First Insight into the Natural Attenuation of Emerging Contaminants Using a Metagenomics Approach from Drinking Water Sources in the Free State
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Water Sample Collection
2.3. Physico-Chemical Characterisation
2.4. Environmental DNA Extraction and Quality Assessment
2.5. Metagenomic Shotgun Sequencing
2.6. Bioinformatic Analysis
2.6.1. Quality Check, Filtering, Assembling, and DiTing
2.6.2. Taxonomic Assignment and Identification of Biodegradation Pathways
2.6.3. Binning, MAGs, and Functional Annotations
3. Results
3.1. Physico-Chemical Characterization
3.2. Bacterial Diversity
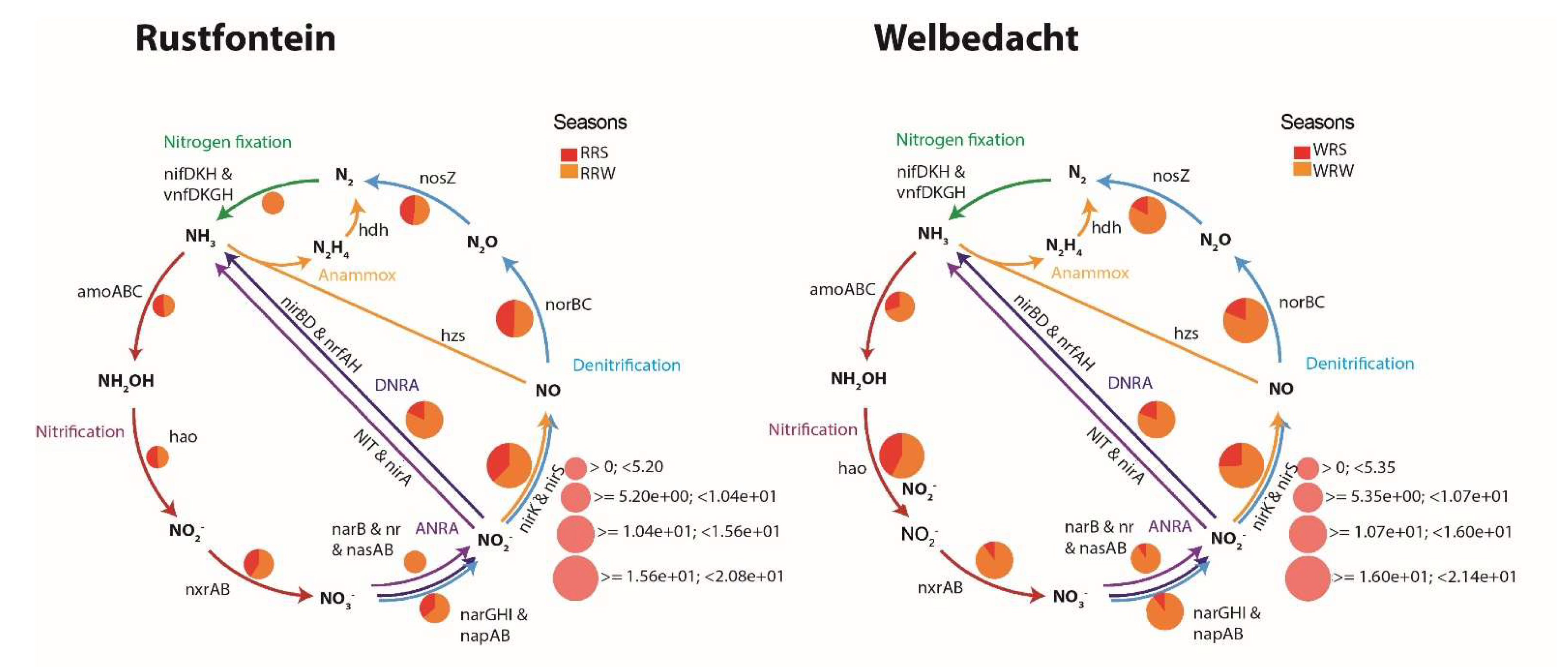
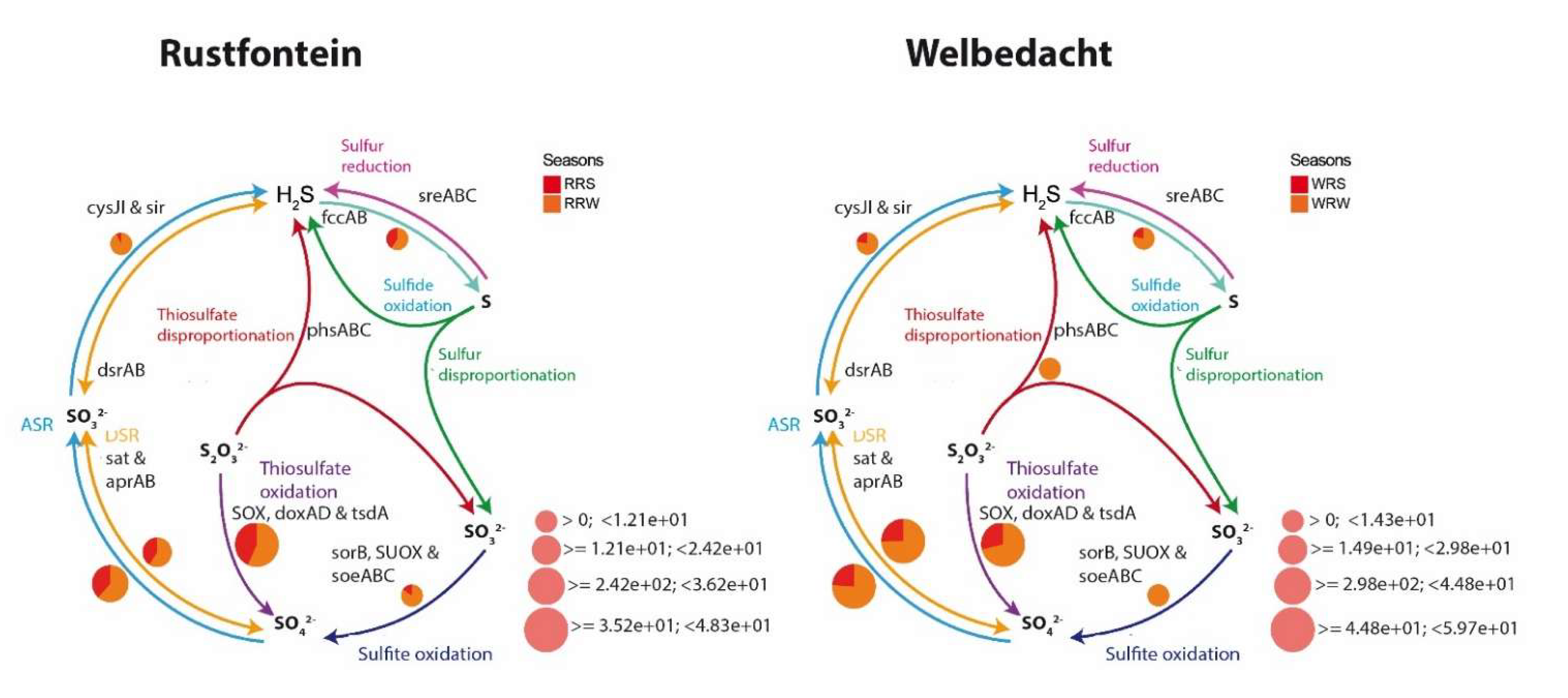
3.3. Main Biogeochemical Pathways Associated with the Co-Metabolism of ECs
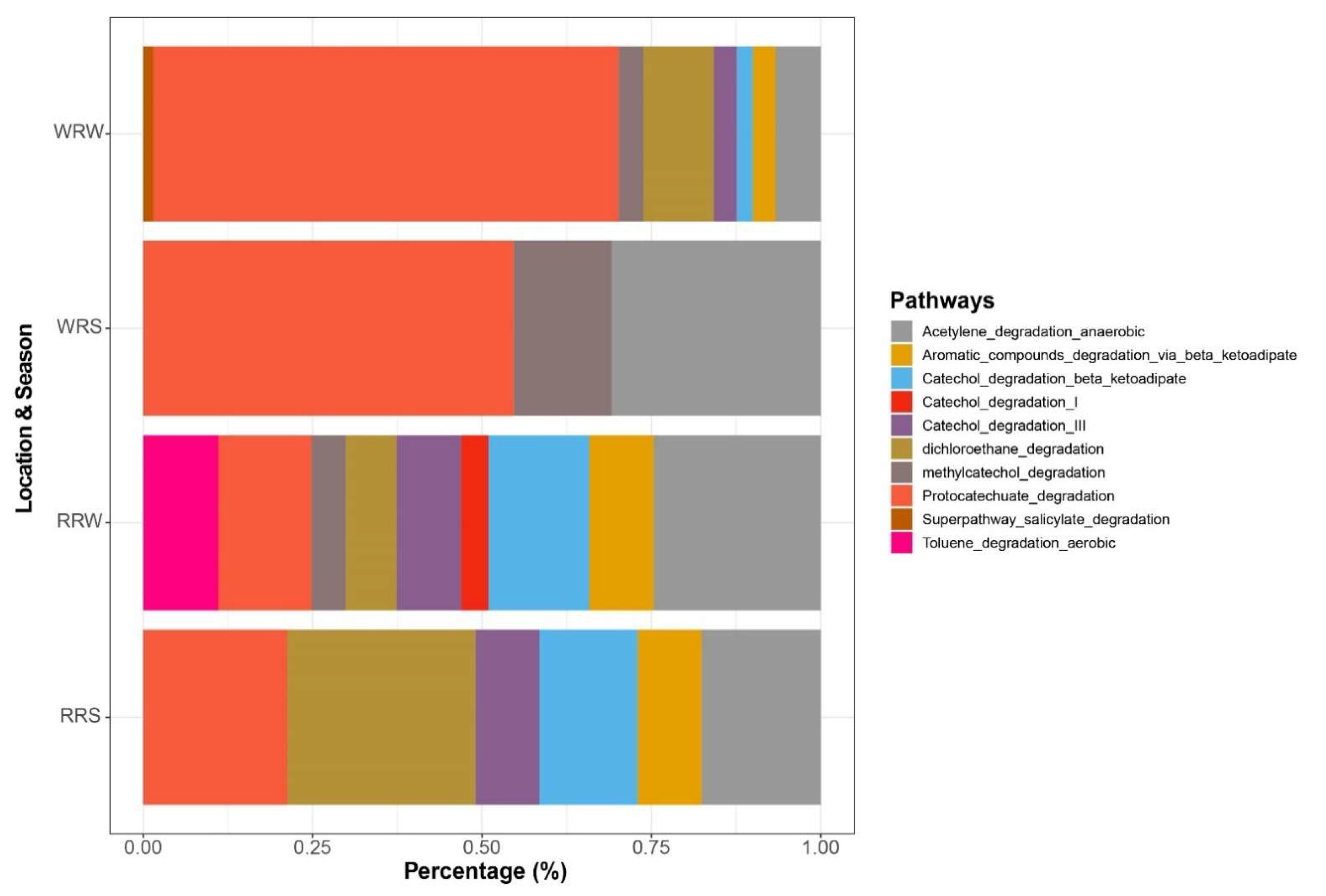
3.4. Presence of Xenobiotic Degradation Pathways
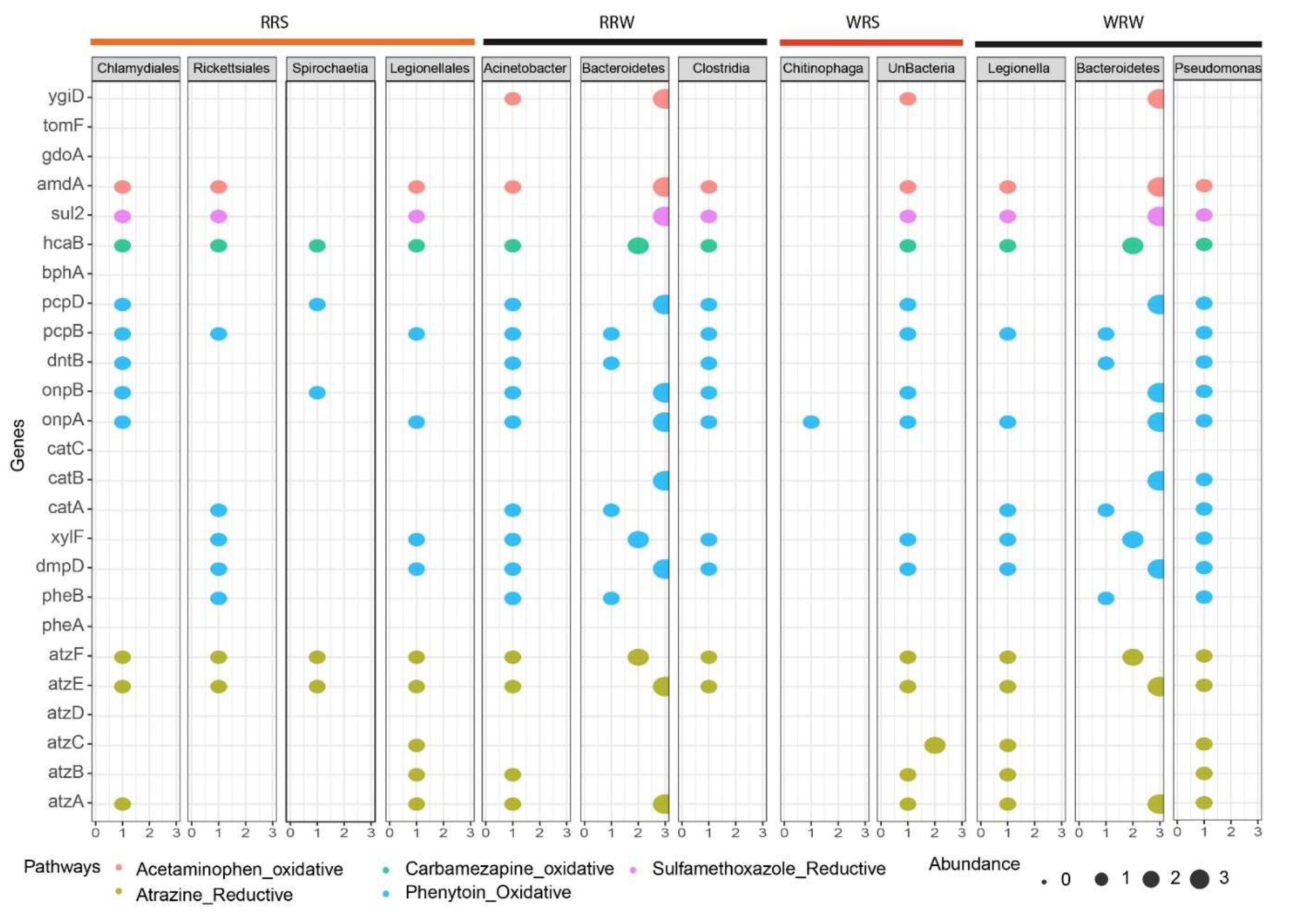
3.5. The Presence of Genes Associated with the Emerging Contaminant Degradation
4. Discussion
4.1. Physico-Chemical Characterization
4.2. Bacterial Diversity and Main Biogeochemical Pathways Associated with Co-Metabolism of ECs
4.3. Presence of Xenobiotic Degradation Pathways and Genes Associated with Degradation in Relation to Seasonal Change
4.4. Effect of Seasonal Change on the Degradation Efficiency and Ecological Implications of Finding Genes in Indigenous Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gogoi, A.; Mazumder, P.; Tyagi, V.K.; Tushara Chaminda, G.G.; An, A.K.; Kumar, M. Occurrence and fate of emerging contaminants in water environment: A review. Groundw. Sustain. Dev. 2018, 6, 169–180. [Google Scholar] [CrossRef]
- Pal, A.; Gin, K.Y.H.; Lin, A.Y.C.; Reinhard, M. Impacts of emerging organic contaminants on freshwater resources: Review of recent occurrences, sources, fate and effects. Sci. Total Environ. 2010, 408, 6062–6069. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Majewsky, M. Cometabolic degradation of organic wastewater micropollutants by activated sludge and sludge-inherent microorganisms. Appl. Microbiol. Biotechnol. 2014, 9, 6583–6597. [Google Scholar] [CrossRef] [PubMed]
