Effective Inhibitor Removal from Wastewater Samples Increases Sensitivity of RT-dPCR and Sequencing Analyses and Enhances the Stability of Wastewater-Based Surveillance
Abstract
1. Introduction
2. Materials and Methods
2.1. Wastewater Sampling, Total Nucleic Acid Extraction, Inhibitor Removal, and Quantification of SARS-CoV-2 RNA Copies/L in Wastewater by dPCR
2.2. Inhibition Assessment by Dilution Assays
2.3. Sequencing of SARS-CoV-2 RNA of Wastewater Samples and Data Analysis
2.4. Statistical Analysis: Flow Normalization and Stability Estimation
3. Results
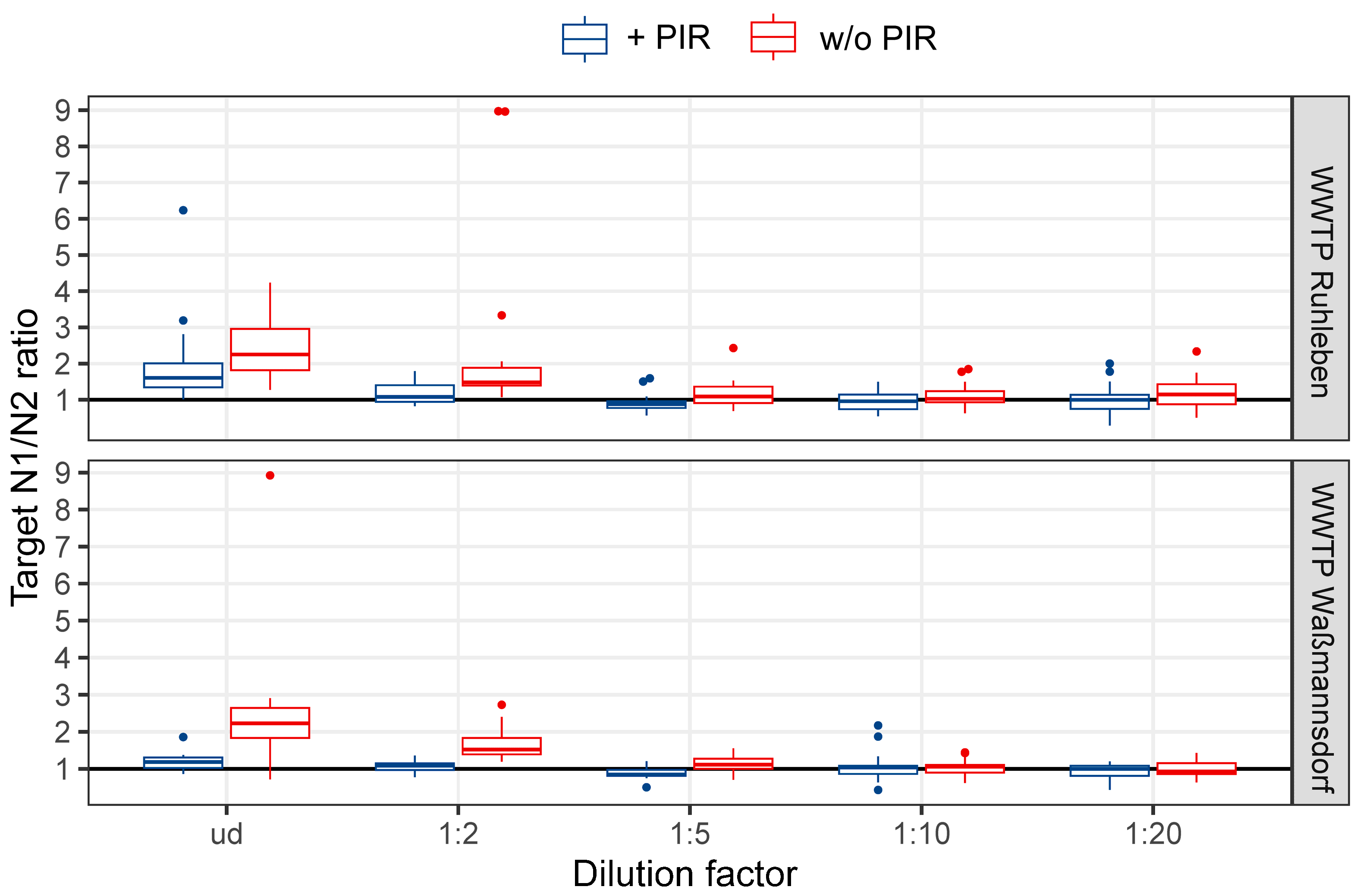
