Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Lactic Acid Bacteria from Swine Feces
2.2. Taxonomic Identification of LAB
2.3. Antagonistic Activity against Salmonella
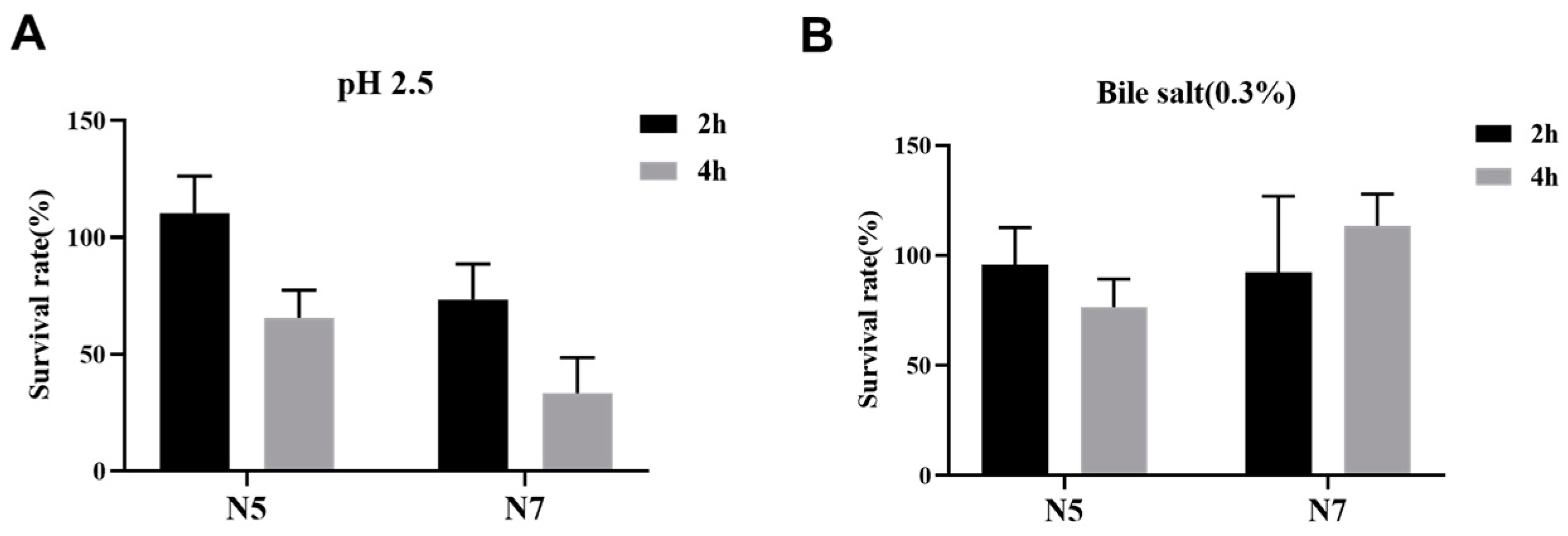
2.4. Bile and Acidic Tolerance
2.5. Analytical Profile Index (API) Experiment
2.6. Whole Genome Sequencing and Genome Assembly
2.7. Comparative Genomics Analysis
3. Results and Discussion
3.1. Probiotic Properties of L. johnsonii Strains Isolated from Heat-Resistant Weaned Piglets
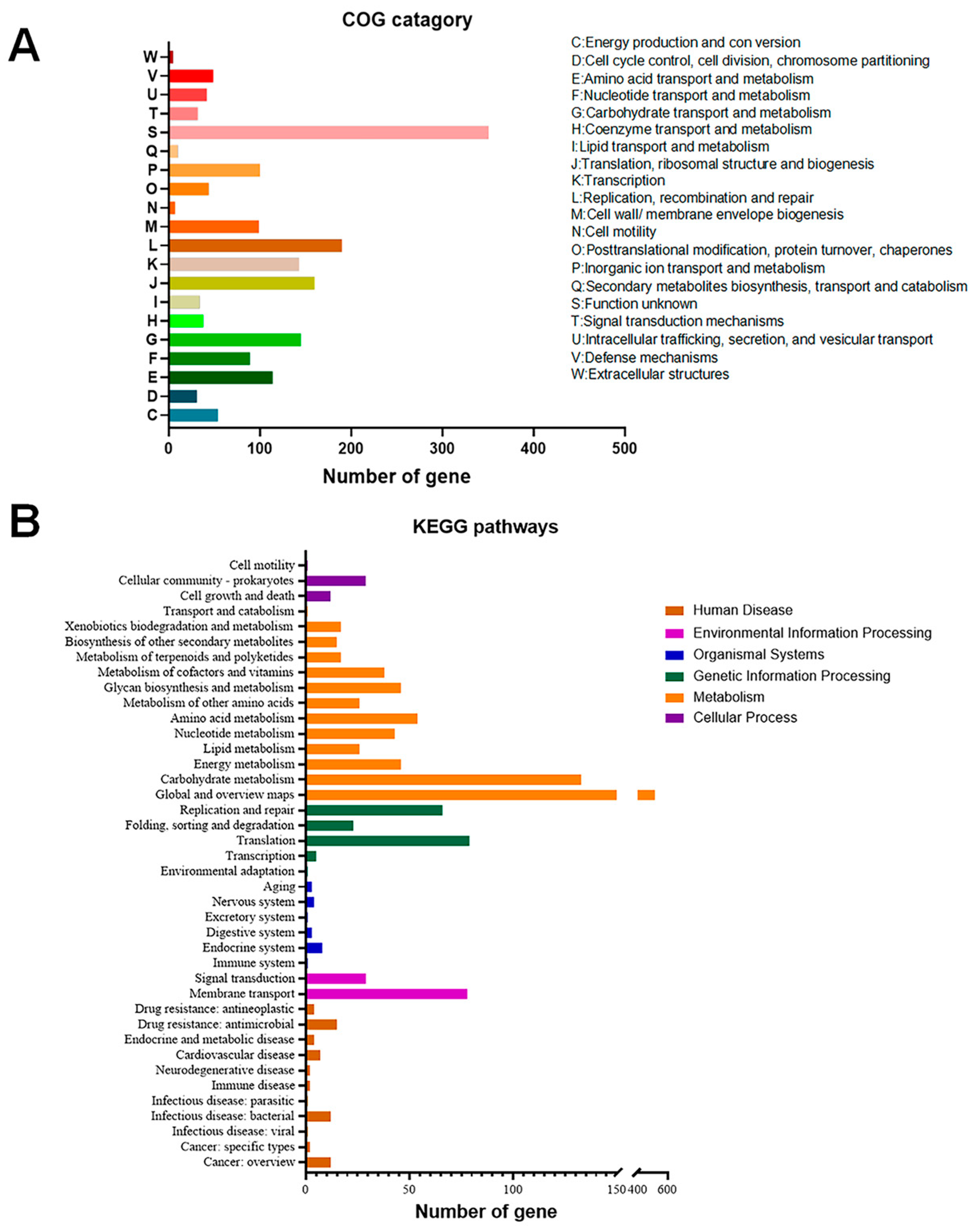
3.2. General Genomic Features of the L. johnsonii N5 and N7 Strains
3.3. Stress Resistance-Associated Genes
3.4. Anti-Inflammation-Associated Genes
3.5. CAZymes
3.6. Biosynthesis and Transport System
3.7. Genome Comparison of Strains N5 and N7 and Related Complete L. johnsonii Genomes
3.8. Unique Genomic Characteristics of L. johnsonii N5 and N7
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; de Melo Franco, B.D. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef]
- Camargo, A.C.; Todorov, S.D.; Chihib, N.E.; Drider, D.; Nero, L.A. Lactic Acid Bacteria (LAB) and Their Bacteriocins as Alternative Biotechnological Tools to Control Listeria monocytogenes Biofilms in Food Processing Facilities. Mol. Biotechnol. 2018, 60, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Goto, R.; Fujimoto, M.; Okada, N.; Hardt, W.-D. The Bactericidal Lectin RegIIIβ Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell Host Microbe 2017, 21, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Rintahaka, J.; Yu, X.; Sigvart-Mattila, P.; Paulin, L.; Mecklin, J.-P.; Saarela, M.; Palva, A.; von Ossowski, I. A Comparative Pan-Genome Perspective of Niche-Adaptable Cell-Surface Protein Phenotypes in Lactobacillus rhamnosus. PLoS ONE 2014, 9, e102762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, J.; Zhang, D.; Liu, H.; Wang, S.; Wang, Y.; Ji, H. Complete Genome Sequencing and Comparative Genome Characterization of Lactobacillus johnsonii ZLJ010, a Potential Probiotic with Health-Promoting Properties. Front. Genet. 2019, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Masuoka, H.; Shimada, K.; Kiyosue-Yasuda, T.; Kiyosue, M.; Oishi, Y.; Kimura, S.; Yamada, A.; Hirayama, K. Transition of the intestinal microbiota of dogs with age. Biosci. Microbiota Food Health 2017, 36, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Miller, E.A.; Weber, B.; Figueroa, C.F.; Aguayo, J.M.; Johny, A.K.; Noll, S.; Brannon, J.; Kozlowicz, B.; Johnson, T.J. Evidence of host specificity in Lactobacillus johnsonii genomes and its influence on probiotic potential in poultry. Poult. Sci. 2023, 102, 102858. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Nie, C.; Wu, Y.; Luo, R.; Chen, C.; Niu, J.; Zhang, W. Effects of two strains of Lactobacillus isolated from the feces of calves after fecal microbiota transplantation on growth performance, immune capacity, and intestinal barrier function of weaned calves. Front. Microbiol. 2023, 14, 1249628. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Cho, M.-J.; Cho, S.; Lee, Y.; Byun, S.J.; Lee, S. Complete Genome Sequence of Lactobacillus johnsonii Strain Byun-jo-01, Isolated from the Murine Gastrointestinal Tract. Microbiol. Resour. Announc. 2018, 7, e00985-18. [Google Scholar] [CrossRef]
- Audisio, M.C.; Benítez-Ahrendts, M.R. Lactobacillus johnsonii CRL1647, isolated from Apis mellifera L. bee-gut, exhibited a beneficial effect on honeybee colonies. Benefic. Microbes 2011, 2, 29–34. [Google Scholar] [CrossRef]
- Zhang, X.; Mushajiang, S.; Luo, B.; Tian, F.; Ni, Y.; Yan, W. The Composition and Concordance of Lactobacillus Populations of Infant Gut and the Corresponding Breast-Milk and Maternal Gut. Front. Microbiol. 2020, 11, 597911. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; O’toole, P.W. The Genomic Basis of Lactobacilli as Health-Promoting Organisms. Microbiol. Spectr. 2017, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 15: Suitability of taxonomic units notified to EFSA until September 2021. EFSA J. 2022, 20, e07045. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ni, X.; Qing, X.; Zeng, D.; Luo, M.; Liu, L.; Li, G.; Pan, K.; Jing, B. Live Probiotic Lactobacillus johnsonii BS15 Promotes Growth Performance and Lowers Fat Deposition by Improving Lipid Metabolism, Intestinal Development, and Gut Microflora in Broilers. Front. Microbiol. 2017, 8, 1073. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, K.; Liu, Y.; Li, Y.; He, F.; Yin, J.; Tang, W. Lactobacillus johnsonii Improves Intestinal Barrier Function and Reduces Post-Weaning Diarrhea in Piglets: Involvement of the Endocannabinoid System. Animals 2024, 14, 493. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Albertsen, M.; Anslan, S.; Callahan, B. Perspectives and Benefits of High-Throughput Long-Read Sequencing in Microbial Ecology. Appl. Environ. Microbiol. 2021, 87, e00626-21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Chen, H.; Yang, F.; Xu, B.; Wang, K.; Liu, Q.; Liang, G.; Zhang, R.; Jiao, X.; et al. Assessment of beneficial effects and identification of host adaptation-associated genes of Ligilactobacillus salivarius isolated from badgers. BMC Genom. 2023, 24, 530. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, R.M.; Kim, S.H.; Vasquez, R.; Hwang, I.-C.; Park, Y.-S.; Paik, H.-D.; Moon, G.-S.; Kang, D.-K. Bioinformatics and its role in the study of the evolution and probiotic potential of lactic acid bacteria. Food Sci. Biotechnol. 2023, 32, 389–412. [Google Scholar] [CrossRef]
