SARS-CoV-2 Infection and Anemia—A Focus on RBC Deformability and Membrane Proteomics—Integrated Observational Prospective Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Enrollment
2.2. Hematological and Biochemical Parameters
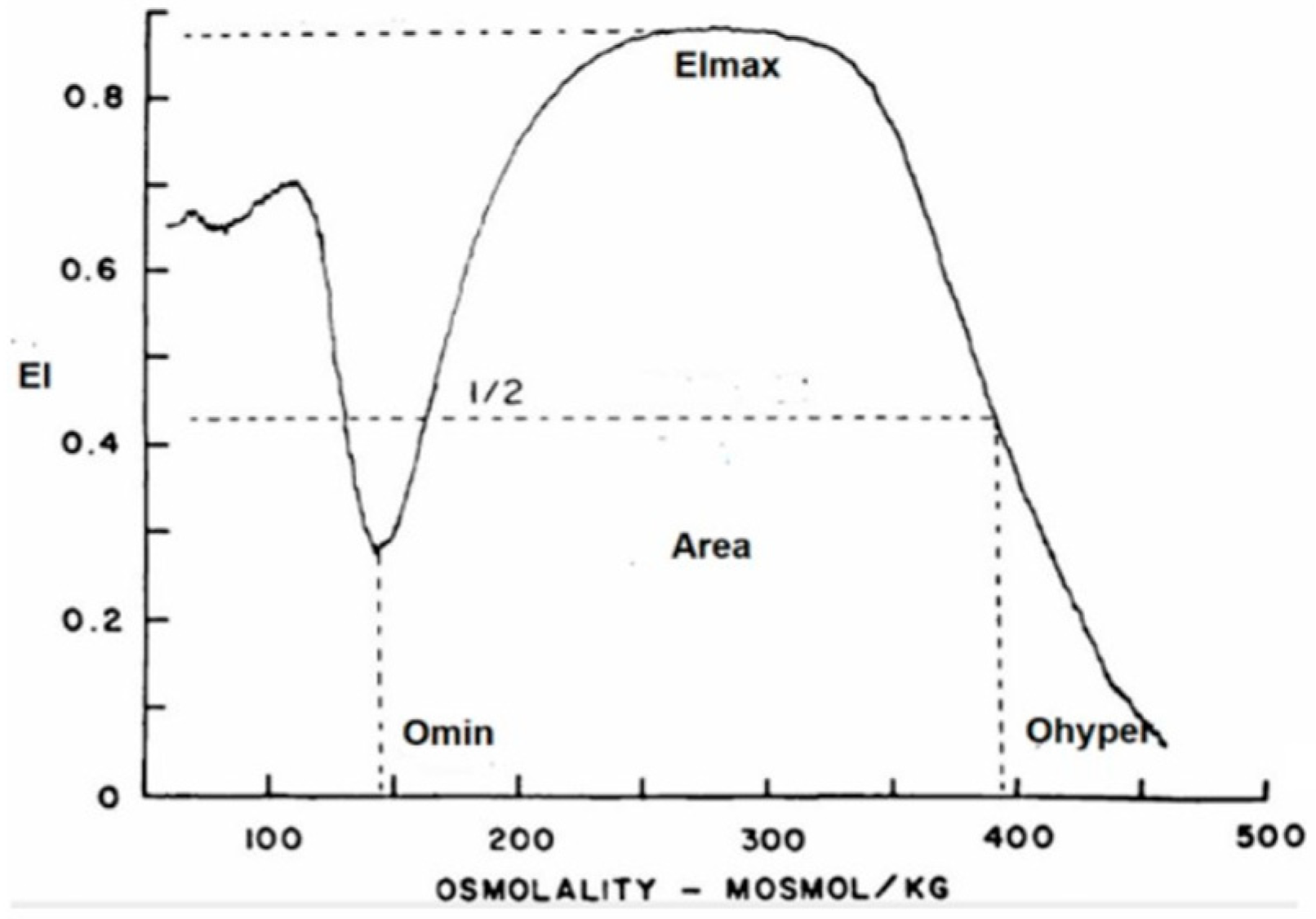
2.3. RBC Deformability
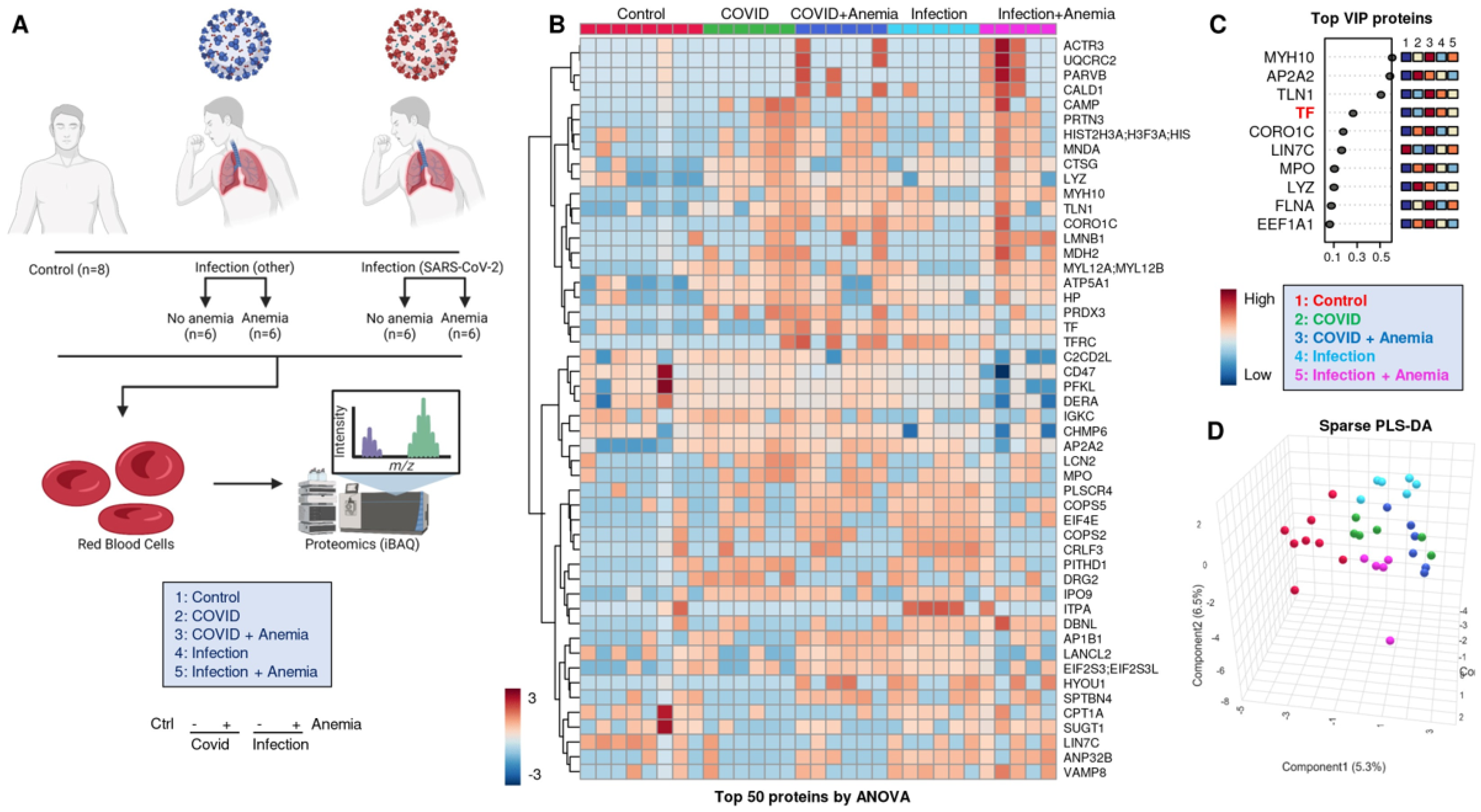
2.4. RBC Membrane Proteins (Proteomics)
3. Results
3.1. General Hematological Data, Iron Metabolism and Inflammation Parameters
3.2. Hemoglobin Stability
3.3. RBC Enzyme Activities
3.4. RBC Deformability
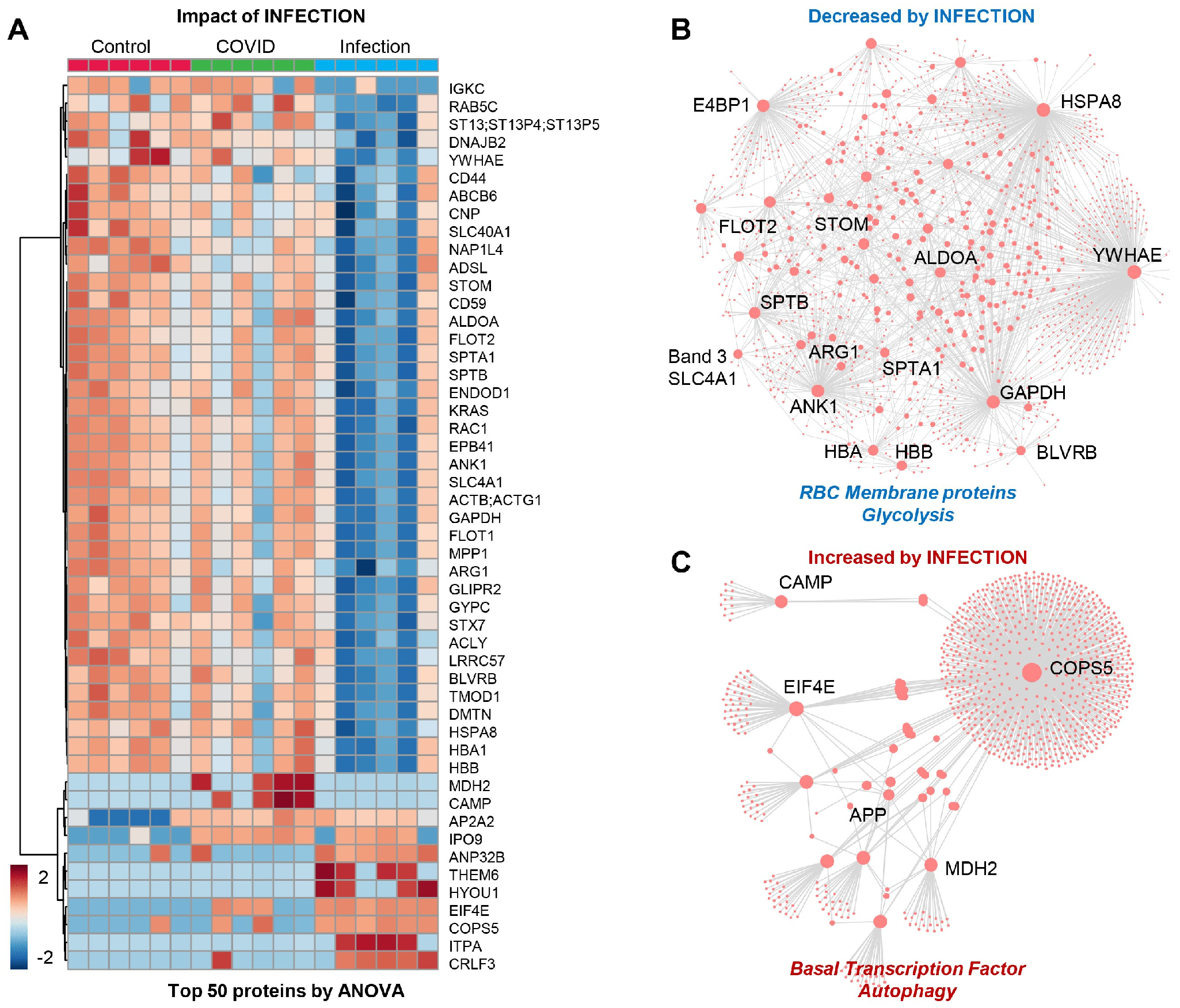
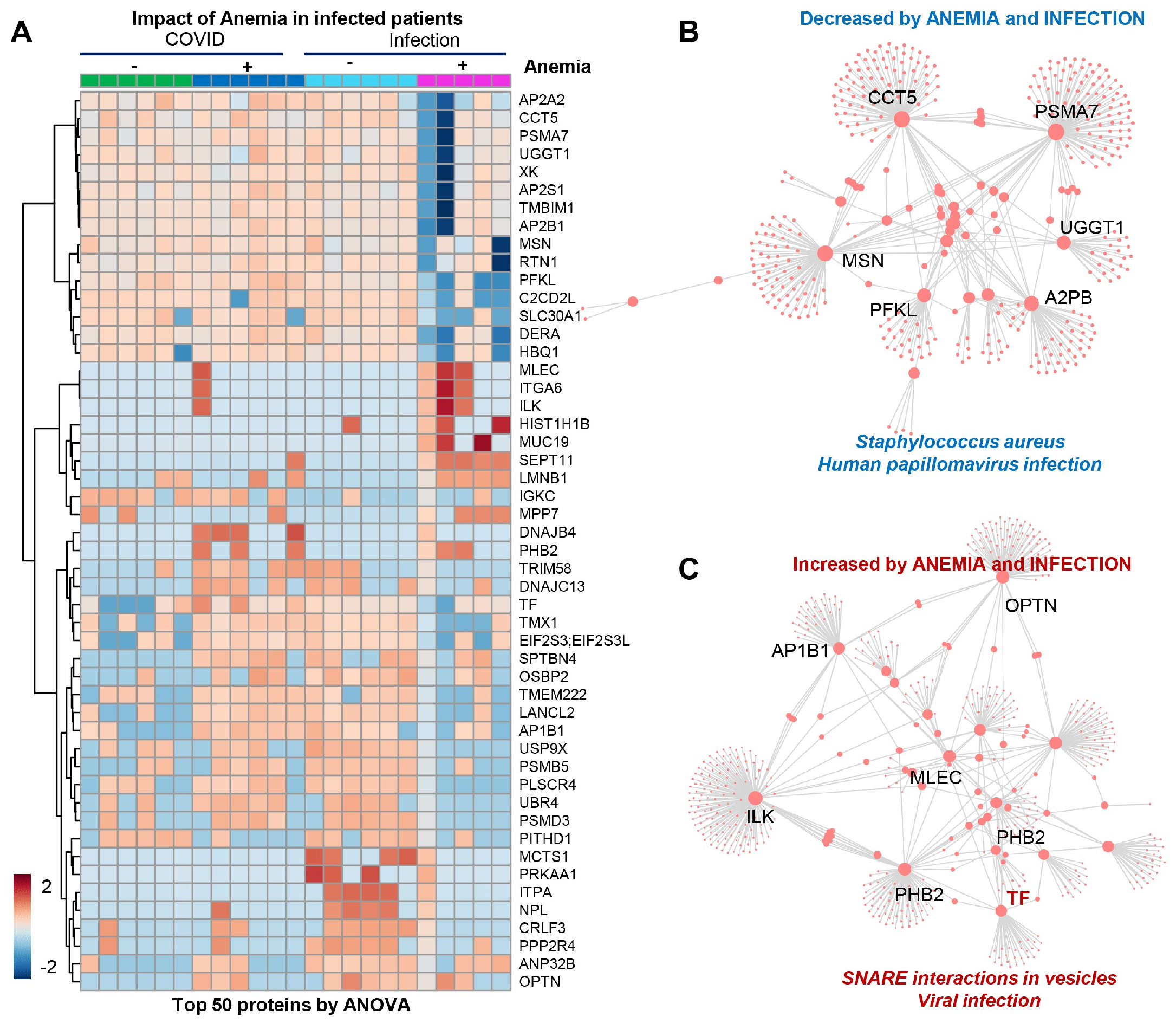
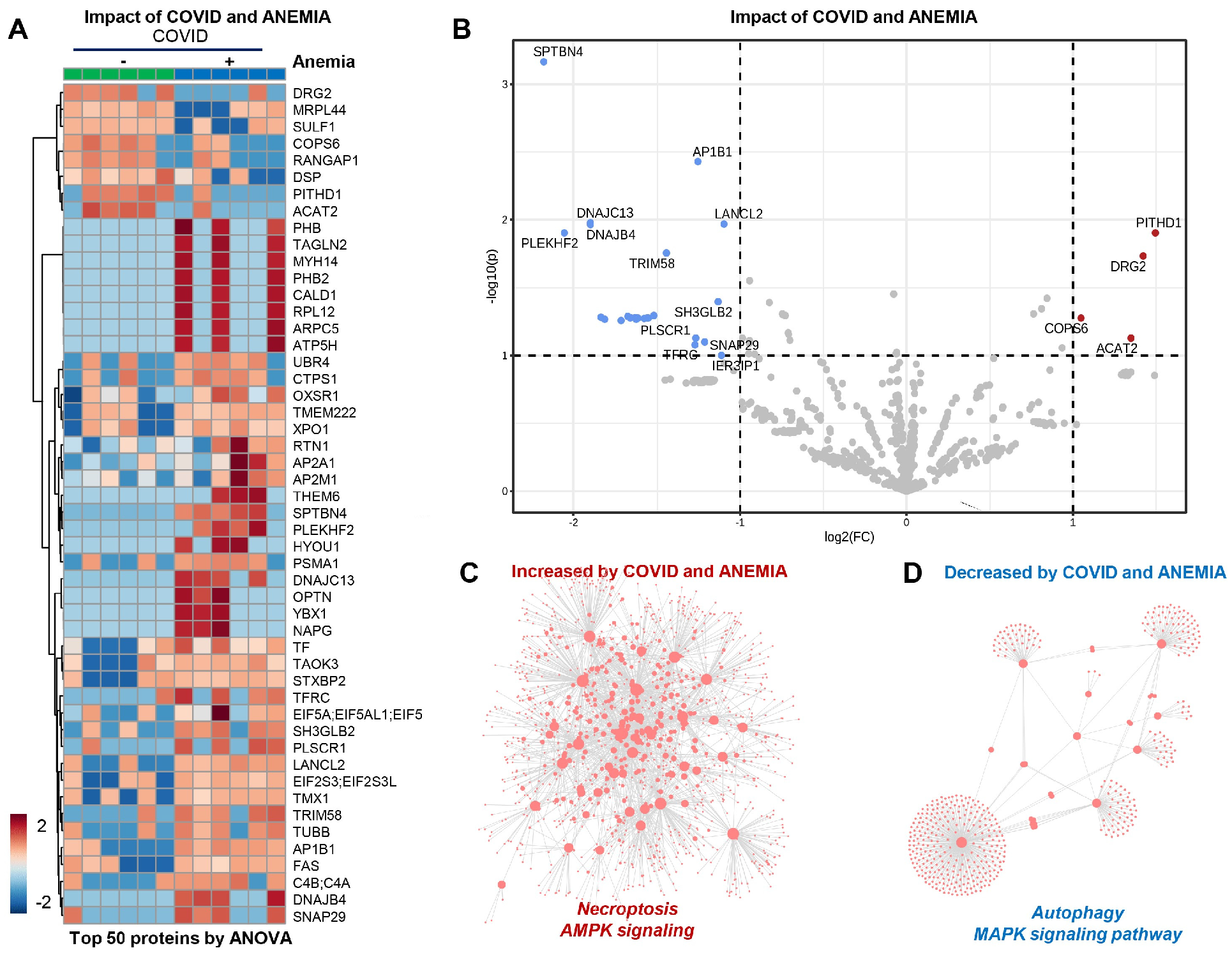
3.5. RBC Membrane Proteins (Proteomics)
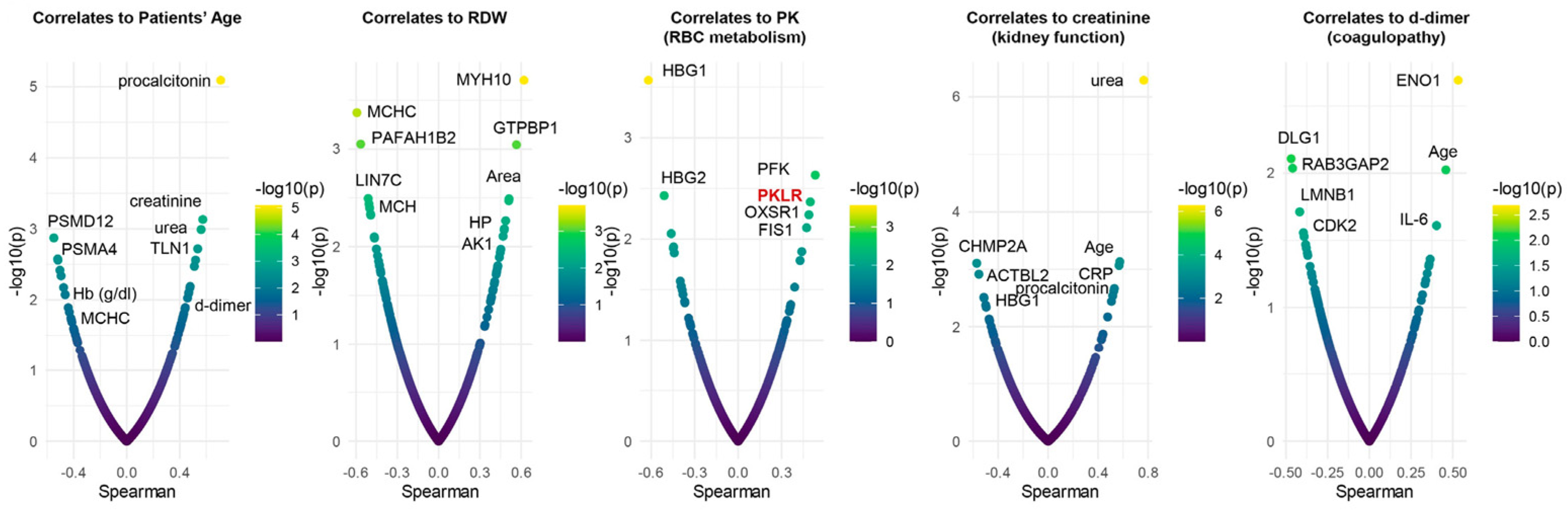
3.6. Omics Correlates to Clinical, Hematological, and Deformability Parameters
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, T.; Stefanoni, D.; Dzieciatkowska, M.; Issaian, A.; Nemkov, T.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Buehler, P.W.; Zimring, J.C.; et al. Evidence of Structural Protein Damage and Membrane Lipid Remodeling in Red Blood Cells from COVID-19 Patients. J. Proteome Res. 2020, 19, 4455–4469. [Google Scholar] [CrossRef] [PubMed]
- Rinalducci, S.; Ferru, E.; Blasi, B.; Turrini, F.; Zolla, L. Oxidative stress and caspase-mediated fragmentation of cytoplasmic domain of erythrocyte band 3 during blood storage. Blood Transfus. 2012, 10 (Suppl. S2), S55–S62. [Google Scholar]
- Issaian, A.; Hay, A.; Dzieciatkowska, M.; Roberti, D.; Perrotta, S.; Darula, Z.; Redzic, J.; Busch, M.P.; Page, G.P.; Rogers, S.C.; et al. The interactome of the N-terminus of band 3 regulates red blood cell metabolism and storage quality. Haematologica 2021, 106, 2971–2985. [Google Scholar] [CrossRef] [PubMed]
- Recktenwald, S.M.; Simionato, G.; Lopes, M.G.M.; Gamboni, F.; Dzieciatkowska, M.; Meybohm, P.; Zacharowski, K.; von Knethen, A.; Wagner, C.; Kaestner, L.; et al. Cross-talk between red blood cells and plasma influences blood flow and omics phenotypes in severe COVID-19. eLife 2022, 11, e81316. [Google Scholar] [CrossRef]
- Marchi, G.; Bozzini, C.; Bertolone, L.; Dima, F.; Busti, F.; Castagna, A.; Stranieri, C.; Fratta Pasini, A.M.; Friso, S.; Lippi, G.; et al. Red Blood Cell Morphologic Abnormalities in Patients Hospitalized for COVID-19. Front. Physiol. 2022, 13, 932013. [Google Scholar] [CrossRef]
- Ramachandran, P.; Gajendran, M.; Perisetti, A.; Elkholy, K.O.; Chakraborti, A.; Lippi, G.; Goyal, H. Red Blood Cell Distribution Width in Hospitalized COVID-19 Patients. Front. Med. 2021, 8, 582403. [Google Scholar] [CrossRef] [PubMed]