- Sililo, O.T.N.; Saayman, I.C.; Fey, M.V. Groundwater vulnerability to pollution in urban catchments. In Water Research Commission Report, (1008/1); University of Cape Town: Cape Town, South Africa, 2001; Volume 56. [Google Scholar]
- Oke, S.A.; Mugudamani, I.; Gumede, T.P.; Senbore, S. Environmental risk assessment of exposure to selected emerging contaminants in dams and wastewater effluent around the city of Bloemfontein, South Africa. J. Harbin Eng. Univ. 2023, 44, 1077–1082. [Google Scholar] [CrossRef]
- Adu, D.; Emmanuel, B.G. 6th International Symposium on Water Resource and Environmental Management, WREM 2023; Springer: Berlin/Heidelberg, Germany, 2024; ISBN 9783031559884. [Google Scholar]
- Gani, K.M.; Hlongwa, N.; Abunama, T.; Kumari, S.; Bux, F. Emerging contaminants in South African water environment—A critical review of their occurrence, sources and ecotoxicological risks. Chemosphere 2021, 269, 128737. [Google Scholar] [CrossRef]
- Wood, T.P.; Duvenage, C.S.J.; Rohwer, E. The occurrence of anti-retroviral compounds used for HIV treatment in South African surface water. Environ. Pollut. 2015, 199, 235–243. [Google Scholar] [CrossRef]
- Rimayi, C.; Odusanya, D.; Weiss, J.M.; de Boer, J.; Chimuka, L. Contaminants of emerging concern in the Hartbeespoort Dam catchment and the uMngeni River estuary 2016 pollution incident, South Africa. Sci. Total Environ. 2018, 627, 1008–10017. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Michael, C.; Duan, X.; He, X.; Dionysiou, D.D.; Mills, M.A.; Fatta-Kassinos, D. Dissolved effluent organic matter: Characteristics and potential implications in wastewater treatment and reuse applications. Water Res. 2015, 77, 213–248. [Google Scholar] [CrossRef]
