Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation, and Identification of Bacteria Strain
2.2. Morphological and Biochemical Analysis
2.3. Bacterial Spore-Forming Analysis
2.4. Tolerance of Isolates to Acids and Bile Salts
2.5. Hemolytic Activity
2.6. Antibiotic Susceptibility Testing
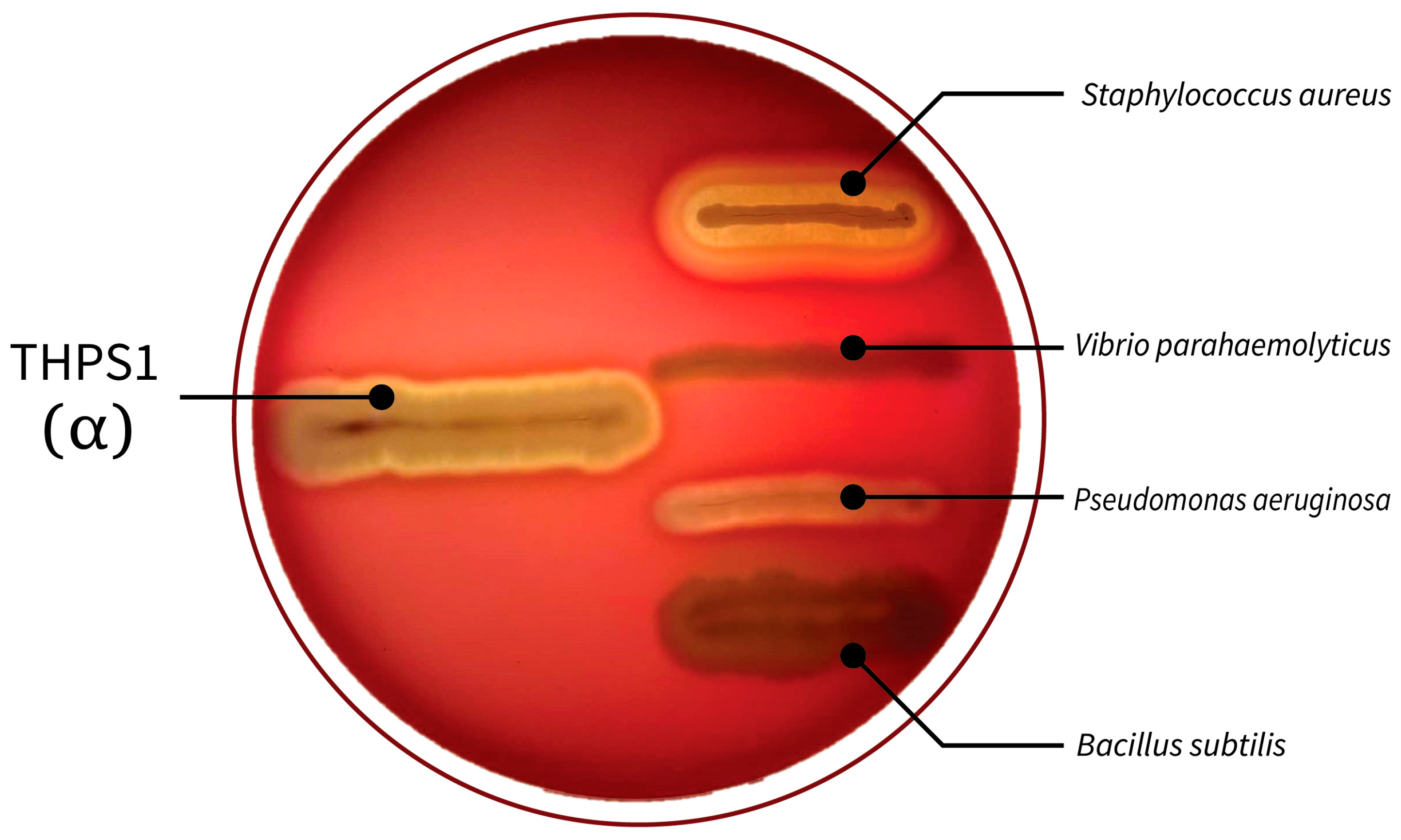
2.7. Antibiofilm Activity Assay
2.8. Storage and Preservation of THPS1 Strain
2.9. DNA Extraction and Whole Genome Sequencing
2.10. Genome Assembly, Classification, and Annotation Analysis
2.11. Taxonomic Classification
2.12. Genome Annotation
2.13. Comparative Genomic Analysis
3. Results
3.1. Morphological and Biochemical Testing
3.2. Antibiotic Susceptibility Analysis
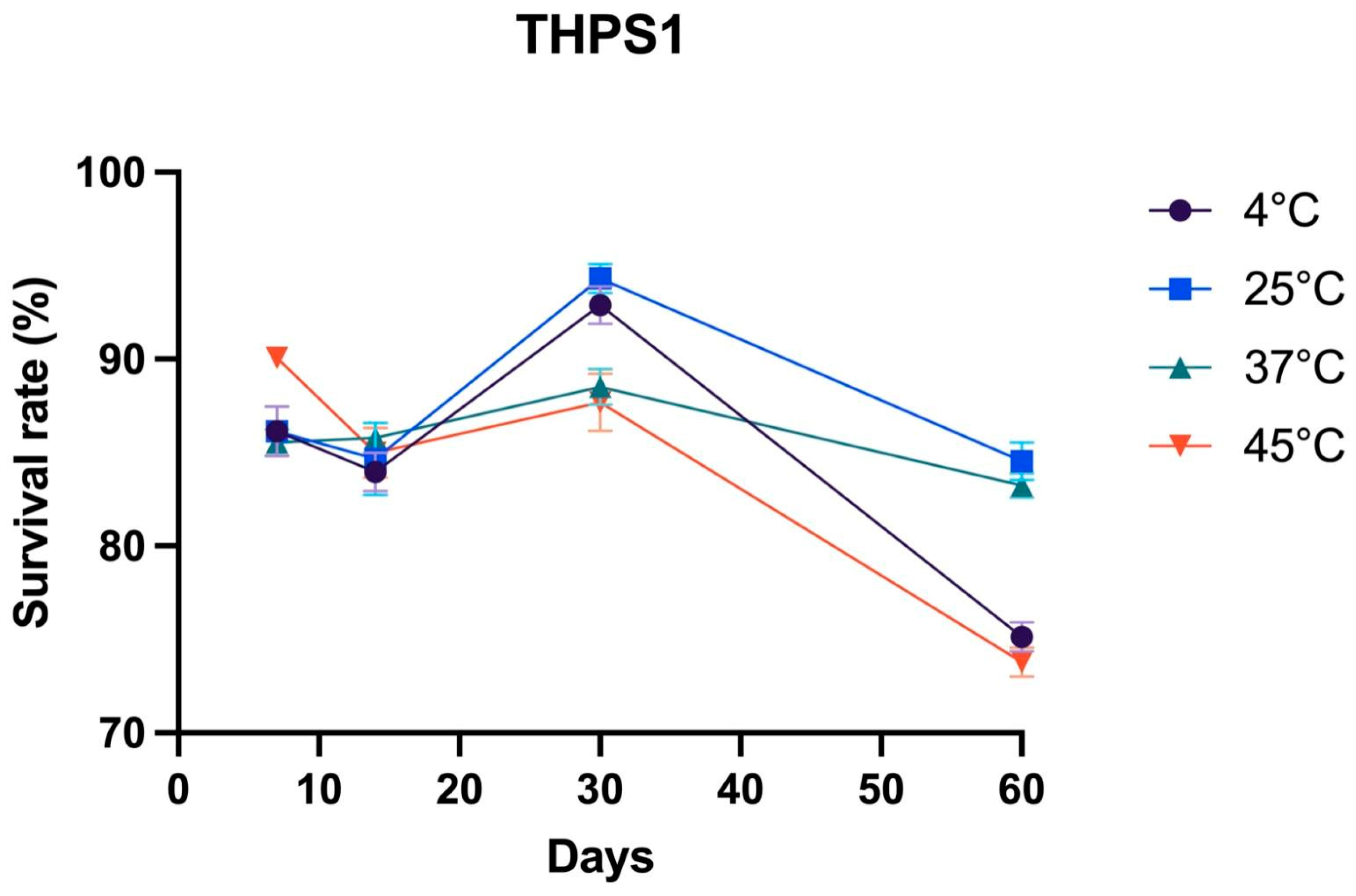
3.3. Storage Stability of Bacillus sp. THPS1 Spores
3.4. Molecular Identification and Genomic Feature Characterization
3.4.1. Molecular Identification
3.4.2. Genomic Features
3.5. Functional Annotation
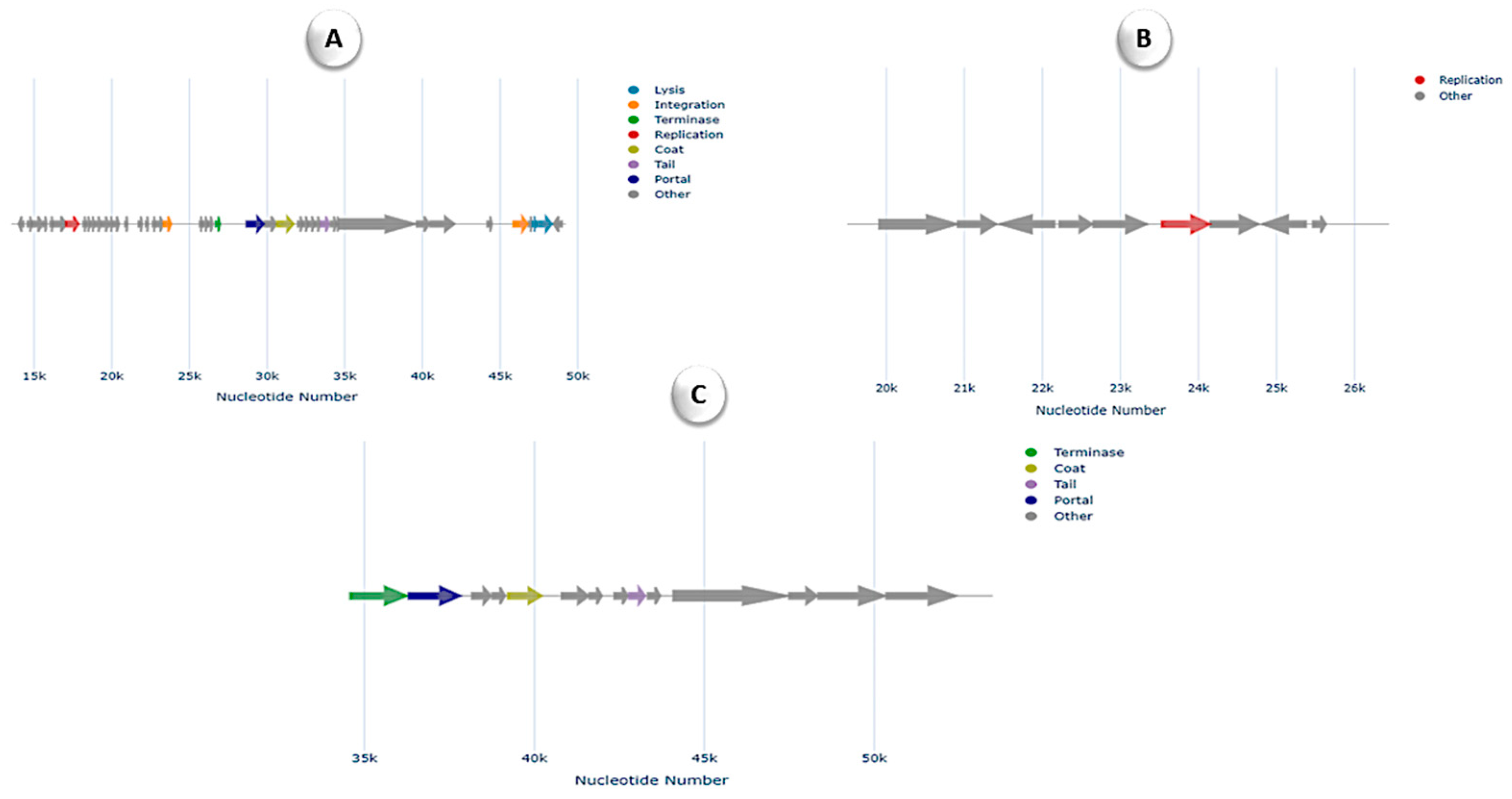
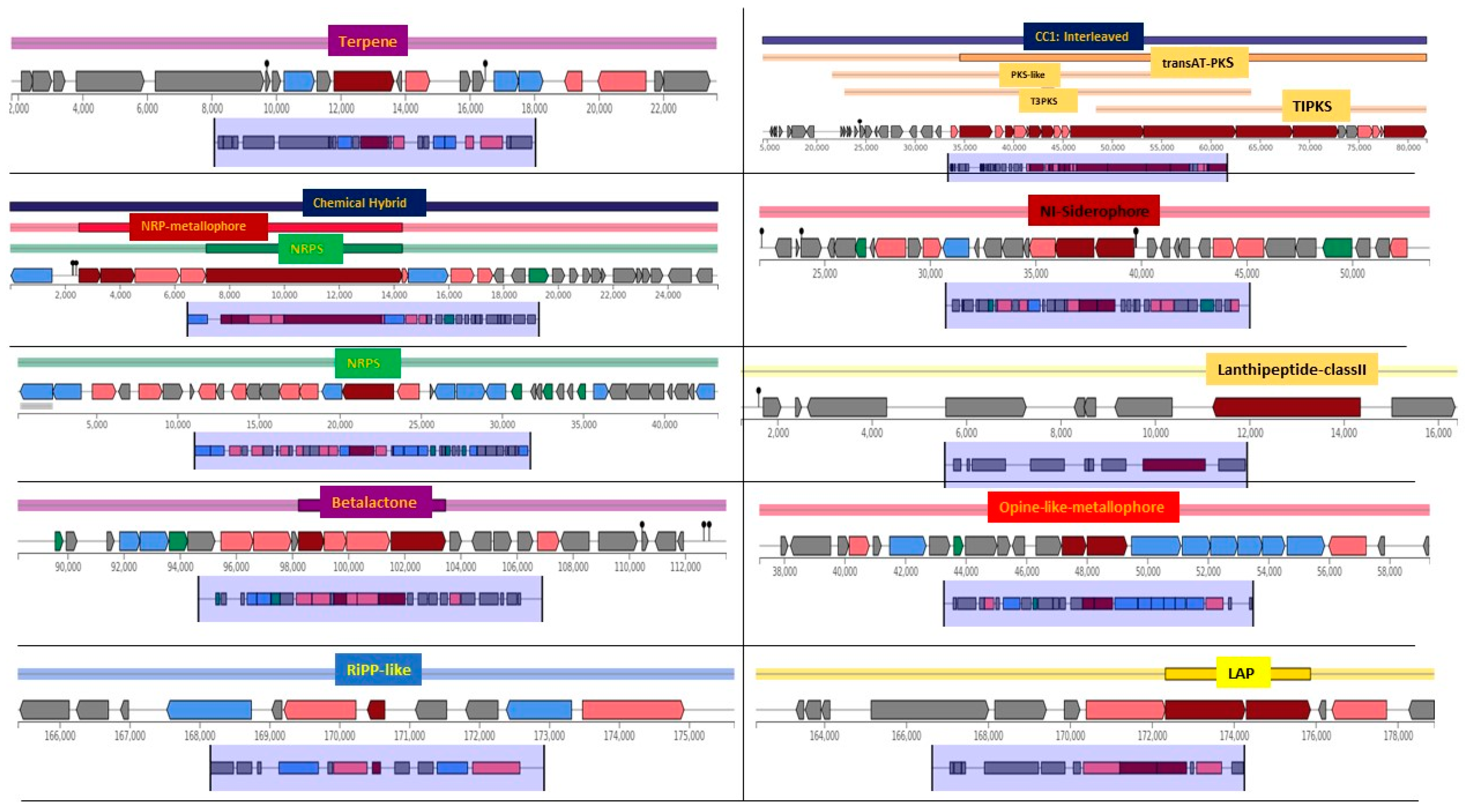
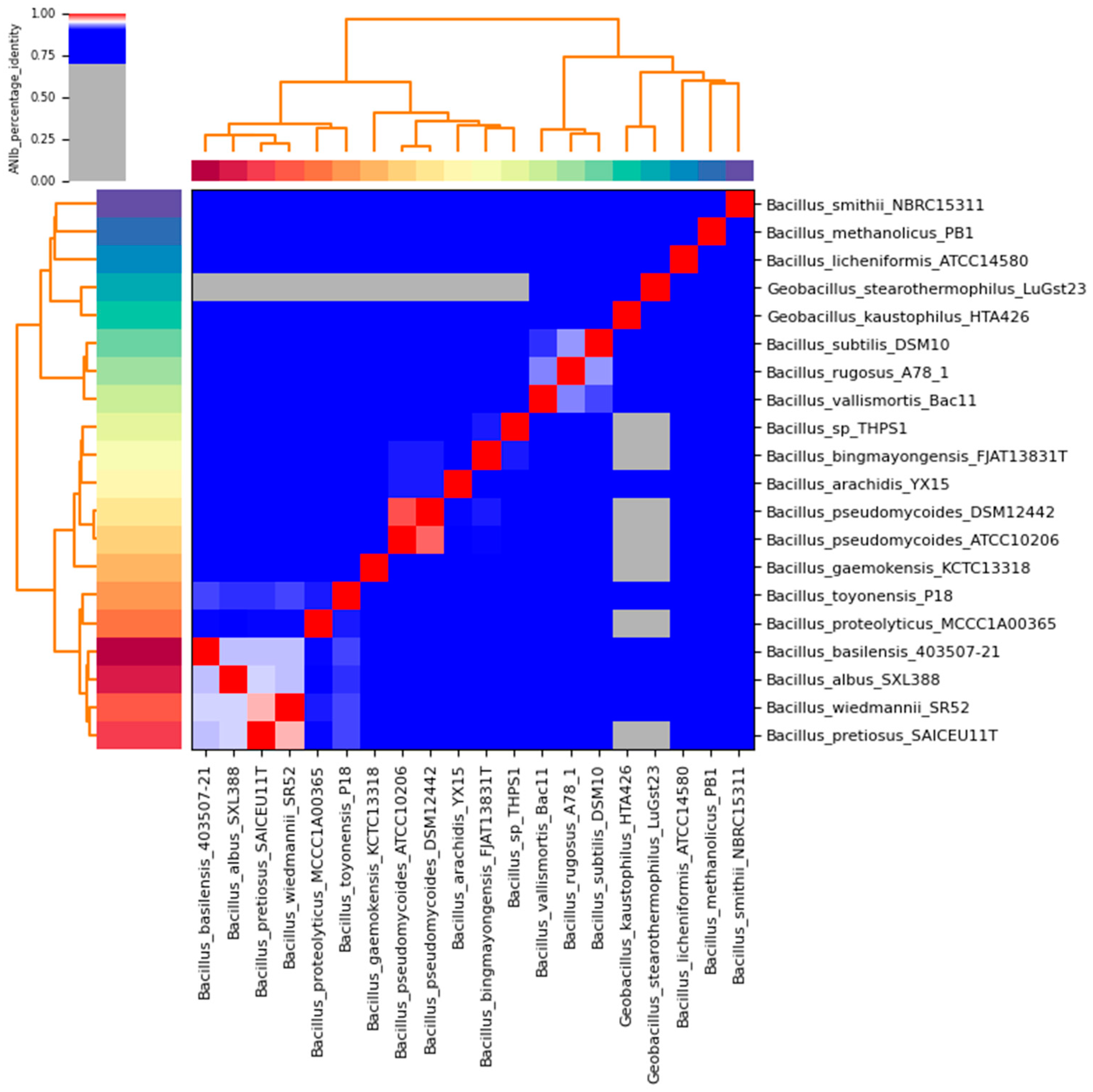
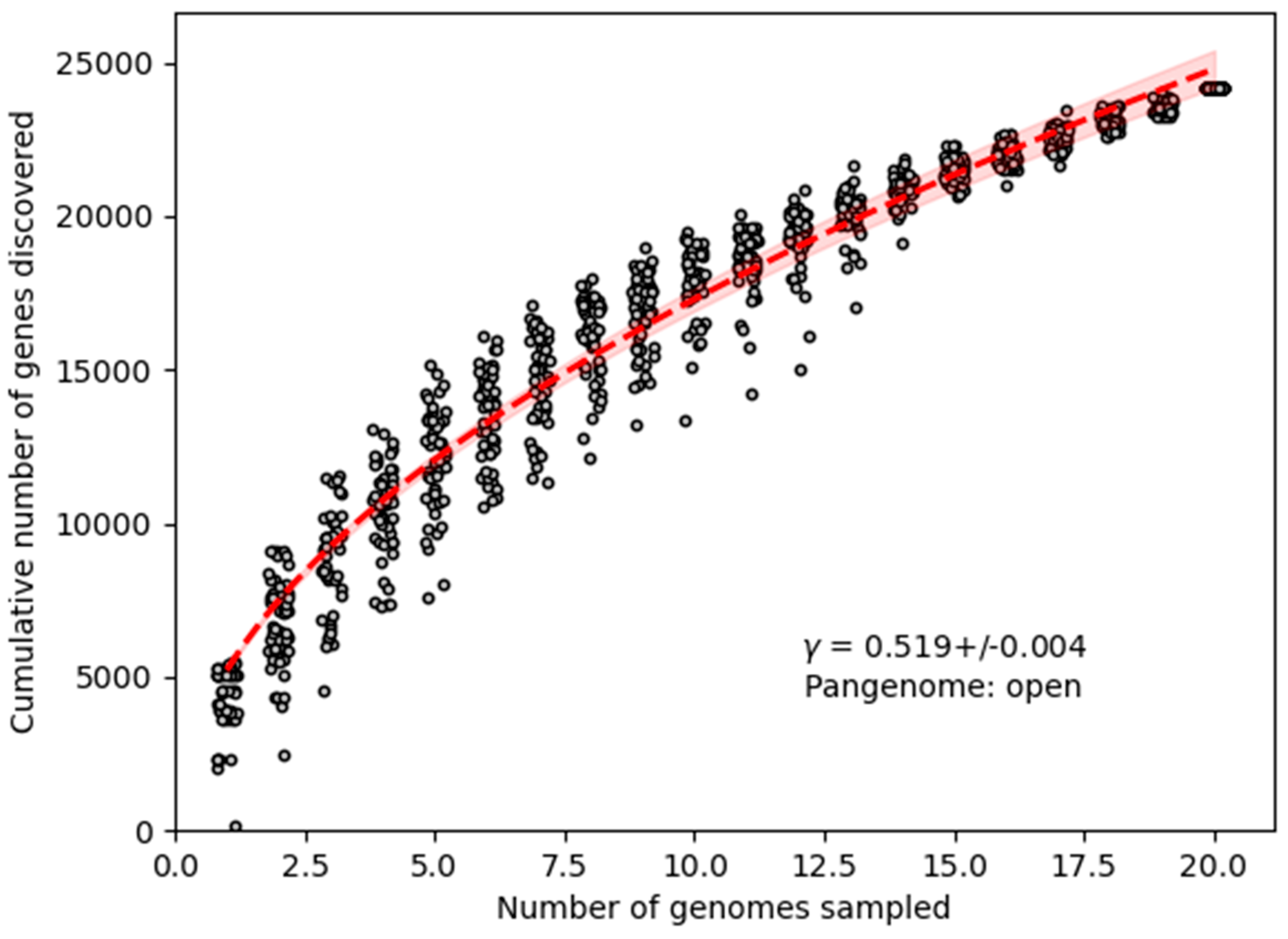
3.6. Comparative Genomics
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minnaard, J.; Rolny, I.S.; Pérez, P.F. Bacillus. In Laboratory Models for Foodborne Infections; CRC Press: Boca Raton, FL, USA, 2017; pp. 131–154. [Google Scholar]
- Hernández-González, I.L.; Moreno-Hagelsieb, G.; Olmedo-Álvarez, G. Environmentally-driven gene content convergence and the Bacillus phylogeny. BMC Evol. Biol. 2018, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Kuan, K.B.; Othman, R.; Abdul Rahim, K.; Shamsuddin, Z.H. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS ONE 2016, 11, e0152478. [Google Scholar] [CrossRef] [PubMed]
- Prashar, P.; Kapoor, N.; Sachdeva, S. Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Bio/Technol. 2014, 13, 63–77. [Google Scholar] [CrossRef]