3.1. Dilutions of the TNA Extracts Reveal a High Inhibitory Effect of the TNAs Derived from Berlin’s WWTPs
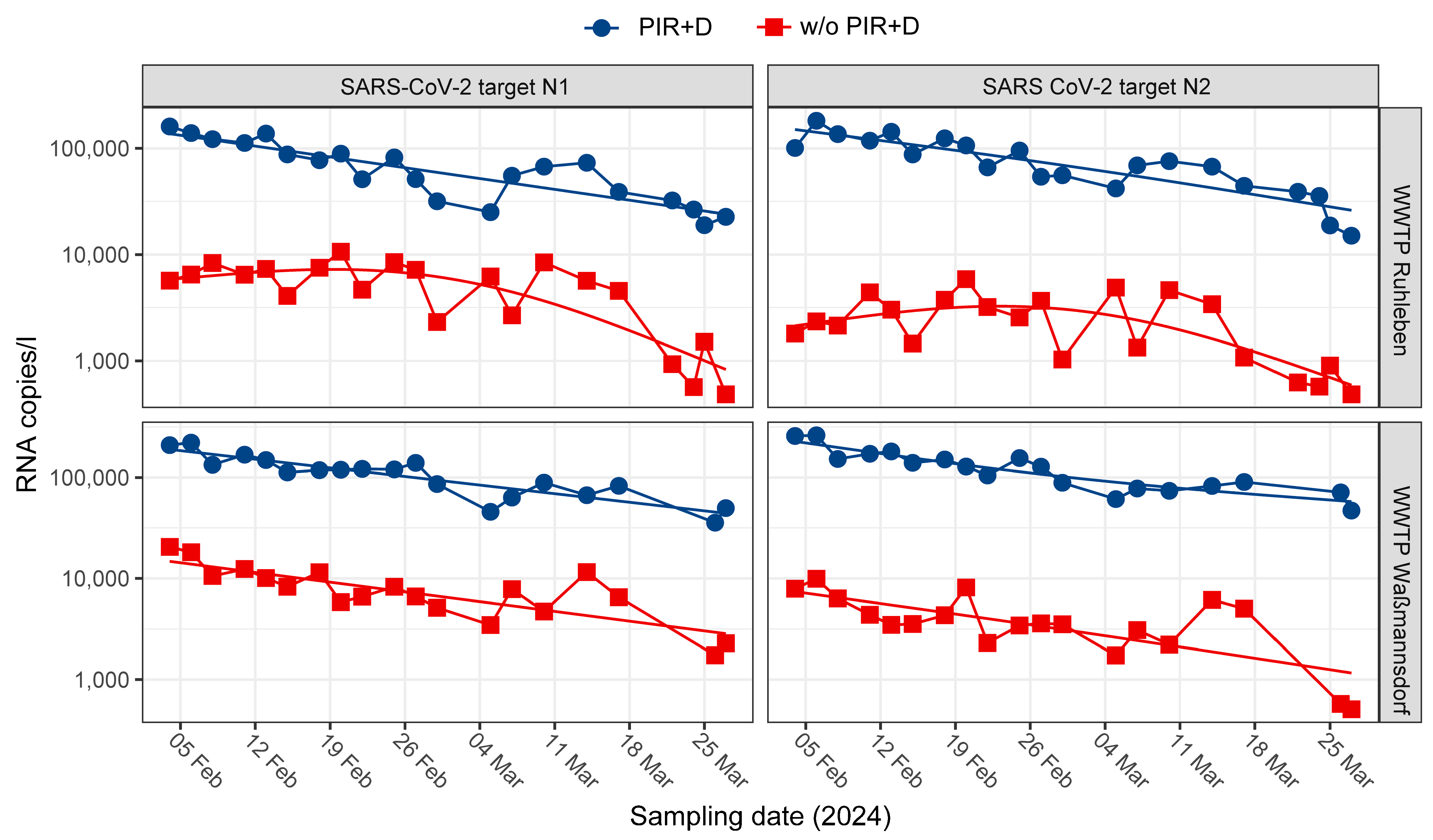
3.2. The Combined Usage of Inhibitor Removal and Dilution (PIR+D) Lead to Enhanced Amplification and an Equal N1/N2 Target Ratio
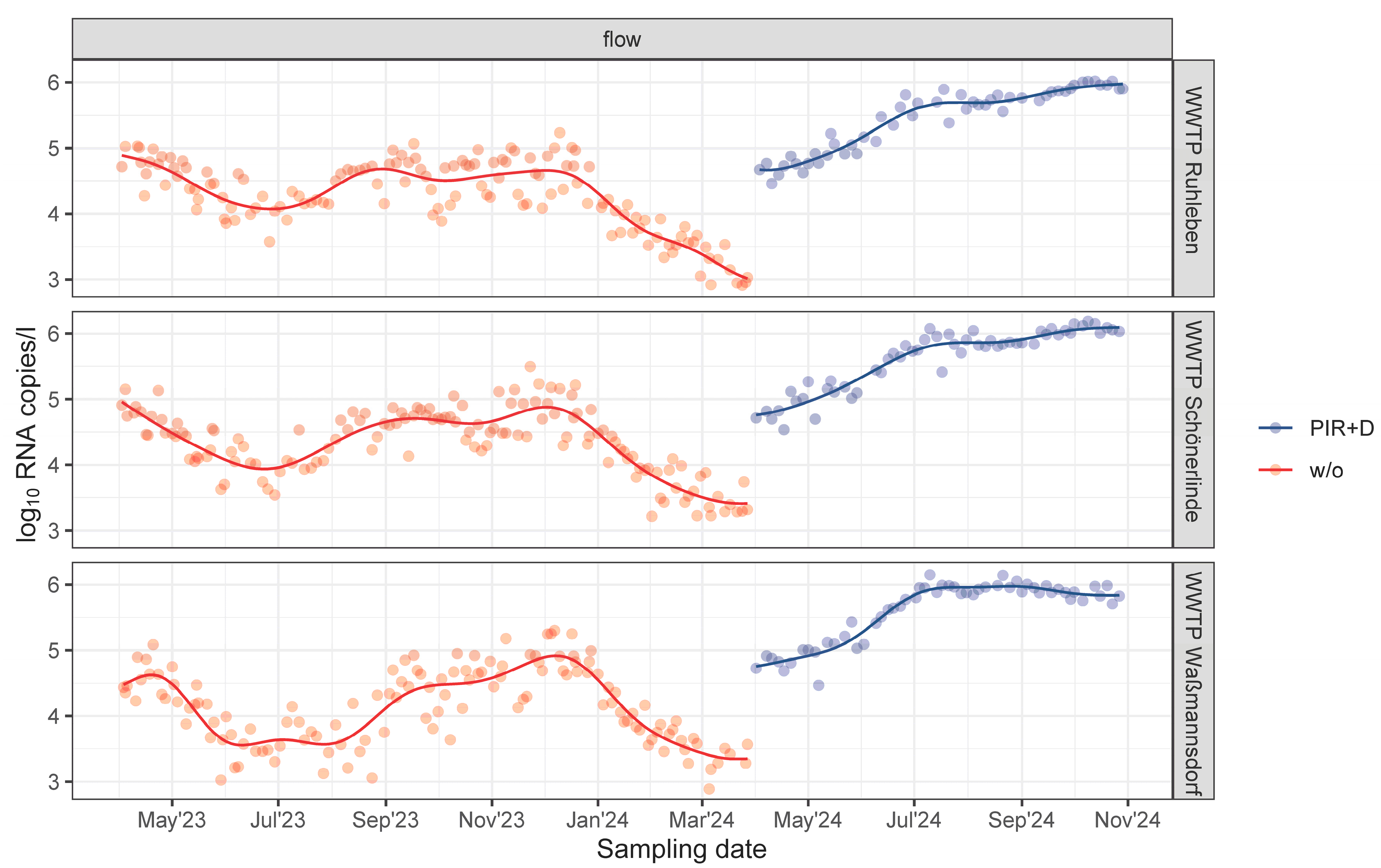