- Proia, L.; Lupini, G.; Osorio, V.; Pérez, S.; Barceló, D.; Schwartz, T.; Amalfitano, S.; Fazi, S.; Romaní, A.M.; Sabater, S. Response of biofilm bacterial communities to antibiotic pollutants in a Mediterranean River. Chemosphere 2013, 92, 1126–1135. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Zhao, Z.; Chen, J.; Lu, H.; Liu, G.; Zhou, J.; Guan, X. PAHs accelerate the propagation of antibiotic resistance genes in coastal water microbial community. Environ. Pollut. 2017, 231, 1145–1152. [Google Scholar] [CrossRef]
- Wang, H.; Hu, C.; Shen, Y.; Shi, B.; Zhao, D.; Xing, X. Response of microorganisms in biofilm to sulfadiazine and ciprofloxacin in drinking water distribution systems. Chemosphere 2019, 218, 197–204. [Google Scholar] [CrossRef]
- Isidori, M.; Lavorgna, M.; Nardelli, A.; Pascarella, L.; Parrella, A. Toxic and genotoxic evaluation of six antibiotics on non-target organisms. Sci. Total Environ. 2005, 346, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.D.; Kraus, P.F.; Lawson, P.A.; Drake, G.R.; Balkwill, D.L.; Tanner, R.S. Desulfovibrio carbinoliphilus sp. nov., a benzyl alcohol-oxidizing, sulfate-reducing bacterium isolated from a gas condensate-contaminated aquifer. Int. J. Syst. Evol. Microbiol. 2008, 58, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Keen, P.L.; Patrick, D.M. Tracking change: A look at the ecological footprint of antibiotics and antimicrobial resistance. Antibiotics 2013, 2, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, V.; Rodríguez, Y. Influence of seasonality and vegetation on the attenuation of emerging contaminants in wastewater effluent-dominated streams. A preliminary study. Chemosphere 2017, 186, 269–277. [Google Scholar] [CrossRef]