- Pouladzadeh, M.; Safdarian, M.; Choghakabodi, P.M.; Amini, F.; Sokooti, A. Validation of red cell distribution width as a COVID-19 severity screening tool. Future Sci. OA 2021, 7, FSO712. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Thomas, T.; Akpan, I.J.; Reisz, J.A.; Cendali, F.I.; Gamboni, F.; Nemkov, T.; Thangaraju, K.; Katneni, U.; Tanaka, K.; et al. Biological and Clinical Factors Contributing to the Metabolic Heterogeneity of Hospitalized Patients with and without COVID-19. Cells 2021, 10, 2293. [Google Scholar] [CrossRef]
- Bergamaschi, G.; Borrelli de Andreis, F.; Aronico, N.; Lenti, M.V.; Barteselli, C.; Merli, S.; Pellegrino, I.; Coppola, L.; Cremonte, E.M.; Croce, G.; et al. Anemia in patients with COVID-19: Pathogenesis and clinical significance. Clin. Exp. Med. 2021, 21, 239–246. [Google Scholar] [CrossRef]
- Lippi, G.; Mattiuzzi, C. Hemoglobin value may be decreased in patients with severe coronavirus disease 2019. Hematol. Transfus. Cell Ther. 2020, 42, 116–117. [Google Scholar] [CrossRef]
- McCullough, J. RBCs as targets of infection. Am. Soc. Hematol. Educ. Program 2014, 2014, 404–409. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Domizio, J.; Di Gulen, M.F.; Saidoune, F.; Thacker, V.V.; Yatim, A.; Sharma, K.; Nass, T.; Guenova, E.; Schaller, M.; Conrad, C.; et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature 2022, 603, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, M.D.; Kinning, K.T.; Sullivan, K.D.; Araya, P.; Smith, K.P.; Granrath, R.E.; Shaw, J.R.; Baxter, R.; Jordan, K.R.; Russell, S.; et al. Specialized interferon action in COVID-19. Proc. Natl. Acad. Sci. USA 2022, 119, e2116730119. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Wei, W.-Y.; Li, H.-C.; Chen, C.-Y.; Yang, C.-H.; Lee, S.-K.; Wang, C.-W.; Ma, H.C.; Juang, Y.L.; Lo, S.Y. SARS-CoV nucleocapsid protein interacts with cellular pyruvate kinase protein and inhibits its activity. Arch. Virol. 2012, 157, 635–645. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. ECDC Assessment of the COVID-19 Situation in Europe as of 2 March 2020. Available online: https://www.ecdc.europa.eu/en/news-events/ecdc-assessment-covid-19-situationeurope-2-march-2020 (accessed on 28 January 2024).
- WHO-China. Joint Mission on Coronavirus Disease 2019 (COVID-19); WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Vives-Corrons, J.L.; Aguilar Bascompte, J.L. Manual of Hematology Laboratory Diagnostic Techniques, 4th ed.; Elssevier-Masson: Paris, France, 2014. (In Spanish) [Google Scholar]