- Yuan, L.; Zhu, C.; Gu, F.; Zhu, M.; Yao, J.; Zhu, C.; Li, S.; Wang, K.; Hu, P.; Zhang, Y.; et al. Lactobacillus johnsonii N5 from heat stress-resistant pigs improves gut mucosal immunity and barrier in dextran sodium sulfate-induced colitis. Anim. Nutr. 2023, 15, 210–224. [Google Scholar] [CrossRef]
- Leite, A.M.; Miguel, M.A.; Peixoto, R.S.; Ruas-Madiedo, P.; Paschoalin, V.M.; Mayo, B.; Delgado, S. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J. Dairy Sci. 2015, 98, 3622–3632. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme an-notation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Liu, J.; Hu, D.; Chen, Y.; Huang, H.; Zhang, H.; Zhao, J.; Gu, Z.; Chen, W. Strain-specific properties of Lactobacillus plantarum for prevention of Salmonella infection. Food Funct. 2018, 9, 3673–3682. [Google Scholar] [CrossRef]
- Neidig, A.; Yeung, A.T.; Rosay, T.; Tettmann, B.; Strempel, N.; Rueger, M.; Lesouhaitier, O.; Overhage, J. TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 2013, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Cheng, J.; Meng, X.; Xu, Y.; Mu, Y. Complete Genome and Comparative Genome Analysis of Lactobacillus reuteri YSJL-12, a Potential Probiotics Strain Isolated from Healthy Sow Fresh Feces. Evol. Bioinform. 2020, 16, 1176934320942192. [Google Scholar] [CrossRef]
- O’Sullivan, O.; O’Callaghan, J.; Sangrador-Vegas, A.; McAuliffe, O.; Slattery, L.; Kaleta, P.; Callanan, M.; Fitzgerald, G.F.; Ross, R.P.; Beresford, T. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 2009, 9, 50. [Google Scholar] [CrossRef]
- Dertli, E.; Mayer, M.J.; Narbad, A. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol. 2015, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Werning, M.L.; Hernández-Alcántara, A.M.; Ruiz, M.J.; Soto, L.P.; Dueñas, M.T.; López, P.; Frizzo, L.S. Biological Functions of Exopolysaccharides from Lactic Acid Bacteria and Their Potential Benefits for Humans and Farmed Animals. Foods 2022, 11, 1284. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; Gueimonde, M.; Margolles, A.; de los Reyes-Gavilán, C.G.; Salminen, S. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J. Food Prot. 2006, 69, 2011–2015. [Google Scholar] [CrossRef]