- Wooding, M.; Rohwer, E.R.; Naudé, Y. Determination of endocrine disrupting chemicals and antiretroviral compounds in surface water: A disposable sorptive sampler with comprehensive gas chromatography—Time-of-flight mass spectrometry and large volume injection with ultra-high performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2017, 1496, 122–132. [Google Scholar] [CrossRef]
- Ghattas, A.K.; Fischer, F.; Wick, A.; Ternes, T.A. Anaerobic biodegradation of (emerging) organic contaminants in the aquatic environment. Water Res. 2017, 116, 268–295. [Google Scholar] [CrossRef]
- Bao, Y.; Zhao, S.; Wu, N.; Yuan, Y.; Ruan, L.; He, J. Degradation of atrazine by an anaerobic microbial consortium enriched from soil of an herbicide-manufacturing plant. Curr. Microbiol. 2024, 81, 117. [Google Scholar] [CrossRef]
- Govantes, F.; García-González, V.; Porrúa, O.; Platero, A.I.; Jiménez-Fernández, A.; Santero, E. Regulation of the atrazine-degradative genes in Pseudomonas sp. strain ADP. FEMS Microbiol. Lett. 2010, 310, 1–8. [Google Scholar] [CrossRef]
- Shapir, N.; Osborne, J.P.; Johnson, G.; Sadowsky, M.J.; Wackett, L.P. Purification, substrate range, and metal center of AtzC: The N-Isopropylammelide aminohydrolase involved in bacterial atrazine metabolism. J. Bacteriol. 2002, 184, 5376–5384. [Google Scholar] [CrossRef]
- Udiković-Kolić, N.; Hřak, D.; Devers, M.; Klepac-Ceraj, V.; Petrić, I.; Martin-Laurent, F. Taxonomic and functional diversity of atrazine-degrading bacterial communities enriched from agrochemical factory soil. J. Appl. Microbiol. 2010, 109, 355–367. [Google Scholar] [CrossRef]
- Jiang, B.; Li, A.; Cui, D.; Cai, R.; Ma, F.; Wang, Y. Biodegradation and metabolic pathway of sulfamethoxazole by Pseudomonas psychrophila HA-4, a newly isolated cold-adapted sulfamethoxazole-degrading bacterium. Appl. Microbiol. Biotechnol. 2014, 98, 4671–4681. [Google Scholar] [CrossRef] [PubMed]