- Weissman, S.M. Red Cell Metabolism. A Manual of Biochemical Methods. 2nd Edition. Yale J. Biol. Med. 1976, 49, 310–311. [Google Scholar]
- Vives-Corrons, J.-L.; Krishnevskaya, E.; Rodriguez, I.H.; Ancochea, A. Characterization of hereditary red blood cell membranopathies using combined targeted next-generation sequencing and osmotic gradient ektacytometry. Int. J. Hematol. 2021, 113, 163–174. [Google Scholar] [CrossRef]
- Pesciotta, E.N.; Lam, H.S.; Kossenkov, A.; Ge, J.; Showe, L.C.; Mason, P.J.; Bessler, M.; Speicher, D.W. In-Depth, Label-Free Analysis of the Erythrocyte Cytoplasmic Proteome in Diamond Blackfan Anemia Identifies a Unique Inflammatory Signature. PLoS ONE 2015, 10, e0140036. [Google Scholar] [CrossRef]
- Hayden, S.J.; Albert, T.J.; Watkins, T.R.; Swenson, E.R. Anemia in critical illness: Insights into etiology, consequences, and management. Am. J. Respir. Crit. Care Med. 2012, 185, 1049–1057. [Google Scholar] [CrossRef]
- Oh, S.M.; Skendelas, J.P.; Macdonald, E.; Bergamini, M.; Goel, S.; Choi, J.; Segal, K.R.; Vivek, K.; Nair, S.; Leff, J. On-admission anemia predicts mortality in COVID-19 patients: A single center, retrospective cohort study. Am. J. Emerg. Med. 2021, 48, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Mourad, S.; Rajab, M.; Alameddine, A.; Fares, M.; Ziade, F.; Merhi, B.A. Hemoglobin level as a risk factor for lower respiratory tract infections in Lebanese children. N. Am. J. Med. Sci. 2010, 2, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Taneri, P.E.; Gómez-Ochoa, S.A.; Llanaj, E.; Raguindin, P.F.; Rojas, L.Z.; Roa-Díaz, Z.M.; Salvador, D.; Groothof, D.; Minder, B.; Kopp-Heim, D.; et al. Anemia and iron metabolism in COVID-19: A systematic review and meta-analysis. Eur. J. Epidemiol. 2020, 35, 763–773. [Google Scholar] [CrossRef]
- Fan, B.E.; Chong, V.C.L.; Chan, S.S.W.; Lim, G.H.; Lim, K.G.E.; Tan, G.B.; Mucheli, S.S.; Kuperan, P.; Ong, K.H. Hematologic parameters in patients with COVID-19 infection. Am. J. Hematol. 2020, 95, E131–E134. [Google Scholar] [PubMed]
- Bellmann-Weiler, R.; Lanser, L.; Barket, R.; Rangger, L.; Schapfl, A.; Schaber, M.; Fritsche, G.; Wöll, E.; Weiss, G. Prevalence and Predictive Value of Anemia and Dysregulated Iron Homeostasis in Patients with COVID-19 Infection. J. Clin. Med. 2020, 9, 2429. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, R.; He, G. Hematological findings in coronavirus disease 2019: Indications of progression of disease. Ann. Hematol. 2020, 99, 1421–1428. [Google Scholar] [CrossRef]
