Paraburkholderia suaedae sp. nov., a Potential Plant Growth-Promoting Bacterium Isolated from the Halophyte Suaeda japonica
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Rhizosphere Bacteria
2.2. 16S rRNA Amplification and Phylogenetic Analysis
2.3. Genome Features and Comparative Analysis
2.4. Morphology and Chemotaxonomic Characteristics
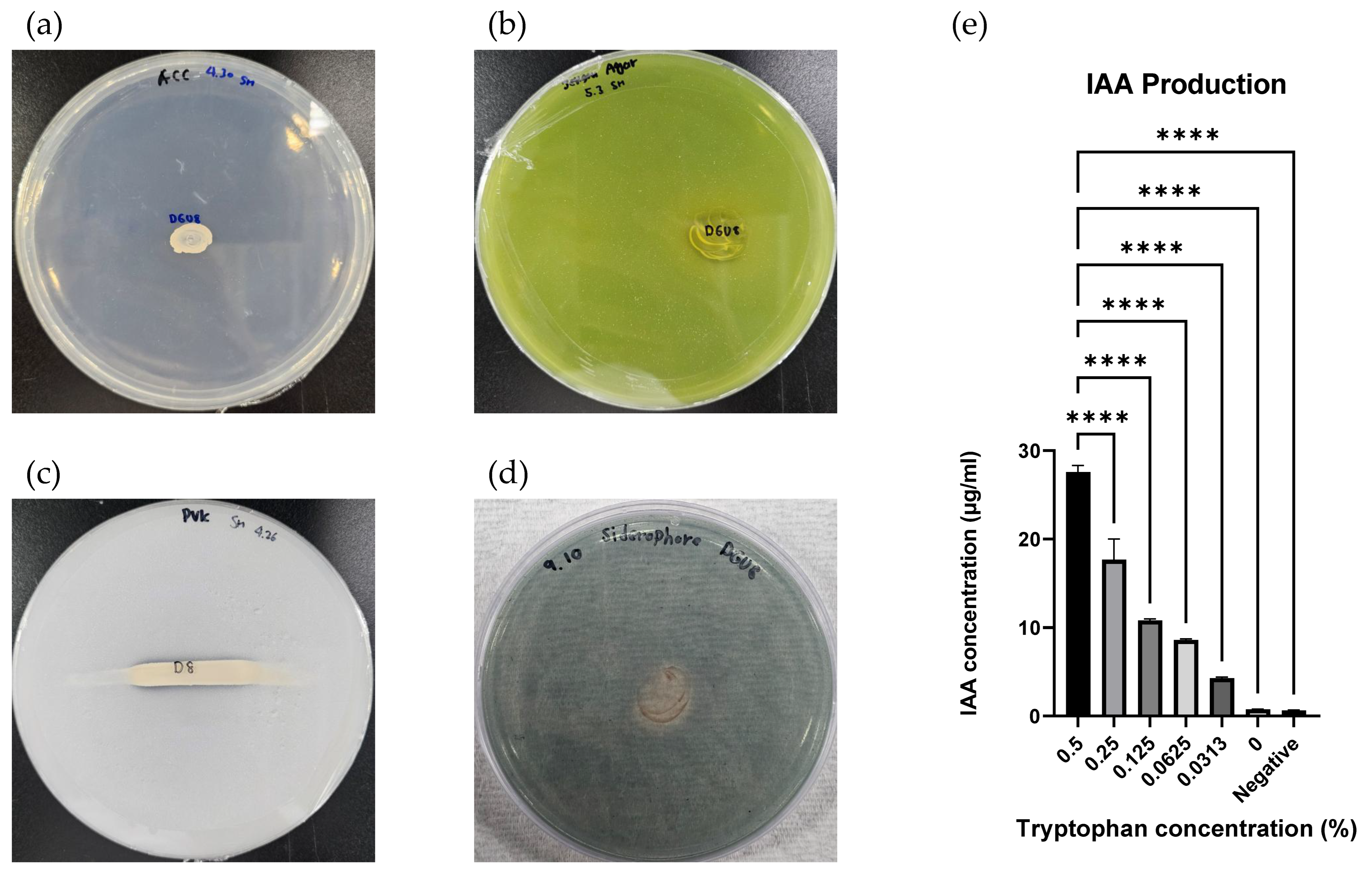
2.5. Functional Traits Related to Plant Growth Promotion
3. Results
3.1. Isolation of Rhizosphere Bacteria
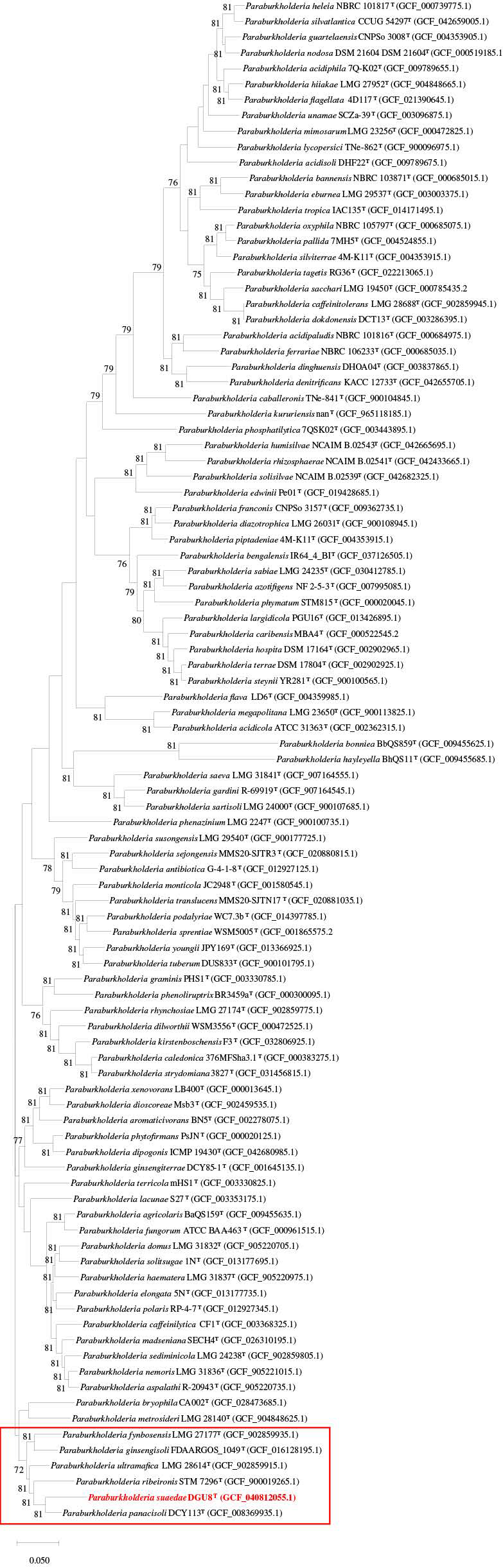
3.2. 16S rRNA Amplification and Phylogenetic Analysis
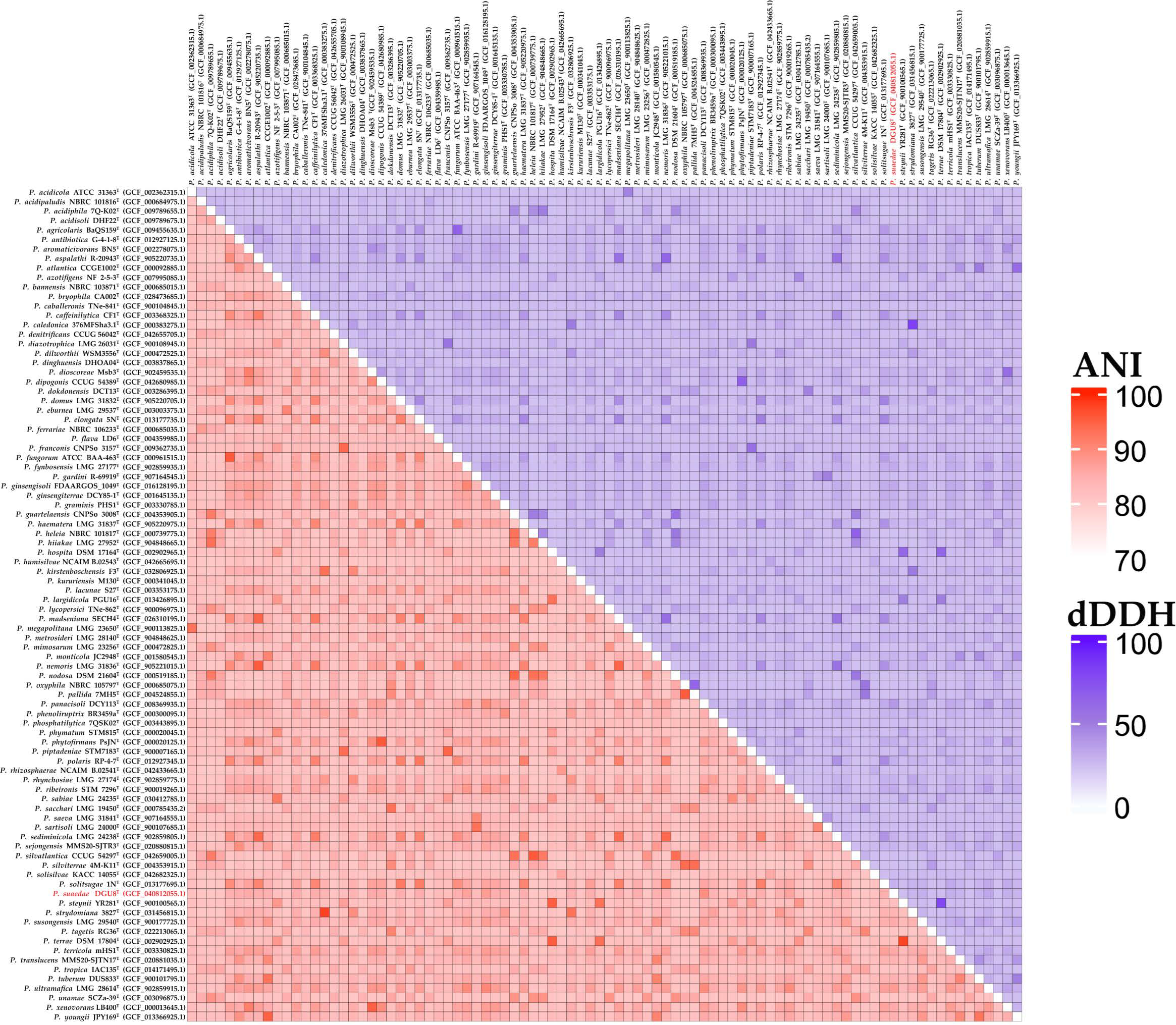
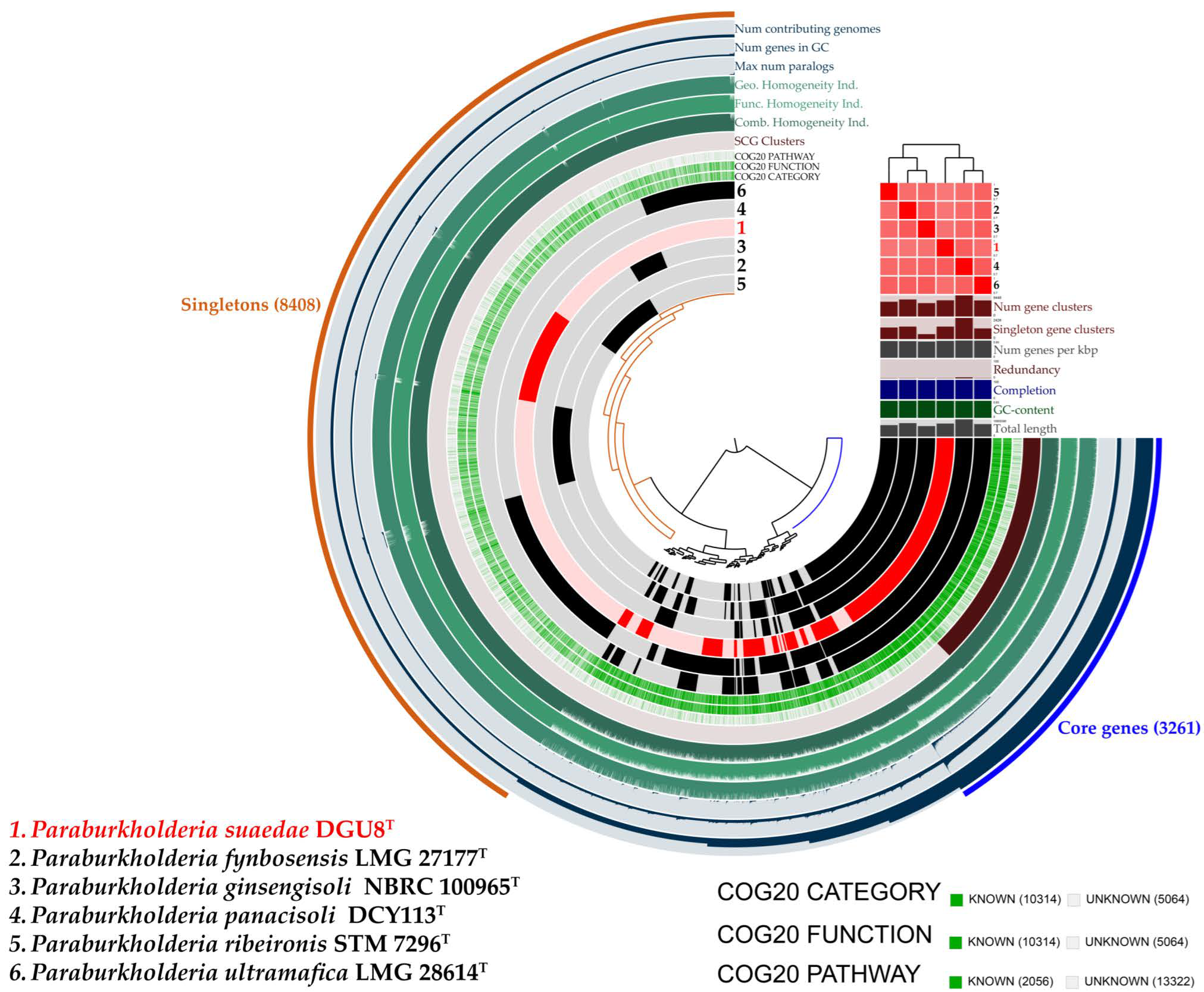
3.3. Genome Features and Comparative Analysis
3.4. Morphology and Chemotaxonomic Characteristics
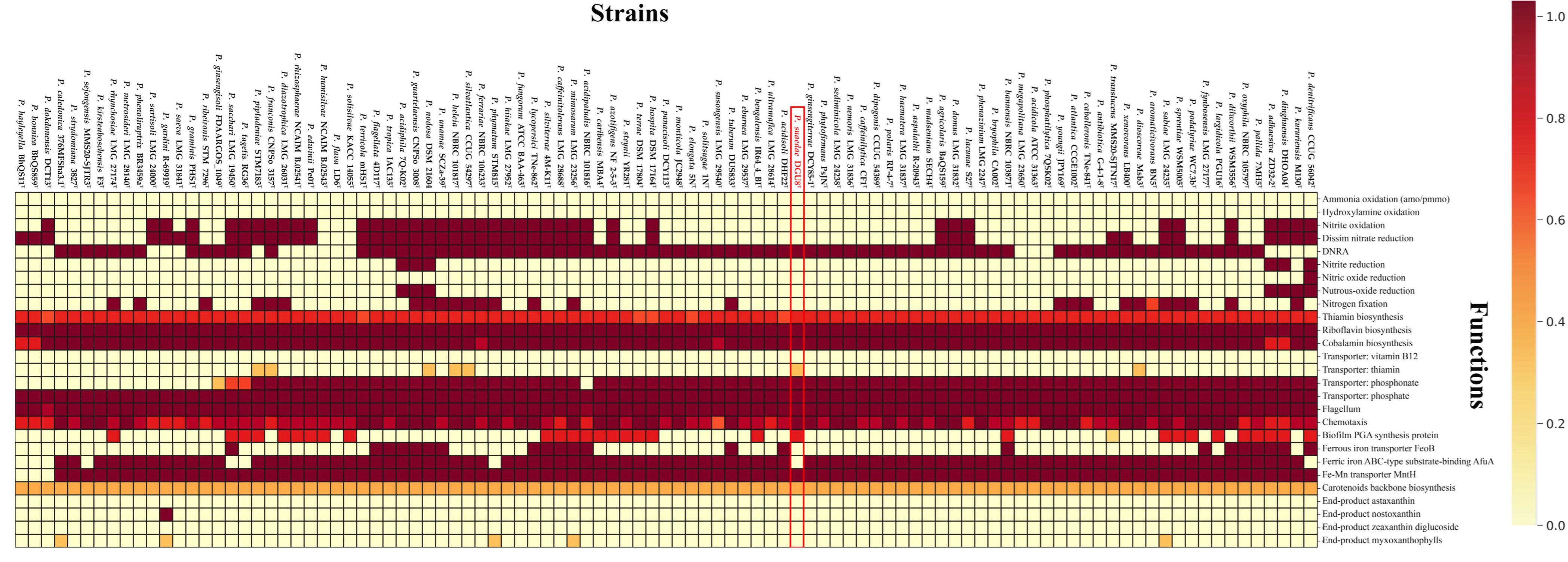
3.5. Functional Traits Related to Plant Growth Promotion
4. Discussion
Description of Paraburkholderia suaedae sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ANI | average nucleotide identity |
| dDDH | digital DNA-DNA hybridization |
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| CAS agar | universal chrome azurol S agar |
References
- Yin, J.; Gentine, P.; Slater, L.; Gu, L.; Pokhrel, Y.; Hanasaki, N.; Guo, S.; Xiong, L.; Schlenker, W. Future socio-ecosystem productivity threatened by compound drought–heatwave events. Nat. Sustain. 2023, 6, 259–272. [Google Scholar] [CrossRef]
- Wahab, A.; Abdi, G.; Saleem, M.H.; Ali, B.; Ullah, S.; Shah, W.; Mumtaz, S.; Yasin, G.; Muresan, C.C.; Marc, R.A. Plants’ Physio-Biochemical and Phyto-Hormonal Responses to Alleviate the Adverse Effects of Drought Stress: A Comprehensive Review. Plants 2022, 11, 1620. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Park, S.; Mir, R.A.; Mushtaq, M.; Bhat, B.; Mahmoudi, H.; Bae, H. Deciphering the Plant Microbiome to Improve Drought Tolerance: Mechanisms and Perspectives. Environ. Exp. Bot. 2022, 201, 104933. [Google Scholar] [CrossRef]
- Kwak, G.-Y.; Han, Y.; Baik, S.; Kong, B.-M.; Yang, D.-C.; Kang, S.-C.; Sukweenadhi, J. Gold Nanoparticles Green-Synthesized by the Suaeda Japonica Leaf Extract and Screening of Anti-Inflammatory Activities on RAW 267.4 Macrophages. Coatings 2022, 12, 460. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.; Chung, Y. The Comparison of the Complete Chloroplast Genome of Suaeda japonica Makino Presenting Different External Morphology (Amaranthaceae). Mitochondrial DNA Part B 2020, 5, 1616–1618. [Google Scholar] [CrossRef]
- Kim, J.M.; Lee, S.H.; Jung, J.Y.; Jeon, C.O. Marinobacterium Lutimaris sp. nov., Isolated from a Tidal Flat. Int. J. Syst. Evol. Microbiol. 2010, 60, 1828–1831. [Google Scholar] [CrossRef]