- Federici, B. Insecticidal protein crystals of Bacillus thuringiensis. In Microbiology Monographs Inclusions in Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Mazzola, M.; Freilich, S. Prospects for biological soilborne disease control: Application of indigenous versus synthetic microbiomes. Phytopathology 2017, 107, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Errington, J.; Aart, L.T.v.d. Microbe Profile: Bacillus subtilis: Model organism for cellular development, and industrial workhorse. Microbiology 2020, 166, 425–427. [Google Scholar] [CrossRef]
- Ortiz, A.; Sansinenea, E. The Industrially important enzymes from bacillus species. In Bacilli in Agrobiotechnology: Plant Stress Tolerance, Bioremediation, and Bioprospecting; Springer: Berlin/Heidelberg, Germany, 2022; pp. 89–99. [Google Scholar]
- Lubis, A.R.; Sumon, M.A.A.; Dinh-Hung, N.; Dhar, A.K.; Delamare-Deboutteville, J.; Kim, D.H.; Shinn, A.P.; Kanjanasopa, D.; Permpoonpattana, P.; Doan, H.V. Review of quorum-quenching probiotics: A promising non-antibiotic-based strategy for sustainable aquaculture. J. Fish Dis. 2024, 47, e13941. [Google Scholar] [CrossRef]
- Bader, J.; Albin, A.; Stahl, U. Spore-forming bacteria and their utilisation as probiotics. Benef. Microbes 2012, 3, 67–75. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sroczyński, M.; Dzitkowska-Zabielska, M.; Bojarczuk, A.; Skowron, P.M.; Cięszczyk, P. Analysis of industrial Bacillus species as potential probiotics for dietary supplements. Microorganisms 2023, 11, 488. [Google Scholar] [CrossRef]
- Popov, I.V.; Algburi, A.; Prazdnova, E.V.; Mazanko, M.S.; Elisashvili, V.; Bren, A.B.; Chistyakov, V.A.; Tkacheva, E.V.; Trukhachev, V.I.; Donnik, I.M. A review of the effects and production of spore-forming probiotics for poultry. Animals 2021, 11, 1941. [Google Scholar] [CrossRef]
- Barák, I. Special Issue “Bacillus subtilis as a Model Organism to Study Basic Cell Processes”. Microorganisms 2021, 9, 2459. [Google Scholar] [CrossRef]
- Earl, A.M.; Losick, R.; Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008, 16, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Aslam, B. Bacillus spp.-Transmission, pathogenesis, host-pathogen interaction, prevention and treatment. Front. Microbiol. 2023, 14, 1307723. [Google Scholar] [CrossRef] [PubMed]