- Larcher, S.; Yargeau, V. Biodegradation of sulfamethoxazole by individual and mixed bacteria. Appl. Microbiol. Biotechnol. 2011, 91, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kassotaki, E.; Buttiglieri, G.; Ferrando-Climent, L.; Rodriguez-Roda, I.; Pijuan, M. Enhanced sulfamethoxazole degradation through ammonia oxidizing bacteria co-metabolism and fate of transformation products. Water Res. 2016, 94, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Ojo, A.O.; Castillo, J.; Cason, E.D.; Valverde, A. Biodegradation of chloroethene compounds under microoxic conditions. Biotechnol. Bioeng. 2024, 121, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Gedalanga, P.B.; Pornwongthong, P.; Mora, R.; Chiang, S.Y.D.; Baldwin, B.; Ogles, D.; Mahendraa, S. Identification of biomarker genes to predict biodegradation of 1,4-Dioxane. Appl. Environ. Microbiol. 2014, 80, 3209–3218. [Google Scholar] [CrossRef]
- Pretorius, E.; Gericke, O.J.; Slabbert, S.W.; Dent, M. Catchment hydrology management using GIS: Case study of the Modder River Basin, South Africa. WIT Trans. Ecol. Environ. 2005, 83, 305–314. [Google Scholar]
- Andrews, S. FastQC—A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinforma. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 September 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate illumina paired-end read mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Xue, C.X.; Lin, H.; Zhu, X.Y.; Liu, J.; Zhang, Y.; Rowley, G.; Todd, J.D.; Li, M.; Zhang, X.H. DiTing: A pipeline to infer and compare biogeochemical pathways from metagenomic and metatranscriptomic data. Front. Microbiol. 2021, 12, 698286. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation. Nat. Commun. 2010, 11, 119. [Google Scholar] [CrossRef]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with Biobakery 3. Elife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- SANS 241-1:2015; South African National Standard (SANS) Drinking Water Specification. South African Bureau of Standards (SABS): Pretoria, South Africa, 2015; Volume 241.