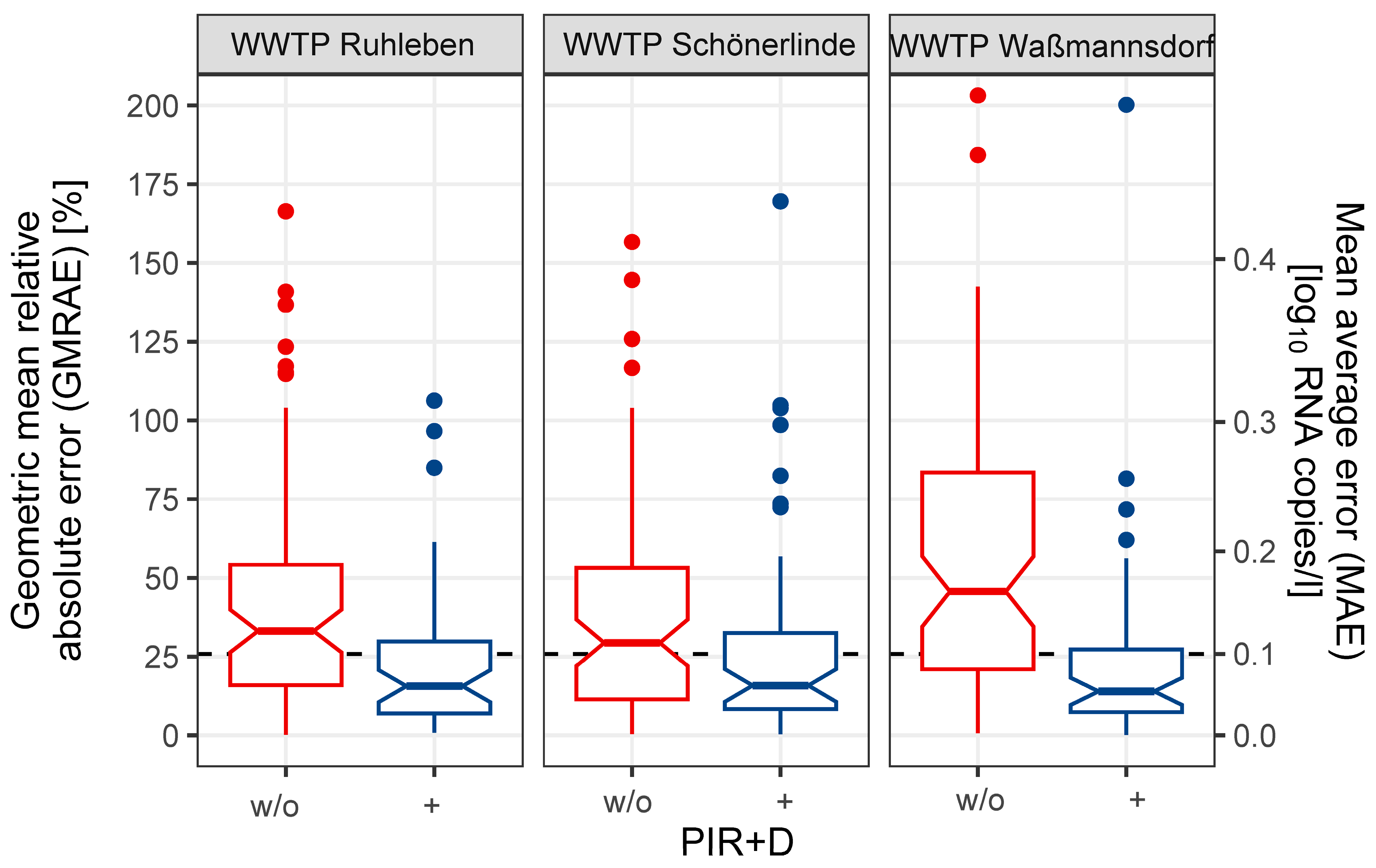
3.3. PIR Led to an Increased Stability of WBS
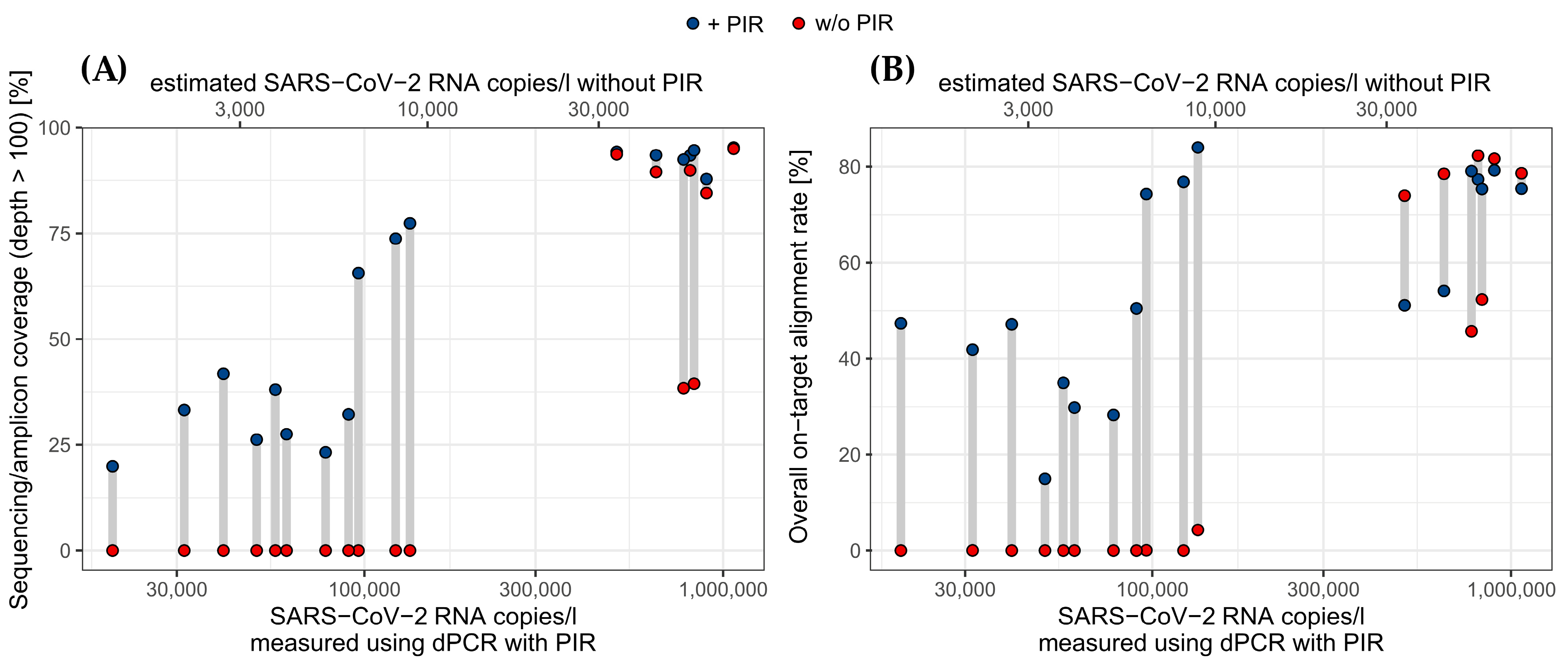
3.4. PIR Improves Coverage and Alignment of NGS
4. Discussion
4.1. Influence of PIR on dPCR and Next Generation Sequencing (NGS) Analyses
4.2. Time Series Stability in Wastewater Surveillance
4.3. Possible Inhibitory Modes of Action in the dPCR Analyses
4.4. Impacts on Inhibitor Concentrations, Workflow Considerations for Wastewater Monitoring, and Possible Inhibitory Modes of Action in Berlin
5. Conclusions
- Inhibitory substances increase the effective detection limit.