- Cavezzi, A.; Troiani, E.; Corrao, S. COVID-19: Hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020, 10, 1271. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Thomas, T.; Dzieciatkowska, M.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hod, E.A.; Spitalnik, S.L.; Hansen, K.C. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J. Proteome Res. 2020, 19, 4417–4427. [Google Scholar] [CrossRef]
- Elahi, S. Hematopoietic responses to SARS-CoV-2 infection. Cell. Mol. Life Sci. 2022, 79, 187. [Google Scholar] [CrossRef]
- Wang, X.; Wen, Y.; Xie, X.; Liu, Y.; Tan, X.; Cai, Q.; Zhang, Y.; Cheng, L.; Xu, G.; Zhang, S.; et al. Dysregulated hematopoiesis in bone marrow marks severe COVID-19. Cell Discov. 2021, 7, 60. [Google Scholar] [CrossRef]
- Kucia, M.; Ratajczak, J.; Bujko, K.; Adamiak, M.; Ciechanowicz, A.; Chumak, V.; Brzezniakiewicz-Janus, K.; Ratajczak, M.Z. An evidence that SARS-CoV-2/COVID-19 spike protein (SP) damages hematopoietic stem/progenitor cells in the mechanism of pyroptosis in Nlrp3 inflammasome-dependent manner. Leukemia 2021, 35, 3026–3029. [Google Scholar] [CrossRef]
- Camaschella, C.; Nai, A.; Silvestri, L. Iron metabolism and iron disorders revisited in the hepcidin era. Haematologica 2020, 105, 260–272. [Google Scholar] [CrossRef]
- Drakesmith, H.; Prentice, A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Bouchla, A.; Kriebardis, A.G.; Georgatzakou, H.T.; Fortis, S.P.; Thomopoulos, T.P.; Lekkakou, L.; Markakis, K.; Gkotzias, D.; Panagiotou, A.; Papageorgiou, E.G.; et al. Red Blood Cell Abnormalities as the Mirror of SARS-CoV-2 Disease Severity: A Pilot Study. Front. Physiol. 2021, 12, 825055. [Google Scholar] [CrossRef]
- Bissinger, R.; Nemkov, T.; D’Alessandro, A.; Grau, M.; Dietz, T.; Bohnert, B.N.; Essigke, D.; Wörn, M.; Schaefer, L.; Xiao, M.; et al. Proteinuric chronic kidney disease is associated with altered red blood cell lifespan, deformability and metabolism. Kidney Int. 2021, 100, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, C.; Zhang, Y.; Dzieciatkowska, M.; Brown, B.C.; Zhang, W.; Xie, T.; Abdulmalik, O.; Song, A.; Tong, C.; et al. Erythrocyte transglutaminase-2 combats hypoxia and chronic kidney disease by promoting oxygen delivery and carnitine homeostasis. Cell Metab. 2022, 34, 299–316.e6. [Google Scholar] [CrossRef] [PubMed]
