023, 2, e0314723. [Google Scholar] [CrossRef]
- Kurotschka, P.K.; Bentivegna, M.; Hulme, C.; Ebell, M.H. Identifying the best initial oral antibiotics for adults with community-acquired pneumonia: A network meta-analysis. J. Gen. Intern. Med. 2024, 39, 1214–1226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beenken, K.E.; Smeltzer, M.S. Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target? Microorganisms 2025, 13, 852. https://doi.org/10.3390/microorganisms13040852
Beenken KE, Smeltzer MS. Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target? Microorganisms. 2025; 13(4):852. https://doi.org/10.3390/microorganisms13040852
Chicago/Turabian StyleBeenken, Karen E., and Mark S. Smeltzer. 2025. "Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target?" Microorganisms 13, no. 4: 852. https://doi.org/10.3390/microorganisms13040852
APA StyleBeenken, K. E., & Smeltzer, M. S. (2025). Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target? Microorganisms, 13(4), 852. https://doi.org/10.3390/microorganisms13040852