- The concentrations and composition of inhibitory substances vary between wastewater samples due to several factors.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Lee, B.E.; Acosta, N.; Bautista, M.; Hubert, C.R.J.; Hrudey, S.E.; Frankowski, K.; Pang, X.-L. Wastewater-Based Surveillance as a Tool for Public Health Action: SARS-CoV-2 and Beyond. Clin. Microbiol. Rev. 2024, 37, e00103-22. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding, and Infectiousness: A Systematic Review and Meta-Analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Crank, K.; Chen, W.; Bivins, A.; Lowry, S.; Bibby, K. Contribution of SARS-CoV-2 RNA Shedding Routes to RNA Loads in Wastewater. Sci. Total Environ. 2022, 806, 150376. [Google Scholar] [CrossRef]
- Toribio-Avedillo, D.; Gómez-Gómez, C.; Sala-Comorera, L.; Rodríguez-Rubio, L.; Carcereny, A.; García-Pedemonte, D.; Pintó, R.M.; Guix, S.; Galofré, B.; Bosch, A.; et al. Monitoring Influenza and Respiratory Syncytial Virus in Wastewater. Beyond COVID-19. Sci. Total Environ. 2023, 892, 164495. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Bofill-Mas, S.; Bosch, A.; Brandão, J.; Choi, P.M.; et al. Minimizing Errors in RT-PCR Detection and Quantification of SARS-CoV-2 RNA for Wastewater Surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef]
- Henze, M.; Concha, L. Wastewater Characterization. In Biological Wastewater Treatment; IWA Publishing: London, UK, 2008; Chapter 3; pp. 33–52. ISBN 978-1-84339-188-3. [Google Scholar]
- Abbaszadegan, M.; Huber, M.S.; Gerba, C.P.; Pepper, I.L. Detection of Enteroviruses in Groundwater with the Polymerase Chain Reaction. Appl. Environ. Microbiol. 1993, 59, 1318–1324. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR Inhibitors—Occurrence, Properties and Removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in qPCR, dPCR and MPS-Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Chianese, S.; Fenti, A.; Iovino, P.; Musmarra, D.; Salvestrini, S. Sorption of Organic Pollutants by Humic Acids: A Review. Molecules 2020, 25, 918. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, J.; Li, L.; Zhen, G.; Lu, X.; Zhang, J.; Liu, H.; Zhou, Z.; Wu, Z.; Zhang, X. Prospects for Humic Acids Treatment and Recovery in Wastewater: A Review. Chemosphere 2023, 312, 137193. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.; Kangro, H.O.; Coates, P.J.; Heath, R.B. Inhibitory Effects of Urine on the Polymerase Chain Reaction for Cytomegalovirus DNA. J. Clin. Pathol. 1991, 44, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, S.; Jaganade, T.; Priyakumar, U.D. Urea-Aromatic Interactions in Biology. Biophys. Rev. 2020, 12, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Lantz, P.-G.; Matsson, M.; Wadström, T.; Rådström, P. Removal of PCR Inhibitors from Human Faecal Samples through the Use of an Aqueous Two-Phase System for Sample Preparation Prior to PCR. J. Microbiol. Methods 1997, 28, 159–167. [Google Scholar] [CrossRef]
- Bartel, A.; Grau, J.H.; Bitzegeio, J.; Werber, D.; Linzner, N.; Schumacher, V.; Garske, S.; Liere, K.; Hackenbeck, T.; Rupp, S.I.; et al. Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses 2024, 16, 102. [Google Scholar] [CrossRef]
- Mondal, S.; Feirer, N.; Brockman, M.; Preston, M.A.; Teter, S.J.; Ma, D.; Goueli, S.A.; Moorji, S.; Saul, B.; Cali, J.J. A Direct Capture Method for Purification and Detection of Viral Nucleic Acid Enables Epidemiological Surveillance of SARS-CoV-2. Sci. Total Environ. 2021, 795, 148834. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Immunization and Respiratory Diseases (U.S.); Division of Viral Diseases. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. Available online: https://stacks.cdc.gov/view/cdc/84525 (accessed on 20 November 2024).