- Granum, P.E.; Lund, T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Vojnovic, S.; Aleksic, I.; Ilic-Tomic, T.; Stevanovic, M.; Nikodinovic-Runic, J. Bacillus and Streptomyces spp. as hosts for production of industrially relevant enzymes. Appl. Microbiol. Biotechnol. 2024, 108, 185. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The genus Geobacillus and their biotechnological potential. Adv. Appl. Microbiol. 2015, 92, 1–48. [Google Scholar]
- Novik, G.; Savich, V.; Meerovskaya, O. Geobacillus Bacteria: Potential commercial applications in industry, bioremediation, and bioenergy production. In Growing and Handling of Bacterial Cultures; IntechOpen: London, UK, 2018; pp. 1–36. [Google Scholar]
- Zhao, N.; Yu, T.; Yan, F. Probiotic role and application of thermophilic Bacillus as novel food materials. Trends Food Sci. Technol. 2023, 138, 1–15. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sobolewski, I.; Skowron, P.M. Bacteriophages of thermophilic ‘Bacillus group’bacteria—A review. Microorganisms 2021, 9, 1522. [Google Scholar] [CrossRef]
- McMullan, G.; Christie, J.; Rahman, T.; Banat, I.; Ternan, N.; Marchant, R. Habitat, applications and genomics of the aerobic, thermophilic genus Geobacillus. Biochem. Soc. Trans. 2004, 32, 214–217. [Google Scholar] [CrossRef]
- Karlowski, W.M.; Varshney, D.; Zielezinski, A. Taxonomically restricted genes in Bacillus may form clusters of homologs and can be traced to a large reservoir of noncoding sequences. Genome Biol. Evol. 2023, 15, evad023. [Google Scholar] [CrossRef]
- Sharma, A.; Satyanarayana, T. Comparative genomics of Bacillus species and its relevance in industrial microbiology. Genom. Insights 2013, 6, GEI.S12732. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J.; Dull, T.J.; Sleeter, D.D.; Noller, H.F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 1981, 148, 107–127. [Google Scholar] [CrossRef]