- Aminianfar, M.; Soleiman-Meigooni, S.; Hamidi-Farahani, R.; Darvishi, M.; Hoseini-Shokouh, S.J.; Asgari, A.; Faraji-Hormozi, S.; Asli, M. Efficacy of Red Blood Cell Exchange as Adjunctive Treatment for Hypoxemia and Survival Rate of Patients with Severe Coronavirus-2 Disease: An Open-Labeled Phase 2 Randomized Clinical Trial. Front. Med. 2022, 9, 899593. [Google Scholar] [CrossRef]
- Park, K.C.; Donovan, K.; McKechnie, S.; Ramamurthy, N.; Klenerman, P.; Swietach, P. Single-cell oxygen saturation imaging shows that gas exchange by red blood cells is not impaired in COVID-19 patients. Br. J. Haematol. 2020, 190, e229–e232. [Google Scholar] [CrossRef]
- Zuin, M.; Rigatelli, G.; Quadretti, L.; Fogato, L.; Zuliani, G.; Roncon, L. Prognostic Role of Anemia in COVID-19 Patients: A Meta-Analysis. Infect. Dis. Rep. 2021, 13, 930–937. [Google Scholar] [CrossRef]
- Liu, W.; Li, H. COVID-19:Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Lechuga, G.C.; Souza-Silva, F.; Sacramento, C.Q.; Trugilho, M.R.O.; Valente, R.H.; Napoleão-Pêgo, P.; Dias, S.S.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Carels, N.; et al. SARS-CoV-2 Proteins Bind to Hemoglobin and Its Metabolites. Int. J. Mol. Sci. 2021, 22, 9035. [Google Scholar] [CrossRef] [PubMed]
- Vives-Corrons, J.-L. The Rare Anaemias. In Rare Diseases; Zhan, H.W., Ed.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar]
- Roy, M.K.; Cendali, F.; Ooyama, G.; Gamboni, F.; Morton, H.; D’Alessandro, A. Red Blood Cell Metabolism in Pyruvate Kinase Deficient Patients. Front. Physiol. 2021, 12, 735543. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.N.S.; Santerre, M.; Arjona, S.P.; Ghaleb, L.J.; Herzi, M.; Llewellyn, M.D.; Shcherbik, N.; Sawaya, B.E. SARS-CoV-2 Causes Lung Inflammation through Metabolic Reprogramming and RAGE. Viruses 2022, 14, 983. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Fu, C.-Y. Adenylate Kinase. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 18–23. ISBN 9780123847331. [Google Scholar]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ní Choileáin, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, M.I. Adenylate Kinase: A Ubiquitous Enzyme Correlated with Medical Conditions. Protein J. 2019, 38, 120–133. [Google Scholar] [CrossRef]
- Foy, B.H.; Carlson, J.C.T.; Reinertsen, E.; Padros, I.; Valls, R.; Pallares Lopez, R.; Palanques-Tost, E.; Mow, C.; Westover, M.B.; Aguirre, A.D.; et al. Association of Red Blood Cell Distribution Width with Mortality Risk in Hospitalized Adults with SARS-CoV-2 Infection. JAMA Netw. Open 2020, 3, e2022058. [Google Scholar] [CrossRef]