- Dertli, E.; Colquhoun, I.J.; Gunning, A.P.; Bongaerts, R.J.; Le Gall, G.; Bonev, B.B.; Mayer, M.J.; Narbad, A. Structure and Biosynthesis of Two Exopolysaccharides Produced by Lactobacillus johnsonii FI9785. J. Biol. Chem. 2013, 288, 31938–31951. [Google Scholar] [CrossRef] [PubMed]
- Boucard, A.-S.; Florent, I.; Polack, B.; Langella, P.; Bermúdez-Humarán, L.G. Genome Sequence and Assessment of Safety and Potential Probiotic Traits of Lactobacillus johnsonii CNCM I-4884. Microorganisms 2022, 10, 273. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Chen, L.; Gu, Q.; Li, P.; Chen, S.; Li, Y. Genomic analysis of Lactobacillus reuteri WHH1689 reveals its probiotic properties and stress resistance. Food Sci. Nutr. 2019, 7, 844–857. [Google Scholar] [CrossRef]
- Serrano, L.M.; Molenaar, D.; Wels, M.; Teusink, B.; A Bron, P.; de Vos, W.M.; Smid, E.J. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell Factories 2007, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Aoudia, N.; Rieu, A.; Briandet, R.; Deschamps, J.; Chluba, J.; Jego, G.; Garrido, C.; Guzzo, J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Rieu, A.; Aoudia, N.; Jego, G.; Chluba, J.; Yousfi, N.; Briandet, R.; Deschamps, J.; Gasquet, B.; Monedero, V.; Garrido, C.; et al. The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell. Microbiol. 2014, 16, 1836–1853. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Hong, T.; Van Pijkeren, J.P.; Hemarajata, P.; Trinh, D.V.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine Derived from Probiotic Lactobacillus reuteri Suppresses TNF via Modulation of PKA and ERK Signaling. PLoS ONE 2012, 7, e31951. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Feng, S.; Yu, L.; Zhao, J.; Tian, F.; Chen, W.; Zhai, Q. Capsular polysaccarides of probiotics and their immunomodulatory roles. Food Sci. Hum. Wellness 2022, 11, 1111–1120. [Google Scholar] [CrossRef]
- Yasuda, E.; Serata, M.; Sako, T. Suppressive Effect on Activation of Macrophages by Lactobacillus casei Strain Shirota Genes Determining the Synthesis of Cell Wall-Associated Polysaccharides. Appl. Environ. Microbiol. 2008, 74, 4746–4755. [Google Scholar] [CrossRef] [PubMed]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef] [PubMed]
- Costa, O.Y.A.; de Hollander, M.; Pijl, A.; Liu, B.; Kuramae, E.E. Cultivation-independent and cultivation-dependent metagenomes reveal genetic and enzymatic potential of microbial community involved in the degradation of a complex microbial polymer. Microbiome 2020, 8, 76. [Google Scholar] [CrossRef]