- Elsadek, M.M.; Wang, S.; Wu, Z.; Wang, J.; Wang, X.; Zhang, Y.; Yu, M.; Guo, Z.; Wang, Q.; Wang, G. Characterization of Bacillus spp. isolated from the intestines of Rhynchocypris lagowskii as a potential probiotic and their effects on fish pathogens. Microb. Pathog. 2023, 180, 106163. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.; Raja, M.; Perumal, P. Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu (Hamilton, 1822). Aquac. Rep. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Nwagu, T.N.; Ugwuodo, C.J.; Onwosi, C.O.; Inyima, O.; Uchendu, O.C.; Akpuru, C. Evaluation of the probiotic attributes of Bacillus strains isolated from traditional fermented African locust bean seeds (Parkia biglobosa),“daddawa”. Ann. Microbiol. 2020, 70, 20. [Google Scholar] [CrossRef]
- Dinh-Hung, N.; Dong, H.T.; Senapin, S.; Pimsannil, K.; Thompson, K.D.; Shinn, A.P.; Soontara, C.; Sirimanapong, W.; Chatchaiphan, S.; Rodkhum, C. Insight into characteristics and pathogenicity of five rapidly growing non-tuberculous Mycobacterium species isolated from the Siamese fighting fish, Betta splendens. Aquaculture 2023, 575, 739822. [Google Scholar] [CrossRef]
- Hamza, F.; Kumar, A.R.; Zinjarde, S. Antibiofilm potential of a tropical marine Bacillus licheniformis isolate: Role in disruption of aquaculture associated biofilms. Aquac. Res. 2016, 47, 2661–2669. [Google Scholar] [CrossRef]
- Coulibaly, W.H.; Kouadio, N.g.R.; Camara, F.; Diguță, C.; Matei, F. Functional properties of lactic acid bacteria isolated from Tilapia (Oreochromis niloticus) in Ivory Coast. BMC Microbiol. 2023, 23, 152. [Google Scholar] [CrossRef]
- Salvà-Serra, F.; Gomila, M.; Svensson-Stadler, L.; Busquets, A.; Jaén-Luchoro, D.; Karlsson, R.; Moore, E.R. A protocol for extraction and purification of high-quality and quantity bacterial DNA applicable for genome sequencing: A modified version of the Marmur procedure. Res. Sq. 2018. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.; Humphris, S.; Elphinstone, J.; Toth, I. Genomics and Taxonomy in Diagnostics for Food Security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Abdella, B.; Abozahra, N.A.; Shokrak, N.M.; Mohamed, R.A.; El-Helow, E.R. Whole spectrum of Aeromonas hydrophila virulence determinants and the identification of novel SNPs using comparative pathogenomics. Sci. Rep. 2023, 13, 7712. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, G.S.; Parkhill, J. Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics 2006, 22, 2196–2203. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-db: A manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, S.; Abreu-Goodger, C. Using MCL to extract clusters from networks. In Bacterial Molecular Networks: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2012; pp. 281–295. [Google Scholar]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed]
- Horsfield, S.T.; Tonkin-Hill, G.; Croucher, N.J.; Lees, J.A. Accurate and fast graph-based pangenome annotation and clustering with ggCaller. Genome Res. 2023, 33, 1622–1637. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ren, G.; Gunning III, W.T.; Weaver, D.A.; Kalinoski, A.L.; Khuder, S.A.; Huntley, J.F. FmvB: A Francisella tularensis magnesium-responsive outer membrane protein that plays a role in virulence. PLoS ONE 2016, 11, e0160977. [Google Scholar] [CrossRef] [PubMed]
- Boneca, I.G.; Dussurget, O.; Cabanes, D.; Nahori, M.-A.; Sousa, S.; Lecuit, M.; Psylinakis, E.; Bouriotis, V.; Hugot, J.-P.; Giovannini, M. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. USA 2007, 104, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rockey, D.D.; Dolan, B.P. Chlamydia lipooligosaccharide has varied direct and indirect roles in evading both innate and adaptive host immune responses. Infect. Immun. 2020, 88, e00198-20. [Google Scholar] [CrossRef] [PubMed]
- Dinh-Hung, N.; Mwamburi, S.M.; Dong, H.T.; Rodkhum, C.; Meemetta, W.; Linh, N.V.; Mai, H.N.; Dhar, A.K.; Hirono, I.; Senapin, S.; et al. Unveiling Insights into the Whole Genome Sequencing of Mycobacterium spp. Isolated from Siamese Fighting Fish (Betta splendens). Animals 2024, 14, 2833. [Google Scholar] [CrossRef] [PubMed]
- Aqel, H.; Farah, H.; Al-Hunaiti, A. Ecological versatility and biotechnological promise: Comprehensive characterization of the isolated thermophilic Bacillus strains. PLoS ONE 2024, 19, e0297217. [Google Scholar] [CrossRef] [PubMed]