- Nappi, F.; Iervolino, A.; Avtaar Singh, S.S. Thromboembolic Complications of SARS-CoV-2 and Metabolic Derangements: Suggestions from Clinical Practice Evidence to Causative Agents. Metabolites 2021, 11, 341. [Google Scholar] [CrossRef]
| PATIENTS | GROUPS | NUMBER of CASES | |
|---|---|---|---|
| COVID-19+ | without ANEMIA | GROUP 1 | 13 (18%) |
| COVID-19+ | with ANEMIA | GROUP 2 | 20 (27%) |
| VIRAL INFECTION | without ANEMIA | GROUP 3 | 10 (13%) |
| VIRAL INFECTION | with ANEMIA | GROUP 4 | 20 (27%) |
| HEALTHY CONTROLS | GROUP 5 | 11 (15%) | |
| GROUP 1 COVID-19 Positive NO ANEMIA (n = 13) | GROUP 2 COVID-19 Positive ANEMIA (n = 20) | GROUP 3 VIRAL INFECTION NO ANEMIA (n = 10) | GROUP 4 VIRAL INFECTION ANEMIA (n = 20) | Normal Controls (n = 11) | p * | ||
|---|---|---|---|---|---|---|---|
| RBCs (×1012/L) | 4.61 (0.67) | 3.86 (0.72) | 4.53 (0.38) | 3.80 (0.74) | 4.51 (0.48) | ||
| Hemoglobin (g/L) | M | 142.7 (21.4) | 121.4 (17.5) | 131.1 (8.2) | 108.1 (18.4) | 139.0 (16.0) | |
| F | 136.0 (13.0) | 107.5 (12.0) | 124.5 (12.0] | 101.5 (20.0) | 131.0 (18.0) | ||
| MCV (fl) | 91.72 (4.68) | 88.98 (6.63) | 88.09 (5.28) | 87.01 (7.51) | 88.28 (3.10) | ||
| MCH (pg) | 30.85 (1.47) | 29.20 (2.68) | 29.04 (1.75) | 29.03 (3.16) | 30.24 (1.24) | ||
| MCHC (g/L) | 338.5 (8.00) | 327.7 (8.90) | 329.2 (5.30) | 329.8 (8.80) | 342.50 (7.20) | ||
| RDW | 13.52 (0.40) | 15.7 (2.33) | 14.88 (1.35) | 15.58 (3.94) | 13.25 (0.52) | ||
| Reticulocytes (%) | 0.87 (0.50) | 1.47 (0.90) | 1.15 (0.56) | 1.51 (1.19) | 1.15 (0.55) | 0.125 | |
| Retis (×109/L) | 40.45 (23.85) | 53.61 (27.38) | 52.23 (25.57) | 55.32 (38.58) | 51.98 (25.16) | 0.537 | |
| WBCs (×109/L) | 6.57 (4.44) | 8.23 (4.62) | 9.55 (3.88) | 10.86 (5.68) | 6.04 (1.61) | 0.002 | |
| Platelets (×109/L) | 170.77 (73.04) | 241.05 (147.99) | 225.2 (67.65) | 259.6 (126.2) | 251.91 (62.9) | 0.121 | |
| MPV (fl) | 8.39 (1.18) | 8.41 (1.15) | 8.41 (1.17) | 8.51 (1.03) | 8.74 (0.85) | 0.703 | |
| Fibrinogen (g/L) | 5.90 (1.32) | 5.80 (1.42) | 5.88 (5.61) | 6.50 (0.97) | 3.58 (1.58) | 0.009 | |
| D-dimer (ng/L) | 1469 (1653.07) | 2858 (2548.69) | 1375 (1438.96) | 1074 (526.26) | 294 (149.46) | 0.046 | |
| Procalcitonin (ng/L) | 2.80 (4.00) | 58.20 (38.70) | 5.30 (9.16) | 16.70 (25.6) | 5.00 (2.5) | 0.001 | |
| Creatinine (mg/L) | 8.70 (3.60) | 16.9 (11.70) | 10.10 (3.40) | 12.20 (6.00) | 6.90 (1.40) | 0.003 | |
| BUN (mg/L) | 403.60 (296.20) | 824.70 (567.70) | 536.70 (212.10) | 505.2 (449.9) | 220 (27.80) | 0.001 | |