- Stevens, H.; Brinkhoff, T.; Rink, B.; Vollmers, J.; Simon, M. Diversity and Abundance of Gram Positive Bacteria in a Tidal Flat Ecosystem. Environ. Microbiol. 2007, 9, 1810–1822. [Google Scholar] [CrossRef]
- Winberg, P.C.; Lynch, T.P.; Murray, A.; Jones, A.R.; Davis, A.R. The Importance of Spatial Scale for the Conservation of Tidal Flat Macrobenthos: An Example from New South Wales, Australia. Biol. Conserv. 2007, 134, 310–320. [Google Scholar] [CrossRef]
- Kang, K.-Y.; Hwang, Y.-H.; Lee, S.-J.; Kim, J.-J.; Nam, S.-J.; Yee, S.-T. Verification of the Antioxidant Activity of a Subterranean Part of Suaeda japonica Makino. Ind. Crops Prod. 2017, 109, 836–842. [Google Scholar] [CrossRef]
- Shim, Y.J.; Soshnikova, V.; Anandapadmanaban, G.; Mathiyalagan, R.; Jimenez Perez, Z.E.; Markus, J.; Ju Kim, Y.; Castro-Aceituno, V.; Yang, D.C. Zinc Oxide Nanoparticles Synthesized by Suaeda japonica Makino and Their Photocatalytic Degradation of Methylene Blue. Optik 2019, 182, 1015–1020. [Google Scholar] [CrossRef]
- Chandrasekaran, A.; Jeon, Y.; Kim, S.-Y.; Seo, D.-H.; Yuk, H.J.; Son, E.; Kim, D.-S.; Kim, S.-H.; Lee, G.-J. Therapeutic Potential of Suaeda japonica Makino Leaf Extract Against Obesity in 3T3-L1 Preadipocytes and HFD-Induced C57BL/6 J Mice. Appl. Biochem. Biotechnol. 2025, 197, 2555–2578. [Google Scholar] [CrossRef]
- Park, S.; Kim, I.; Chhetri, G.; Jung, Y.; Woo, H.; Seo, T. Draft Genome Sequence Analyses of Two Novel Marinobacter suadae sp. nov. and Wenyingzhuangia gilva sp. nov. Isolated from the Root of Suaeda Japonica Makino. Life 2024, 14, 296. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef]
- Beukes, C.W.; Steenkamp, E.T.; Van Zyl, E.; Avontuur, J.; Chan, W.Y.; Hassen, A.I.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K.; et al. Paraburkholderia Strydomiana sp. nov. and Paraburkholderia Steynii sp. nov.: Rhizobial symbionts of the fynbos legume Hypocalyptus sophoroides. Antonie Van Leeuwenhoek 2019, 112, 1369–1385. [Google Scholar] [CrossRef]
- Feng, T.; Jeong, S.E.; Lim, J.J.; Hyun, S.; Jeon, C.O. Paraburkholderia Lacunae sp. nov., isolated from soil near an artificial pond. J. Microbiol. 2019, 57, 232–237. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Murphy, S.J.L.; Feriancek, N.M.; Karasz, D.C.; DeRito, C.M.; Newman, J.D.; Buckley, D.H. Paraburkholderia Madseniana sp. nov., a phenolic acid-degrading bacterium isolated from acidic forest soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 2137–2146. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; Cyle, K.T.; Martinez, C.E.; Karasz, D.C.; Newman, J.D.; Buckley, D.H. Paraburkholderia Solitsugae sp. nov. and Paraburkholderia Elongata sp. nov., a henolic acid-degrading bacteria isolated from forest soil and emended description of Paraburkholderia madseniana. Int. J. Syst. Evol. Microbiol. 2020, 70, 5093–5105. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature (Bacterio.Net), 20 Years On. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Paulitsch, F.; Dall’Agnol, R.F.; Delamuta, J.R.M.; Ribeiro, R.A.; Da Silva Batista, J.S.; Hungria, M. Paraburkholderia Atlantica sp. nov. and Paraburkholderia Franconis sp. nov., two new nitrogen-fixing nodulating species isolated from atlantic forest soils in brazil. Arch. Microbiol. 2020, 202, 1369–1380. [Google Scholar] [CrossRef]
- Vanwijnsberghe, S.; Peeters, C.; De Ridder, E.; Dumolin, C.; Wieme, A.D.; Boon, N.; Vandamme, P. Genomic Aromatic compound degradation potential of novel Paraburkholderia species: Paraburkholderia Domus sp. nov., Paraburkholderia Haematera sp. nov. and Paraburkholderia Nemoris sp. nov. Int. J. Mol. Sci. 2021, 22, 7003. [Google Scholar] [CrossRef]
- Fu, W.; Zhao, D.; Liu, H.; Wu, L.; Hao, L. Inoculation of Paraburkholderia sp. GD17 improves seedling growth and tolerance to cadmium in Chinese cabbage. Environ. Exp. Bot. 2024, 227, 105955. [Google Scholar] [CrossRef]
- Prihatna, C.; Yan, Q. Exopolysaccharide is required by Paraburkholderia Phytofirmans PsJN to confer drought-stress tolerance in Pea. Front. Microbiol. 2024, 15, 1442001. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, G.; Kim, I.; Kim, J.; So, Y.; Park, S.; Jung, Y.; Seo, T. Paraburkholderia Tagetis sp. nov., a novel species isolated from roots of Tagetes patula enhances the growth and yield of Solanum Lycopersicum L. (Tomato). Front. Microbiol. 2023, 14, 1140484. [Google Scholar] [CrossRef] [PubMed]
- Herpell, J.B.; Schindler, F.; Bejtović, M.; Fragner, L.; Diallo, B.; Bellaire, A.; Kublik, S.; Foesel, B.U.; Gschwendtner, S.; Kerou, M.; et al. The Potato Yam Phyllosphere Ectosymbiont Paraburkholderia sp. Msb3 Is a Potent Growth Promotor in Tomato. Front. Microbiol. 2020, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Clewley, J.P. Macintosh Sequence Analysis Software. DNAStar’s LaserGene. Mol. Biotechnol. 1995, 3, 221–224. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and, maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of mucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Schalk, I.J.; Rigouin, C.; Godet, J. An overview of siderophore biosynthesis among fluorescent pseudomonads and new insights into their complex cellular organization. Environ. Microbiol. 2020, 22, 1447–1466. [Google Scholar] [CrossRef]