- Elleuche, S.; Schröder, C.; Sahm, K.; Antranikian, G. Extremozymes—Biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 2014, 29, 116–123. [Google Scholar] [CrossRef]
- Lajis, A.F.B. Realm of thermoalkaline lipases in bioprocess commodities. J. Lipids 2018, 2018, 5659683. [Google Scholar] [CrossRef]
- Mullis, M.M.; Rambo, I.M.; Baker, B.J.; Reese, B.K. Diversity, ecology, and prevalence of antimicrobials in nature. Front. Microbiol. 2019, 10, 2518. [Google Scholar] [CrossRef]
- Grubbs, K.J.; Bleich, R.M.; Santa Maria, K.C.; Allen, S.E.; Farag, S.; Team, A.; Shank, E.A.; Bowers, A.A. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in bacterial physiology. MSystems 2017, 2, e00040-17. [Google Scholar] [CrossRef]
- Yin, Q.-J.; Ying, T.-T.; Zhou, Z.-Y.; Hu, G.-A.; Yang, C.-L.; Hua, Y.; Wang, H.; Wei, B. Species-specificity of the secondary biosynthetic potential in Bacillus. Front. Microbiol. 2023, 14, 1271418. [Google Scholar] [CrossRef] [PubMed]
- Belaouni, H.A.; Yekkour, A.; Zitouni, A.; Meklat, A. Delving into the Bacillus cereus group biosynthetic gene clusters cosmos: A comparative-genomics-based classification framework. bioRxiv 2023. bioRxiv:2025.530005. [Google Scholar]
- Sriaporn, C.; Campbell, K.A.; Van Kranendonk, M.J.; Handley, K.M. Genomic adaptations enabling Acidithiobacillus distribution across wide-ranging hot spring temperatures and pHs. Microbiome 2021, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Dhakan, D.B.; Mittal, P.; Waiker, P.; Chowdhury, A.; Ghatak, A.; Sharma, V.K. Metagenomic analysis of hot springs in Central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Front. Microbiol. 2017, 7, 2123. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, H.B.; Oliverio, A.M. Extreme environments offer an unprecedented opportunity to understand microbial eukaryotic ecology, evolution, and genome biology. Nat. Commun. 2023, 14, 4959. [Google Scholar] [CrossRef] [PubMed]
- Durrant, M.G.; Li, M.M.; Siranosian, B.A.; Montgomery, S.B.; Bhatt, A.S. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 2020, 27, 140–153.e9. [Google Scholar] [CrossRef]