- Wood, S.N. Fast Stable Direct Fitting and Smoothness Selection for Generalized Additive Models. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2008, 70, 495–518. [Google Scholar] [CrossRef]
- Chen, C.; Twycross, J.; Garibaldi, J.M. A New Accuracy Measure Based on Bounded Relative Error for Time Series Forecasting. PLoS ONE 2017, 12, e0174202. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, A.L.W.; Be, N.A.; Mulakken, N.; Nelson, K.L.; Kantor, R.S. Evaluation of the Impact of Concentration and Extraction Methods on the Targeted Sequencing of Human Viruses from Wastewater. Environ. Sci. Technol. 2024, 58, 8239–8250. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Smith, W.J.M.; Metcalfe, S.; Jackson, G.; Choi, P.M.; Morrison, M.; Field, D.; Gyawali, P.; Bivins, A.; Bibby, K.; et al. Comparison of RT-qPCR and RT-dPCR Platforms for the Trace Detection of SARS-CoV-2 RNA in Wastewater. ACS ES T Water 2022, 2, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Belouhova, M.; Peykov, S.; Stefanova, V.; Topalova, Y. Comparison of Two Methods for SARS-CoV-2 Detection in Wastewater: A Case Study from Sofia, Bulgaria. Water 2023, 15, 658. [Google Scholar] [CrossRef]
- Hamza, I.A.; Leifels, M. Assessment of PCR Inhibitor Removal Methods to Monitor Viruses in Environmental Water Samples: DAX-8 Outperforms Competitors. Water Air Soil. Pollut. 2023, 235, 20. [Google Scholar] [CrossRef]
- Assurian, A.; Murphy, H.; Shipley, A.; Cinar, H.N.; Da Silva, A.; Almeria, S. Assessment of Commercial DNA Cleanup Kits for Elimination of Real-Time PCR Inhibitors in the Detection of Cyclospora Cayetanensis in Cilantro. J. Food Prot. 2020, 83, 1863–1870. [Google Scholar] [CrossRef]
- Schriewer, A.; Wehlmann, A.; Wuertz, S. Improving qPCR Efficiency in Environmental Samples by Selective Removal of Humic Acids with DAX-8. J. Microbiol. Methods 2011, 85, 16–21. [Google Scholar] [CrossRef]
- Widjojoatmodjo, M.N.; Fluit, A.C.; Torensma, R.; Verdonk, G.P.; Verhoef, J. The Magnetic Immuno Polymerase Chain Reaction Assay for Direct Detection of Salmonellae in Fecal Samples. J. Clin. Microbiol. 1992, 30, 3195–3199. [Google Scholar] [CrossRef]
- Scipioni, A.; Bourgot, I.; Mauroy, A.; Ziant, D.; Saegerman, C.; Daube, G.; Thiry, E. Detection and Quantification of Human and Bovine Noroviruses by a TaqMan RT-PCR Assay with a Control for Inhibition. Mol. Cell Probes 2008, 22, 215–222. [Google Scholar] [CrossRef]
- Wang, H.; Qi, J.; Xiao, D.; Wang, Z.; Tian, K. A Re-Evaluation of Dilution for Eliminating PCR Inhibition in Soil DNA Samples. Soil Biol. Biochem. 2017, 106, 109–118. [Google Scholar] [CrossRef]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 Wastewater Surveillance in Germany: Long-Term RT-Digital Droplet PCR Monitoring, Suitability of Primer/Probe Combinations and Biomarker Stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef]
- Holcomb, D.A.; Christensen, A.; Hoffman, K.; Lee, A.; Blackwood, A.D.; Clerkin, T.; Gallard-Góngora, J.; Harris, A.; Kotlarz, N.; Mitasova, H.; et al. Estimating Rates of Change to Interpret Quantitative Wastewater Surveillance of Disease Trends. Sci. Total Environ. 2024, 951, 175687. [Google Scholar] [CrossRef] [PubMed]
- Dhiyebi, H.A.; Abu Farah, J.; Ikert, H.; Srikanthan, N.; Hayat, S.; Bragg, L.M.; Qasim, A.; Payne, M.; Kaleis, L.; Paget, C.; et al. Assessment of Seasonality and Normalization Techniques for Wastewater-Based Surveillance in Ontario, Canada. Front. Public Health 2023, 11, 1186525. [Google Scholar] [CrossRef] [PubMed]