- Quadri, L.E.N.; Keating, T.A.; Patel, H.M.; Walsh, C.T. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: In vitro reconstitution of aryl-4,2-Bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 1999, 38, 14941–14954. [Google Scholar] [CrossRef]
- Comai, L.; Kosuge, T. Cloning and characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J. Bacteriol. 1982, 149, 40–46. [Google Scholar] [CrossRef]
- Kim, C.H.; Han, S.H.; Kim, K.Y.; Cho, B.H.; Kim, Y.H.; Koo, B.S.; Kim, Y.C. Cloning and expression of Pyrroloquinoline Quinone (PQQ) genes from a phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 2003, 47, 457–461. [Google Scholar] [CrossRef]
- Dos Santos, P.C.; Johnson, D.C.; Ragle, B.E.; Unciuleac, M.-C.; Dean, D.R. Controlled expression of Nif and Isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 2007, 189, 2854–2862. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kondo, A. Disruption of Poly (3-Hydroxyalkanoate) depolymerase gene and overexpression of three poly (3-Hydroxybutyrate) biosynthetic genes improve poly (3-Hydroxybutyrate) production from nitrogen rich medium by Rhodobacter sphaeroides. Microb. Cell Fact. 2019, 18, 40. [Google Scholar] [CrossRef]
- Wanner, B.L. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 1993, 51, 47–54. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Li, X.; Chen, S. Identification of genes involved in Fe–S cluster biosynthesis of nitrogenase in Paenibacillus Polymyxa WLY78. Int. J. Mol. Sci. 2021, 22, 3771. [Google Scholar] [CrossRef]
- Holden, V.I.; Wright, M.S.; Houle, S.; Collingwood, A.; Dozois, C.M.; Adams, M.D.; Bachman, M.A. Iron acquisition and siderophore release by carbapenem-resistant sequence Type 258 Klebsiella pneumoniae. mSphere 2018, 3, e00125-18. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A Hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Chklovski, A.; Parks, D.H.; Woodcroft, B.J.; Tyson, G.W. CheckM2: A Rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 2023, 20, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Na, S.-I.; Kim, D.; Chun, J. UBCG2: Up-to-Date Bacterial Core Genes and pipeline for phylogenomic analysis. J. Microbiol. 2021, 59, 609–615. [Google Scholar] [CrossRef]
- Kim, D.; Park, S.; Chun, J. Introducing EzAAI: A pipeline for high throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 2021, 59, 476–480. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A Large-Scale Evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, A.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 2018, 6, e4320. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Graham, E.D.; Heidelberg, J.F.; Tully, B.J. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 2018, 12, 1861–1866. [Google Scholar] [CrossRef]