| PCR (mg/L) | 627.50 (47.59) | 767.10 (458.20) | 586.10 (439.20) | 1180 (775.60) | 4.80 (2.10) | 0.006 | |
| Hepcidin (nM/L) | 44.71 (35.26) | 70.93 (91.47) | 194.77 (426.99) | 212 (299.54) | 9.55 (6.32) | 0.009 | |
| Serum iron (mg/L) | 664.02 (410.0) | 544.40 (381.8) | 465.7 (300.2) | 213.0 (118.7) | 1035(339.5) | 0.002 | |
| Serum ferritin (ng/L) | 1550 (522.50) | 5375.3 (4052.2) | 3681.40 (2839.80) | 4520 (4611.4) | 825 (741.7) | 0.012 | |
| Serum transferrin (mg/L) | 2136 (592.20) | 1572 (561.20) | 1747.10 (284.70) | 2011 (468.20) | 2650 (261.5) | 0.002 | |
| Transferrin saturation Index (TSI) | 24.00 (5.89) | 26.67 (17.21) | 21.29 (17.59) | 7.80 (4.71) | 28.5 (10.07) | 0.003 | |
| Serum cobalamin (Vit B12) (pg/L) | 577.0 (191.09) | 908.63 (760.30) | 640.29 (439.93) | 614.60 (517.0) | 486.43 (218.4) | 0.971 | |
| Serum folate (ng/L) | 66.00 (45.7) | 59.40 (27.8) | 69.90 (47.7) | 74.7 (42.0) | 55.0 (12.40) | 0.809 | |
| Serum bilirubin (Total) (mg/L) | 5.10 (1.1) | 6.20 (4.5) | 4.80 (0.19) | 5.60 (2.50) | 7.00 (2.90) | 0.597 | |
| ALT (IU/L) | 19.8 (12.1) | 21.67 (10.32) | 25.13 (19.14) | 24.47 (13.89) | 15.67 (7.23) | 0.162 | |
| Alkaline phosphatase (IU/L) | 86.25 (25.36) | 72.5 (32.53) | 69.86 (34.98) | 118.69 (87.5) | 50.80 (6.91) | 0.085 |
| GROUP 1 COVID Positive NO ANEMIA (n = 13) | GROUP 2 COVID Positive ANEMIA (n = 20) | GROUP 3 COVID Negative NO ANEMIA (n = 10) | GROUP 4 COVID Negative ANEMIA (n = 20) | Reference Values (n = 11) | p * | |
|---|---|---|---|---|---|---|
| Age | 62.15 (21.5) | 71.8 (14.23) | 70.0 (19.50) | 67.8 (18.72) | 58.20 (15.50) | |
| Pyruvate kinase (PK) | 13.1 (1.66) | 14.40 (2.55) | 13.10 (1.68) | 12.55 (2.13) | 11.23 (3.31) | 0.047 * |
| Hexokinase (HK) | 1.10 (0.34) | 1.25 (0.36) | 1.18 (0.16) | 1.14 (0.36) | 0.94 (0.34) | 0.088 |
| Glucose-6-phosphate dehydrogenase (G6PD) | 7.48 (1.15) | 7.62 (1.47) | 8.53 (1.92) | 7.29 (1.24) | 7.53 (1.12) | 0.517 |
| Glucose phosphate isomerase (GPI) | 53.22 (11.8) | 55.28 (9.11) | 52.13 (11.86) | 50.81 (12.20) | 53.05 (7.15) | 0.729 |
| Adenylate kinase (AK) | 226.2 (23.7) | 258.5 (38.25) | 220.9 (37.50) | 249.2 (38.24) | 236.5 (33.32) | 0.049 * |
| Phosphofructokinase (PFK) | 10.45 (2.80) | 11.28 (2.59) | 9.73 (2.63) | 10.53 (1.66) | 9.42 (3.16) | 0.412 |
| Phosphoglycerate kinase (PGK) | 31.57 (4.68) | 32.30 (4.30) | 31.67 (4.87) | 31.62 (4.89) | 30.74 (4.79) | 0.765 |
| Triosephosphate isomerase (TPI) | 2032 (386) | 2185 (435) | 1975.4 (256) | 2051.2 (246) | 1897.1 (292) | 0.291 |
| Glutathione peroxidase (GPx) | 16.3 (4.65) | 18.88 (4.99) | 17.17 (4.45) | 16.66 (3.68) | 16.04 (4.89) | 0.403 |