- Department of Water Affairs and Forestry (DWAF). DWAF South African Water Quality Guidelines: Volume 1 Domestic Use; Department of Water Affairs and Forestry: Pretoria, South Africa, 1996; Volume 1, ISBN 0798853387. [Google Scholar]
- World Health Organization (WHO). Guidelines for Drinking-Water Quality; World Health Organization: Geneva, Switzerland, 2017; Volume 55, ISBN 9789241549950. [Google Scholar]
- Tran, N.H.; Urase, T.; Ngo, H.H.; Hu, J.; Ong, S.L. Insight into metabolic and cometabolic activities of autotrophic and heterotrophic microorganisms in the biodegradation of emerging trace organic contaminants. Bioresour. Technol. 2013, 146, 721–731. [Google Scholar] [CrossRef]
- Tadkaew, N.; Hai, F.I.; McDonald, J.A.; Khan, S.J.; Nghiem, L.D. Removal of trace organics by MBR treatment: The role of molecular properties. Water Res. 2011, 45, 2439–2451. [Google Scholar] [CrossRef]
- Czajka, C.P.; Londry, K.L. Anaerobic biotransformation of estrogens. Sci. Total Environ. 2006, 367, 932–941. [Google Scholar] [CrossRef]
- Ledesma, J.L.J.; Köhler, S.J.; Futter, M.N. Long-term dynamics of dissolved organic carbon: Implications for drinking water supply. Sci. Total Environ. 2012, 432, 1–11. [Google Scholar] [CrossRef]
- Bilal, M.; Adeel, M.; Rasheed, T.; Zhao, Y.; Iqbal, H.M.N. Emerging contaminants of high concern and their enzyme-assisted biodegradation—A review. Environ. Int. 2019, 124, 336–353. [Google Scholar] [CrossRef]
- Su, Q.; Schittich, A.R.; Jensen, M.M.; Ng, H.; Smets, B.F. Role of ammonia oxidation in organic micropollutant transformation during wastewater treatment: Insights from molecular, cellular, and community level observations. Environ. Sci. Technol. 2021, 55, 2173–2188. [Google Scholar] [CrossRef]
- Nsenga Kumwimba, M.; Meng, F. Roles of ammonia-oxidizing bacteria in improving metabolism and cometabolism of trace organic chemicals in biological wastewater treatment processes: A review. Sci. Total Environ. 2019, 659, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Nzila, A. Update on the cometabolism of organic pollutants by bacteria. Environ. Pollut. 2013, 178, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Kahlon, R.S. Pseudomonas: Molecular and Applied Biology; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9783319311982. [Google Scholar]
- Zhang, S.; Courtois, S.; Gitungo, S.; Raczko, R.F.; Dyksen, J.E.; Li, M.; Axe, L. Microbial community analysis in biologically active filters exhibiting efficient removal of emerging contaminants and impact of operational conditions. Sci. Total Environ. 2018, 640–641, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Geng, J.; Xu, K.; Ren, H. Removal of pharmaceuticals by ammonia oxidizers during nitrification. Appl. Microbiol. Biotechnol. 2021, 105, 909–921. [Google Scholar] [CrossRef]
- Benner, J.; De Smet, D.; Ho, A.; Kerckhof, F.M.; Vanhaecke, L.; Heylen, K.; Boon, N. Exploring methane-oxidizing communities for the co-metabolic degradation of organic micropollutants. Appl. Microbiol. Biotechnol. 2015, 99, 3609–3618. [Google Scholar] [CrossRef]
- Pandey, V.C.; Singh, J.S.; Singh, D.P.; Singh, R.P. Methanotrophs: Promising bacteria for environmental remediation. Int. J. Environ. Sci. Technol. 2014, 11, 241–250. [Google Scholar] [CrossRef]
- Murreil, J.C.; Gilbert, B.; McDonald, I.R. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef]
- Wang, S.; Yun, Y.; Tian, X.; Su, Z.; Liao, Z.; Li, G.; Ma, T. HMDB: A curated database of genes involved in hydrocarbon monooxygenation reaction with homologous genes as background. J. Hazard. Mater. 2023, 460, 132397. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; Dispirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed]
- Salcher, M.M.; Schaefle, D.; Kaspar, M.; Neuenschwander, S.M.; Ghai, R. Evolution in action: Habitat transition from sediment to the pelagial leads to genome streamlining in Methylophilaceae. ISME J. 2019, 13, 2764–2777. [Google Scholar] [CrossRef]
- Salcher, M.M.; Neuenschwander, S.M.; Posch, T.; Pernthaler, J. The ecology of pelagic freshwater methylotrophs assessed by a high-resolution monitoring and isolation campaign. ISME J. 2015, 9, 2442–2453. [Google Scholar] [CrossRef]