- Park, S.; Kim, I.; Woo, H.; Lee, H.; Yook, S.; Seo, T. Flavobacterium Arundinis sp. nov., Flavobacterium Calami sp. nov., Flavobacterium Helocola sp. nov. and Flavobacterium Flavipallidum sp. nov., isolated from stagnant water in a clump of Phragmites Japonica Steud. Int. J. Syst. Evol. Microbiol. 2025, 75, 006667. [Google Scholar] [CrossRef]
- Baek, C.; Kim, I.; Park, S.; Woo, H.; Lee, H.; Yook, S.; Kwak, S.; Kim, J.; Seo, T. Isolation and characterization of Fibrella aquatica sp. nov. from freshwater, Fibrella musci sp. nov. from moss on soil and Fibrella arboris sp. nov. from moss on a tree. Int. J. Syst. Evol. Microbiol. 2025, 75, 006842. [Google Scholar] [CrossRef]
- Gregersen, T. Rapid method for distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 1978, 5, 123–127. [Google Scholar] [CrossRef]
- Kuykendall, L.D.; Roy, M.A.; O’neill, J.J.; Devine, T.E. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Evol. Microbiol. 1988, 38, 358–361. [Google Scholar] [CrossRef]
- Minnikin, D.E.; Patel, P.V.; Alshamaony, L.; Goodfellow, M. Polar Lipid Composition in the Classification of Nocardia and Related Bacteria. Int. J. Syst. Evol. Microbiol. 1977, 27, 104–117. [Google Scholar] [CrossRef]
- Ali, S.Z.; Sandhya, V.; Venkateswar Rao, L. Isolation and characterization of drought-tolerant ACC Deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann. Microbiol. 2014, 64, 493–502. [Google Scholar] [CrossRef]
- Milagres, A.M.F.; Machuca, A.; Napoleão, D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 1999, 37, 1–6. [Google Scholar] [CrossRef]
- Myo, E.M.; Ge, B.; Ma, J.; Cui, H.; Liu, B.; Shi, L.; Jiang, M.; Zhang, K. Indole-3-acetic acid production by Streptomyces fradiae NKZ-259 and its formulation to enhance plant growth. BMC Microbiol. 2019, 19, 155. [Google Scholar] [CrossRef]
- Kumar, H.; Khan, U.; Gond, S.K. Functional characterization and alleviation of cadmium stress by endophytic bacteria isolated from Cynodon dactylon (L.) Pers. Total Environ. Microbiol. 2025, 1, 100015. [Google Scholar] [CrossRef]
- Palacios, O.A.; Bashan, Y.; de-Bashan, L.E. Proven and potential involvement of vitamins in interactions of plants with plant growth-promoting bacteria—An overview. Biol. Fertil. Soils 2014, 50, 415–432. [Google Scholar] [CrossRef]
- Yu, Y.; Yan, F.; Chen, Y.; Jin, C.; Guo, J.-H.; Chai, Y. Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front. Microbiol. 2016, 7, 1811. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Abo Elsoud, M.M.; Hasan, S.F.; Elhateir, M.M. Optimization of indole-3-acetic acid production by Bacillus velezensis isolated from Pyrus rhizosphere and its effect on plant growth. Biocatal. Agric. Biotechnol. 2023, 50, 102714. [Google Scholar] [CrossRef]