- Khedkar, S.; Smyshlyaev, G.; Letunic, I.; Maistrenko, O.M.; Coelho, L.P.; Orakov, A.; Forslund, S.K.; Hildebrand, F.; Luetge, M.; Schmidt, T.S. Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes. Nucleic Acids Res. 2022, 50, 3155–3168. [Google Scholar] [CrossRef]
- O’Meara, D.; Nunney, L. A phylogenetic test of the role of CRISPR-Cas in limiting plasmid acquisition and prophage integration in bacteria. Plasmid 2019, 104, 102418. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhang, Y.; Liu, Z.; Dong, Z.; Xie, C.; Bravo, A.; Soberón, M.; Mahillon, J.; Sun, M.; Peng, D. The CRISPR-Cas systems were selectively inactivated during evolution of Bacillus cereus group for adaptation to diverse environments. ISME J. 2020, 14, 1479–1493. [Google Scholar] [CrossRef]
- Zhang, J.; White, M.F. Hot and crispy: CRISPR–Cas systems in the hyperthermophile Sulfolobus solfataricus. Biochem. Soc. Trans. 2013, 41, 1422–1426. [Google Scholar] [CrossRef] [PubMed][Green Version]




| Source | Isolate | Colony Characteristics | Shape | Gram |
|---|---|---|---|---|
| Hot spring sediments | THPS1 | 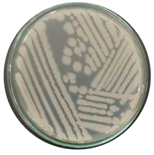 |  | + |
| Parameter Analysis | Isolate THPS1 | |
|---|---|---|
| Motility | + | |
| Catalase | + | |
| Oxidase | + | |
| Indole | − | |
| Starch hydrolysis | + | |
| Spore formation efficiency (%) | 92.21 | |
| Hemolytic activity | α | |
| Morphology | Color | Yellow |
| Gram staining | + | |
| Shape | Bacilli | |
| % survival rate in acids | pH 2 | 72.50 ± 5.29 |
| pH 3 | 87.20 ± 6.54 | |
| pH 4 | 84.30 ± 2.09 | |
| % survival rate in bile salts | 1.0% | 71.48 ± 1.50 |
| 2.5% | 69.79 ± 1.44 | |
| 5.0% | 68.49 ± 0.52 | |
| % Inhibit biofilm formation | Vibrio parahaemolyticus | 70.78 |
| Pseudomonas aeruginosa | 50.08 | |
| Staphylococcus aureus | 39.25 | |
| Bacillus subtilis | 6.81 | |
| Antibiotic | Concentration (µg/disk) | Susceptibility | Zone Diameter (mm) | Classification |
|---|---|---|---|---|
| Amoxicillin | 30 | Sensitive (S) | ≥20 | Effective |
| Ampicillin | 10 | Sensitive (S) | ≥20 | Effective |
| Chloramphenicol | 30 | Sensitive (S) | ≥20 | Effective |