| Glutathione reductase (GR) | 8.22 (1.82) | 8.91 (2.57) | 9.08 (2.61) | 9.11 (2.74) | 8.15 (1.97) | 0.941 |
| Reduced glutathione (GSH) | 77.89 (14.7) | 75.96 (14.27) | 73.73 (7.20) | 74.96 (18.79) | 81.45 (13.04) | 0.500 |
| GSH Stability (+APH) | 61.59 (17.3) | 65.77 (12.50) | 59.15 (18.54) | 63.40 (25.26) | 65.08 (20.24) | 0.86 2 |
| COVID | ANEMIA | VIRAL INFECTION | O Min (X ± SD) | EI Max (X ± SD) | O Max (X ±SD) | O Hyper (X ± SD) | Area (X ± SD) | |
|---|---|---|---|---|---|---|---|---|
| Group 1 | positive | NO | 145 (5.7) | 0.612 (0.01) | 311 (8.9) | 457 (16.7) | 162 (7.45) | |
| Group 2 | positive | YES | 137 (11.2) | 0.607 (0.01) | 300 (21.9) | 464 (14.9) * | 168 (7.5) * | |
| Group 3 | negative | NO | YES | 136 (8.8) * | 0.612 (0.01) | 297 (22.6) | 463 (13.2) * | 172 (3.7) * |
| Group 4 | negative | YES | YES | 143 (11.1) | 0.611 (0.01) | 309 (21.1) | 466 (13.7) * | 168 (6.2) * |
| Normal Controls | 146 (6.6) | 0.613 (0.01) | 309 (14.1) | 449 (15.1) | 160 (6.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Alessandro, A.; Krisnevskaya, E.; Leguizamon, V.; Hernández, I.; de la Torre, C.; Bech, J.-J.; Navarro, J.-T.; Vives-Corrons, J.-L. SARS-CoV-2 Infection and Anemia—A Focus on RBC Deformability and Membrane Proteomics—Integrated Observational Prospective Study. Microorganisms 2024, 12, 453. https://doi.org/10.3390/microorganisms12030453
D’Alessandro A, Krisnevskaya E, Leguizamon V, Hernández I, de la Torre C, Bech J-J, Navarro J-T, Vives-Corrons J-L. SARS-CoV-2 Infection and Anemia—A Focus on RBC Deformability and Membrane Proteomics—Integrated Observational Prospective Study. Microorganisms. 2024; 12(3):453. https://doi.org/10.3390/microorganisms12030453
Chicago/Turabian StyleD’Alessandro, Angelo, Elena Krisnevskaya, Valentina Leguizamon, Ines Hernández, Carolina de la Torre, Joan-Josep Bech, Josep-Tomàs Navarro, and Joan-Lluis Vives-Corrons. 2024. "SARS-CoV-2 Infection and Anemia—A Focus on RBC Deformability and Membrane Proteomics—Integrated Observational Prospective Study" Microorganisms 12, no. 3: 453. https://doi.org/10.3390/microorganisms12030453
APA StyleD’Alessandro, A., Krisnevskaya, E., Leguizamon, V., Hernández, I., de la Torre, C., Bech, J.-J., Navarro, J.-T., & Vives-Corrons, J.-L. (2024). SARS-CoV-2 Infection and Anemia—A Focus on RBC Deformability and Membrane Proteomics—Integrated Observational Prospective Study. Microorganisms, 12(3), 453. https://doi.org/10.3390/microorganisms12030453