- Pedraza, R.O. Recent advances in nitrogen-fixing acetic acid bacteria. Int. J. Food Microbiol. 2008, 125, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Vega-Celedón, P.; Castillo-Novales, D.; Bravo, G.; Cárdenas, F.; Romero-Silva, M.J.; Seeger, M. Synthesis and Degradation of the Phytohormone Indole-3-Acetic Acid by the Versatile Bacterium Paraburkholderia Xenovorans LB400 and Its Growth Promotion of Nicotiana Tabacum Plant. Plants 2024, 13, 3533. [Google Scholar] [CrossRef]
- Meher; Shivakrishna, P.; Ashok Reddy, K.; Manohar Rao, D. Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J. Biol. Sci. 2018, 25, 285–289. [Google Scholar] [CrossRef]
- Pishchik, V.N.; Chizhevskaya, E.P.; Chebotar, V.K.; Mirskaya, G.V.; Khomyakov, Y.V.; Vertebny, V.E.; Kononchuk, P.Y.; Kudryavtcev, D.V.; Bortsova, O.A.; Lapenko, N.G.; et al. PGPB Isolated from Drought-Tolerant Plants Help Wheat Plants to Overcome Osmotic Stress. Plants 2024, 13, 3381. [Google Scholar] [CrossRef]



| Function | Start | End | Product | Gene |
|---|---|---|---|---|
| Phosphate solubilization | 203,877 | 205,151 | Phosphonoacetate hydrolase | phnA |
| 268,010 | 268,771 | Phosphate-import ATP-binding protein PhnC | phnC_1 | |
| 281,941 | 282,801 | Phosphate-import ATP-binding protein PhnC | phnC_2 | |
| 267,015 | 267,914 | Putative ABC transporter phosphonate/phosphite binding protein PhnD2 | phnD2 | |
| 268,768 | 269,568 | Phosphate-import permease protein PhnE | phnE_1 | |
| 280,104 | 280,928 | Phosphate-import permease protein PhnE | phnE_2 | |
| 277,734 | 278,492 | putative transcriptional regulator PhnF | phnF | |
| 277,076 | 277,558 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate synthase subunit PhnG | phnG | |
| 276,426 | 277,076 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate synthase subunit PhnH | phnH | |
| 275,287 | 276,426 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate synthase subunit PhnI | phnI | |
| 274,382 | 275,290 | Alpha-D-ribose 1-methyl phosphonate 5-phosphate C-P lyase | phnJ | |
| 273,615 | 274,385 | Putative phosphonates utilization ATP-binding protein PhnK | phnK | |
| 272,839 | 273,606 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate synthase subunit PhnL | phnL | |
| 265,661 | 266,794 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate diphosphatase | phnM_1 | |
| 271,672 | 272,805 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate diphosphatase | phnM_2 | |