- Ardèvol, A.; Rovira, C. Reaction Mechanisms in Carbohydrate-Active Enzymes: Glycoside Hydrolases and Glycosyltransferases. Insights from ab Initio Quantum Mechanics/Molecular Mechanics Dynamic Simulations. J. Am. Chem. Soc. 2015, 137, 7528–7547. [Google Scholar] [CrossRef]
- Kumar, S.; Bansal, K.; Sethi, S.K.; Bindhani, B.K. Phylo-taxonogenomics of 182 strains of genus Leuconostoc elucidates its robust taxonomy and biotechnological importance. BioRxiv 2021. [Google Scholar] [CrossRef]
- Mao, B.; Yin, R.; Li, X.; Cui, S.; Zhang, H.; Zhao, J.; Chen, W. Comparative Genomic Analysis of Lactiplantibacillus plantarum Isolated from Different Niches. Genes 2021, 12, 241. [Google Scholar] [CrossRef] [PubMed]
- Pridmore, R.D.; Berger, B.; Desiere, F.; Vilanova, D.; Barretto, C.; Pittet, A.-C.; Zwahlen, M.-C.; Rouvet, M.; Altermann, E.; Barrangou, R.; et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 2004, 101, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J.; Francke, C.; Postma, P.W. How Phosphotransferase System-Related Protein Phosphorylation Regulates Carbohydrate Metabolism in Bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. [Google Scholar] [CrossRef]
- Liu, C.-J.; Wang, R.; Gong, F.-M.; Liu, X.-F.; Zheng, H.-J.; Luo, Y.-Y.; Li, X.-R. Complete genome sequences and comparative genome analysis of Lactobacillus plantarum strain 5-2 isolated from fermented soybean. Genomics 2015, 106, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, X.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Comparative Genomics and Specific Functional Characteristics Analysis of Lactobacillus acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef]
- Guinane, C.M.; Kent, R.M.; Norberg, S.; Hill, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Host Specific Diversity in Lactobacillus johnsonii as Evidenced by a Major Chromosomal Inversion and Phage Resistance Mechanisms. PLoS ONE 2011, 6, e18740. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ventura, M.; Canchaya, C.; Pridmore, R.D.; Brüssow, H. The prophages of Lactobacillus johnsonii NCC 533: Comparative genomics and transcription analysis. Virology 2004, 320, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Grover, S.; Kaushik, J.K.; Batish, V.K. IS30-related transposon mediated insertional inactivation of bile salt hydrolase (bsh1) gene of Lactobacillus plantarum strain Lp20. Microbiol. Res. 2014, 169, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.H.; Wray, L.V., Jr. Bacillus subtilis 168 contains two differentially regulated genes encoding L-asparaginase. J. Bacteriol. 