| Cloxacillin | 1 | Sensitive (S) | ≥20 | Effective |
| Kanamycin | 30 | Sensitive (S) | ≥20 | Effective |
| Penicillin | 10 | Moderate (M) | 12–19 | Partial sensitivity |
| Features | Isolate THPS1 | |
|---|---|---|
| GenBank accession | - | |
| Species | Bacillus sp. | |
| Size (bp) | 5,379,921 | |
| Minimum sequence length | 223 | |
| Maximum sequence length | 458,254 | |
| Contigs | 144 | |
| Species | Bacillus sp. | |
| L50 | 16 | |
| N50 (bp) | 83,797 | |
| Completeness (CheckM) (%) | 99.12 | |
| Contamination (%) | 0.10 | |
| Genes | 5606 | |
| CDSs | 5492 | |
| Genes (coding) | 5259 | |
| Genes (RNA) | 114 | |
| rRNA | 5S | 5 |
| 16S | 4 | |
| 23S | 3 | |
| tRNAs | 97 | |
| ncRNAs | 5 | |
| Pseudo Genes | 233 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mwamburi, S.M.; Islam, S.I.; Dinh-Hung, N.; Dangsawat, O.; Sowanpreecha, R.; Khang, L.T.P.; Montha, N.; Therdtatha, P.; Dwinanti, S.H.; Permpoonpattana, P.; et al. Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential. Microorganisms 2024, 12, 2476. https://doi.org/10.3390/microorganisms12122476
Mwamburi SM, Islam SI, Dinh-Hung N, Dangsawat O, Sowanpreecha R, Khang LTP, Montha N, Therdtatha P, Dwinanti SH, Permpoonpattana P, et al. Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential. Microorganisms. 2024; 12(12):2476. https://doi.org/10.3390/microorganisms12122476
Chicago/Turabian StyleMwamburi, Samuel Mwakisha, Sk Injamamul Islam, Nguyen Dinh-Hung, Orathai Dangsawat, Rapeewan Sowanpreecha, Luu Tang Phuc Khang, Napatsorn Montha, Phatthanaphong Therdtatha, Sefti Heza Dwinanti, Patima Permpoonpattana, and et al. 2024. "Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential" Microorganisms 12, no. 12: 2476. https://doi.org/10.3390/microorganisms12122476
APA StyleMwamburi, S. M., Islam, S. I., Dinh-Hung, N., Dangsawat, O., Sowanpreecha, R., Khang, L. T. P., Montha, N., Therdtatha, P., Dwinanti, S. H., Permpoonpattana, P., & Linh, N. V. (2024). Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential. Microorganisms, 12(12), 2476. https://doi.org/10.3390/microorganisms12122476