| 67,312 | 68,208 | Alpha-D-ribose 1-methyl phosphonate 5-triphosphate diphosphatase | phnM_3 | |
| 279,238 | 279,795 | Ribose 1,5-bisphosphate phosphokinase PhnN | phnN | |
| 198,905 | 199,996 | Putative 2-aminoethyl phosphonate-binding periplasmic protein | phnS | |
| 200,094 | 201,194 | Putative 2-aminoethyl phosphonate import ATP-binding protein PhnT | phnT | |
| 202,112 | 202,975 | Putative 2-aminoethyl phosphonate transport system permease protein PhnV | phnV | |
| 18,652 | 18,798 | Phosphonoacetaldehyde dehydrogenase | phnY_1 | |
| 205,148 | 206,599 | Phosphonoacetaldehyde dehydrogenase | phnY_2 | |
| 206,604 | 207,164 | 2-Amino-1-hydroxymethyl phosphonate dioxygenase (glycine-forming) | phnZ | |
| 332,262 | 332,963 | Phosphate regulon transcriptional regulatory protein PhoB | phoB_1 | |
| 125,536 | 126,231 | Phosphate regulon transcriptional regulatory protein PhoB | phoB_2 | |
| 124,094 | 124,777 | Virulence transcriptional regulatory protein PhoP | phoP | |
| 333,075 | 334,388 | Phosphate regulon sensor protein PhoR | phoR | |
| 331,531 | 332,235 | Phosphate-specific transport system accessory protein PhoU | phoU_1 | |
| 34,391 | 35,098 | Phosphate-specific transport system accessory protein PhoU | phoU_2 | |
| 45,975 | 46,682 | Phosphate-specific transport system accessory protein PhoU | phoU_3 | |
| 334,488 | 336,551 | Polyphosphate kinase | ppk | |
| 336,733 | 338,367 | Exopolyphosphatase | ppx | |
| 329,732 | 330,628 | Phosphate transport system permease protein PstA | pstA | |
| 330,646 | 331,503 | Phosphate import ATP-binding protein PstB | pstB | |
| 328,737 | 329,735 | Phosphate transport system permease protein PstC | pstC | |
| 327534 | 328565 | Phosphate-binding protein PstS | pstS | |
| 180,222 | 181,160 | Phosphate acetyltransferase | pta | |
| 196,720 | 198,183 | Glucose-6-phosphate 1-dehydrogenase | zwf_1 | |
| Pyrroloquinoline quinone (PQQ) production | 207,713 | 208,651 | Coenzyme PQQ synthesis protein B | pqqB |
| 208,714 | 209,481 | Pyrroloquinoline-quinone synthase | pqqC | |
| 209,478 | 209,768 | PqqA binding protein | pqqD | |
| 209,773 | 210,993 | PqqA peptide cyclase | pqqE | |
| Nitrogen fixation | 123,632 | 124,045 | Iron-sulfur cluster assembly scaffold protein IscU | iscU |
| 140,604 | 141,059 | Zinc-dependent sulfurtransferase SufU | sufU | |
| 92,432 | 94,072 | Nitrogen fixation protein VnfA | vnfA | |
| Production of IAA | 5454 | 7304 | Tryptophan 2-monooxygenase | iaaM |
| 154,601 | 155,413 | Tryptophan synthase alpha chain | trpA | |
| 152,447 | 153,640 | Tryptophan synthase beta chain | trpB | |
| 14,333 | 15,118 | Indole-3-glycerol phosphate synthase | trpC | |
| 13,271 | 14,302 | Anthranilate phosphoribosyltransferase | trpD | |