- Martínez-Quintela, M.; Arias, A.; Alvarino, T.; Suarez, S.; Garrido, J.M.; Omil, F. Cometabolic removal of organic micropollutants by enriched nitrite-dependent anaerobic methane oxidizing cultures. J. Hazard. Mater. 2021, 402, 123450. [Google Scholar] [CrossRef]
- Boopathy, R. Anaerobic Degradation of atrazine. Int. Biodeterior. Biodegrad. 2017, 119, 626–630. [Google Scholar] [CrossRef]
- Yang, Y.; Li, N.; Zhao, Q.; Yang, M.; Wu, Z.; Xie, S.; Liu, Y. Ammonia-oxidizing archaea and bacteria in water columns and sediments of a highly eutrophic plateau freshwater lake. Environ. Sci. Pollut. Res. 2016, 23, 15358–15369. [Google Scholar] [CrossRef]
- Zeng, J.; Zhao, D.; Yu, Z.; Huang, R.; Wu, Q.L. Temperature responses of ammonia-oxidizing prokaryotes in freshwater sediment microcosms. PLoS ONE 2014, 9, e100653. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Bao, L.; Xie, S. Abundance and community structure of ammonia-oxidizing microorganisms in reservoir sediment and adjacent soils. Appl. Microbiol. Biotechnol. 2014, 98, 1883–1892. [Google Scholar] [CrossRef]
- Bollmann, A.; Bullerjahn, G.S.; Mckay, R.M. Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS ONE 2014, 9, e97068. [Google Scholar] [CrossRef]
- Zhang, S.; Karthikeyan, R.; Fernando, S.D. Low-temperature biological activation of methane: Structure, function and molecular interactions of soluble and particulate methane monooxygenases. Rev. Environ. Sci. Biotechnol. 2017, 16, 611–623. [Google Scholar] [CrossRef]
- Merkx, M.; Kopp, D.A.; Sazinsky, M.H.; Blazyk, J.L.; Müller, J.; Lippard, S.J. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: A tale of two irons and three proteins. Angew. Chem. Int. Ed. 2001, 40, 2782–2807. [Google Scholar] [CrossRef]
- Colby, B.J.; Stirling, D.I.; Dalton, H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 1977, 165, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Miglani, R.; Parveen, N.; Kumar, A.; Ansari, M.A.; Khanna, S.; Rawat, G.; Panda, A.K.; Bisht, S.S.; Upadhyay, J.; Ansari, M.N. Degradation of xenobiotic pollutants: An environmentally sustainable approach. Metabolites 2022, 12, 818. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Das, S. Bacterial enzymatic degradation of recalcitrant organic pollutants: Catabolic pathways and genetic regulations. Environ. Sci. Pollut. Res. 2023, 30, 79676–79705. [Google Scholar] [CrossRef]
- Fan, X.; Song, F. Bioremediation of atrazine: Recent advances and promises. J. Soils Sediments 2014, 14, 1727–1737. [Google Scholar] [CrossRef]
- Chopra, S.; Kumar, D. Characterization, optimization and kinetics study of acetaminophen degradation by Bacillus drentensis strain S1 and waste water degradation analysis. Bioresour. Bioprocess. 2020, 7, 9. [Google Scholar] [CrossRef]
- Pandey, B.; Pandey, A.K.; Tripathi, K.; Dubey, S.K. Biodegradation of acetaminophen: Microcosm centric genomic-proteomic-metabolomics evidences. Bioresour. Technol. 2024, 401, 130732. [Google Scholar] [CrossRef]
- Enguita, F.J.; Pereira, S.; Leitão, A.L. Transcriptomic analysis of acetaminophen biodegradation by Penicillium chrysogenum var. Halophenolicum and insights into energy and stress response pathways. J. Fungi 2023, 9, 408. [Google Scholar] [CrossRef]
- Rios-Miguel, A.B.; Smith, G.J.; Cremers, G.; van Alen, T.; Jetten, M.S.M.; Op den Camp, H.J.M.; Welte, C.U. Microbial paracetamol degradation involves a high diversity of novel Amidase enzyme Candidates. Water Res. X 2022, 16, 100152. [Google Scholar] [CrossRef] [PubMed]
- Nasir, N.M.; Talib, S.A.; Hashim, S.N.; Tay, C.C. Biodegradation of carbamazepine using fungi and bacteria. J. Fundam. Appl. Sci. 2018, 9, 124–146. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, J.; Zhou, S.; Lian, M. Microbial degradation of carbamazepine by a newly isolated of Gordonia polyophrenivorans. Environ. Technol. Innov. 2023, 32, 103322. [Google Scholar] [CrossRef]
- Aukema, K.G.; Escalante, D.E.; Maltby, M.M.; Bera, A.K.; Aksan, A.; Wackett, L.P. In silico identification of bioremediation potential: Carbamazepine and other recalcitrant personal care products. Environ. Sci. Technol. 2017, 52, 880–888. [Google Scholar] [CrossRef]