2002, 184, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, P.I.; Shulenina, O.; Evfratov, S.; Treshin, V.; Subach, M.F.; Serebryakova, M.V.; Osterman, I.A.; Paleskava, A.; Bogdanov, A.A.; Dontsova, O.A.; et al. Ribosomal protein S18 acetyltransferase RimI is responsible for the acetylation of elongation factor Tu. J. Biol. Chem. 2022, 298, 101914. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y.; Li, L.; Li, Y.; Yuan, L.; E, Y.; Qiao, J. Positive regulation of the DLT operon by TCSR7 enhances acid tolerance of Lactococcus lactis F44. J. Dairy Sci. 2022, 105, 7940–7950. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, B.; Ross, P.; Stanton, C.; Zhang, H.; Zhao, J.; Chen, W. Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches. Genes 2020, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.J.; D’amato, A.; Colquhoun, I.J.; Le Gall, G.; Narbad, A. Identification of Genes Required for Glucan Exopolysaccharide Production in Lactobacillus johnsonii Suggests a Novel Biosynthesis Mechanism. Appl. Environ. Microbiol. 2020, 86, e02808-19. [Google Scholar] [CrossRef]
- Altermann, E.; Klaenhammer, T.R. Group-specific comparison of four lactobacilli isolated from human sources using differential blast analysis. Genes Nutr. 2011, 6, 319–340. [Google Scholar] [CrossRef]




| Strain | RefSeq Assembly Accession | GC% | Size (Mb) | CDS | Plasmid | Isolation Source | Host | Country |
|---|---|---|---|---|---|---|---|---|
| N5 (this study) | GCF_032463685.1 | 35 | 1.88 | 1863 | 0 | Feces | Swine | China |
| N7 (this study) | GCF_032463545.1 | 35 | 1.88 | 1870 | 0 | Feces | Swine | China |
| GHZ10a | GCF_014841035.1 | 34.9 | 2.01 | 1921 | 2 | Feces | Swine | China |
| 7409N31 | GCF_022810665.1 | 35 | 2.19 | 2146 | 0 | Feces | Bovine | NA |
| BS15 | GCF_001714745.1 | 34.9 | 2.16 | 2076 | 1 | Yoghurt | - | China |
| ZLJ010 | GCF_004011315.1 | 34.9 | 2.00 | 1959 | 0 | Intestine | Swine | China |
| UMNLJ22 | GCF_002176835.1 | 34.6 | 1.99 | 1922 | 2 | Ileum | Poultry | USA |
| UMNLJ21 | GCF_002176855.1 | 35.12 | 2.01 | 1888 | 2 | Ileum | Poultry | USA |
| Byun-jo-01 | GCF_003316915.1 | 34.7 | 1.96 | 1781 | 0 | Jejunum | Mouse | South Korea |
| MR1 | GCF_022213385.1 | 34.7 | 1.95 | 1805 | 0 | Mice cecum | Mouse | USA |
| NCK2677 | GCF_014058685.1 | 34.7 | 1.95 | 1792 | 0 | Mouse | Mouse | USA |
| G2A | GCF_010586925.1 | 34.6 | 2.17 | 2045 | 2 | Mouse | Mouse | USA |
| DC22.2 | GCF_009769185.1 | 34.5 | 1.94 | 1858 | 4 | Birds | Junglefowl | UK |
| IDCC9203 | GCF_003428395.1 | 34.7 | 1.90 | 1882 | 0 | Infant feces | Humans | South Korea |
| NCC 533 | GCF_000008065.1 | 34.6 | 1.99 | 1821 | 0 | Humans | Humans | NA |
| DPC 6026 | GCF_000204985.1 | 34.8 | 1.97 | 1900 | 0 | Small intestine | Swine | NA |