| 11,120 | 12,613 | Anthranilate synthase component 1 | trpE | |
| 151,687 | 152,421 | N-(5′-phosphoribosyl)anthranilate isomerase | trpF | |
| 12,627 | 13,253 | Anthranilate synthase component 2 | trpG | |
| 5983 | 6861 | HTH-type transcriptional regulator TrpI | trpI | |
| 91,037 | 91,969 | Tryptophan 2,3-dioxygenase | kynA | |
| Production of siderophore | 46,942 | 48,519 | 4-Cresol dehydrogenase [hydroxylating] flavoprotein subunit | pchF |
| 5918 | 7165 | Enterobactin exporter EntS | entS_1 | |
| 161,718 | 163,079 | Enterobactin exporter EntS | entS_2 | |
| 137,907 | 139,169 | Enterobactin exporter EntS | entS_3 | |
| 105,651 | 108,170 | Acyl-homoserine lactone acylase PvdQ | pvdQ |
| Characteristic | Strain | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Colony color | Ivory | White | Cream | Yellow | White | White |
| Optimum growth temperature (°C) | 30 | 25 | 30 | 30 | 30 | 30 |
| Optimum growth pH | 7 | 8 | 7 | 7 | 6 | 6 |
| Catalase activity | + | + | + | + | + | + |
| Oxidase activity | – | + | – | + | – | + |
| Hydrolysis of: | ||||||
| Casein | – | – | + | + | – | – |
| Chitin | – | – | – | – | – | – |
| Tween 20 | – | – | – | – | + | – |
| Carboxymethyl cellulose | + | – | + | – | – | + |
| Assimilation of (API 20 NE): | ||||||
| D-maltose | + | – | – | – | – | – |
| 4-Nitrophenyl-β-d-galactopyranoside | + | + | + | – | + | + |
| Aesculin | + | + | + | + | – | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Lee, H.; Yook, S.; Baek, C.; Kim, J.; Kwak, S.; Na, T.; Seo, T. Paraburkholderia suaedae sp. nov., a Potential Plant Growth-Promoting Bacterium Isolated from the Halophyte Suaeda japonica. Microorganisms 2025, 13, 2498. https://doi.org/10.3390/microorganisms13112498
Park S, Lee H, Yook S, Baek C, Kim J, Kwak S, Na T, Seo T. Paraburkholderia suaedae sp. nov., a Potential Plant Growth-Promoting Bacterium Isolated from the Halophyte Suaeda japonica. Microorganisms. 2025; 13(11):2498. https://doi.org/10.3390/microorganisms13112498
Chicago/Turabian StylePark, Sunho, Hyunji Lee, Subin Yook, Chunghwan Baek, Jisu Kim, Seunghui Kwak, Taeho Na, and Taegun Seo. 2025. "Paraburkholderia suaedae sp. nov., a Potential Plant Growth-Promoting Bacterium Isolated from the Halophyte Suaeda japonica" Microorganisms 13, no. 11: 2498. https://doi.org/10.3390/microorganisms13112498
APA StylePark, S., Lee, H., Yook, S., Baek, C., Kim, J., Kwak, S., Na, T., & Seo, T. (2025). Paraburkholderia suaedae sp. nov., a Potential Plant Growth-Promoting Bacterium Isolated from the Halophyte Suaeda japonica. Microorganisms, 13(11), 2498. https://doi.org/10.3390/microorganisms13112498