- Wang, J.; Wang, S. Microbial degradation of sulfamethoxazole in the environment. Appl. Microbiol. Biotechnol. 2018, 102, 3573–3582. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hua, T.; Yuan, C.S.; Li, B.; Zhu, X.; Li, F. Degradation pathways, microbial community and electricity properties analysis of antibiotic sulfamethoxazole by bio-electro-Fenton system. Bioresour. Technol. 2020, 298, 122501. [Google Scholar] [CrossRef]
- Rucká, L.; Nešvera, J.; Pátek, M. Biodegradation of phenol and its derivatives by engineered bacteria: Current knowledge and perspectives. World J. Microbiol. Biotechnol. 2017, 33, 174. [Google Scholar] [CrossRef]
- Nešvera, J.; Rucká, L.; Pátek, M. Catabolism of phenol and its derivatives in bacteria: Genes, their regulation, and use in the biodegradation of toxic pollutants. Adv. Appl. Microbiol. 2015, 93, 107–160. [Google Scholar] [CrossRef]
- Wang, Z.; Ouyang, W.; Tysklind, M.; Lin, C.; Wang, B. Seasonal variations in Atrazine degradation in a typical semi enclosed bay of the Northwest Pacific ocean. Environ. Pollut. 2021, 283, 117072. [Google Scholar] [CrossRef]
- Muhammad, F.; Yusuf, F.; Ahmad, F.A.; Shehu, U.; Yakasai, H.M. Optimizing the effect of pH and temperature on atrazine degradation by Bacillus safensis strain BUK _ BCH _ BTE6 an efficient atrazine tolerating bacteria from an agricultural soil in Kura Local government area of Kano State, Nigeria. Niger. J. Biotechnol. 2021, 38, 92–100. [Google Scholar] [CrossRef]
- Govantes, F.; Porrúa, O.; García-González, V.; Santero, E. Atrazine biodegradation in the lab and in the field: Enzymatic activities and gene regulation. Microb. Biotechnol. 2009, 2, 178–185. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J. Biodegradation and metabolic pathway of sulfamethoxazole by a novel strain Acinetobacter sp. Appl. Microbiol. Biotechnol. 2018, 102, 425–432. [Google Scholar] [CrossRef]
- Liu, J.; Amemiya, T.; Chang, Q. Toluene dioxygenase expression correlates with trichloroethylene degradation capacity in Pseudomonas putida F1 cultures. Biodegradation 2012, 23, 683–691. [Google Scholar] [CrossRef]


| Emerging Contaminant | Summer | Winter |
|---|---|---|
| Carbamazepine (ug/L) | 0.21 | 0.19 |
| Atrazine (mg/L) | 0.04 | 0.02 |
| Metolachlor (mg/L) | 0.04 | 0.02 |
| Terbuthylazine (mg/L) | 0.08 | 0.03 |
| 17-alpha-ethinyl-estradiol (mg/L) | 3.40 | 14.80 |
| Physicochemical Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| pH | Eh (mV) | EC (mS/m) | DO (mg/L) | DOC (mg/L) | SO42− (mg/L) | NO3− (mg/L) | NH4+ (mg/L) | Cu (mg/L) | |
| Summer | |||||||||
| Rustfontein Dam | 7.83 | 202.50 | 17.81 | 8.00 | 2.71 | 8.99 | 0.44 | 0.18 | 0.07 |
| Weldebacht Dam | 7.58 | 176.30 | 42.61 | 4.00 | 3.62 | 19.93 | 0.29 | 0.20 | 0.006 |
| Winter | |||||||||
| Rustfontein Dam | 7.72 | 206.50 | 16.45 | 12.50 | 2.80 | 7.99 | 0.43 | <0.10 | 0.053 |
| Weldebacht Dam | 7.58 | 160.50 | 40.61 | 5.00 | 4.20 | 20.13 | 0.30 | <0.10 | 0.014 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mqambalala, A.; Maleke, M.; Deysel, L.-M.; Osman, J.R.; Gomez-Arias, A.; Valverde, A.; Hernandez, J.C. First Insight into the Natural Attenuation of Emerging Contaminants Using a Metagenomics Approach from Drinking Water Sources in the Free State. Microorganisms 2025, 13, 2349. https://doi.org/10.3390/microorganisms13102349
Mqambalala A, Maleke M, Deysel L-M, Osman JR, Gomez-Arias A, Valverde A, Hernandez JC. First Insight into the Natural Attenuation of Emerging Contaminants Using a Metagenomics Approach from Drinking Water Sources in the Free State. Microorganisms. 2025; 13(10):2349. https://doi.org/10.3390/microorganisms13102349
Chicago/Turabian StyleMqambalala, Avela, Maleke Maleke, Lore-Mari Deysel, Jorge R. Osman, Alba Gomez-Arias, Angel Valverde, and Julio Castillo Hernandez. 2025. "First Insight into the Natural Attenuation of Emerging Contaminants Using a Metagenomics Approach from Drinking Water Sources in the Free State" Microorganisms 13, no. 10: 2349. https://doi.org/10.3390/microorganisms13102349
APA StyleMqambalala, A., Maleke, M., Deysel, L.-M., Osman, J. R., Gomez-Arias, A., Valverde, A., & Hernandez, J. C. (2025). First Insight into the Natural Attenuation of Emerging Contaminants Using a Metagenomics Approach from Drinking Water Sources in the Free State. Microorganisms, 13(10), 2349. https://doi.org/10.3390/microorganisms13102349