| N6.2 | GCF_000498675.1 | 34.2 | 1.89 | 1718 | 0 | Rat | Rat | NA |
| FI9785 | GCF_000091405.1 | 34.5 | 1.76 | 1765 | 2 | Poultry | Poultry | UK |
| pf01 | GCA_000219475.3 | 34.6 | 1.93 | 1846 | 2 | Piglet feces | Swine | Korea |
| Substrate | Result | Substrate | Result | ||
|---|---|---|---|---|---|
| N5 | N7 | N5 | N7 | ||
| Control | − | − | Esculine | − | − |
| Glycerol | − | − | Salicine | + | + |
| Erythritol | − | − | D-cellobiose | + | + |
| D-arabinose | − | − | D-maltose | + | + |
| L-arabinose | − | − | D-lactose | − | − |
| D-ribose | − | − | D-melibiose | + | + |
| D-xylose | − | − | D-saccharose | + | + |
| L-xylose | − | − | D-threalose | − | − |
| D-adonitol | − | − | Inulin | − | − |
| Methyl-β-dxylopyranoside | + | + | D-melezitose | + | + |
| D-galactose | + | + | D-raffinose | − | − |
| D-glucose | + | + | Starch | − | − |
| D-fructose | − | − | Glycogene | − | − |
| D-mannose | − | − | Xylitol | + | + |
| L-sorbose | − | − | Gentiobiose | − | − |
| L-rhamnose | − | − | D-turanose | − | − |
| Dulcitol | − | − | D-lyxose | − | − |
| Inositol | − | − | D-tagatose | − | − |
| D-mannitol | − | − | D-fucose | − | − |
| D-sorbitol | − | − | L-fucose | − | − |
| Methyl-αD-mannopyranoside | − | − | D-arabitol | − | − |
| Methyl-αD-glucopyranoside | + | + | L-arabitol | − | − |
| N-acetylglucosamine | − | − | Potassium gluconate | − | − |
| Amygdaline | − | − | Potassium 2-cetogluconate | − | − |
| Arbutine | + | + | Potassium 5-cetogluconate | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Wang, Y.; Gu, L.; Yu, J.; Liu, Q.; Zhang, R.; Liang, G.; Chen, H.; Gu, F.; Liu, H.; et al. Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine. Microorganisms 2024, 12, 672. https://doi.org/10.3390/microorganisms12040672
Wang K, Wang Y, Gu L, Yu J, Liu Q, Zhang R, Liang G, Chen H, Gu F, Liu H, et al. Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine. Microorganisms. 2024; 12(4):672. https://doi.org/10.3390/microorganisms12040672
Chicago/Turabian StyleWang, Kun, Yu Wang, Lifang Gu, Jinyan Yu, Qianwen Liu, Ruiqi Zhang, Guixin Liang, Huan Chen, Fang Gu, Haoyu Liu, and et al. 2024. "Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine" Microorganisms 12, no. 4: 672. https://doi.org/10.3390/microorganisms12040672
APA StyleWang, K., Wang, Y., Gu, L., Yu, J., Liu, Q., Zhang, R., Liang, G., Chen, H., Gu, F., Liu, H., Jiao, X., & Zhang, Y. (2024). Characterization of Probiotic Properties and Whole-Genome Analysis of Lactobacillus johnsonii N5 and N7 Isolated from Swine. Microorganisms, 12(4), 672. https://doi.org/10.3390/microorganisms12040672