- Van Boven, M.; Hetebrij, W.A.; Swart, A.; Nagelkerke, E.; Van Der Beek, R.F.; Stouten, S.; Hoogeveen, R.T.; Miura, F.; Kloosterman, A.; Van Der Drift, A.-M.R.; et al. Patterns of SARS-CoV-2 Circulation Revealed by a Nationwide Sewage Surveillance Programme, the Netherlands, August 2020 to February 2022. Eurosurveillance 2023, 28, 2200700. [Google Scholar] [CrossRef] [PubMed]
- Rauch, W.; Schenk, H.; Insam, H.; Markt, R.; Kreuzinger, N. Data Modelling Recipes for SARS-CoV-2 Wastewater-Based Epidemiology. Environ. Res. 2022, 214, 113809. [Google Scholar] [CrossRef]
- Aberi, P.; Arabzadeh, R.; Insam, H.; Markt, R.; Mayr, M.; Kreuzinger, N.; Rauch, W. Quest for Optimal Regression Models in SARS-CoV-2 Wastewater Based Epidemiology. Ijerph 2021, 18, 10778. [Google Scholar] [CrossRef]
- Hedman, J.; Rådström, P. Overcoming Inhibition in Real-Time Diagnostic PCR. Methods Mol. Biol. 2013, 943, 17–48. [Google Scholar] [CrossRef]
- Opel, K.L.; Chung, D.; McCord, B.R. A Study of PCR Inhibition Mechanisms Using Real Time PCR. J. Forensic Sci. 2010, 55, 25–33. [Google Scholar] [CrossRef]
- Zipper, H.; Buta, C.; Lämmle, K.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Mechanisms Underlying the Impact of Humic Acids on DNA Quantification by SYBR Green I and Consequences for the Analysis of Soils and Aquatic Sediments. Nucleic Acids Res. 2003, 31, e39. [Google Scholar] [CrossRef]
- Sidstedt, M.; Hedman, J.; Romsos, E.L.; Waitara, L.; Wadsö, L.; Steffen, C.R.; Vallone, P.M.; Rådström, P. Inhibition Mechanisms of Hemoglobin, Immunoglobulin G, and Whole Blood in Digital and Real-Time PCR. Anal. Bioanal. Chem. 2018, 410, 2569–2583. [Google Scholar] [CrossRef]
- da Silva, A.K.; Le Saux, J.-C.; Parnaudeau, S.; Pommepuy, M.; Elimelech, M.; Le Guyader, F.S. Evaluation of Removal of Noroviruses during Wastewater Treatment, Using Real-Time Reverse Transcription-PCR: Different Behaviors of Genogroups I and II. Appl. Environ. Microbiol. 2007, 73, 7891–7897. [Google Scholar] [CrossRef]
- Ansari, S.A.; Farrah, S.R.; Chaudhry, G.R. Presence of Human Immunodeficiency Virus Nucleic Acids in Wastewater and Their Detection by Polymerase Chain Reaction. Appl. Environ. Microbiol. 1992, 58, 3984–3990. [Google Scholar] [CrossRef] [PubMed]
- Ijzerman, M.M.; Dahling, D.R.; Fout, G.S. A Method to Remove Environmental Inhibitors Prior to the Detection of Waterborne Enteric Viruses by Reverse Transcription-Polymerase Chain Reaction. J. Virol. Methods 1997, 63, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Mitranescu, A.; Uchaikina, A.; Kau, A.-S.; Stange, C.; Ho, J.; Tiehm, A.; Wurzbacher, C.; Drewes, J.E. Wastewater-Based Epidemiology for SARS-CoV-2 Biomarkers: Evaluation of Normalization Methods in Small and Large Communities in Southern Germany. ACS ES T Water 2022, 2, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.; Kasprzyk-Hordern, B. Future Perspectives of Wastewater-Based Epidemiology: Monitoring Infectious Disease Spread and Resistance to the Community Level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.; Evens, N.; Porter, J.; Walker, D.I. The Inhibition and Variability of Two Different RT-qPCR Assays Used for Quantifying SARS-CoV-2 RNA in Wastewater. Food Environ. Virol. 2023, 15, 71–81. [Google Scholar] [CrossRef]
- Audi, A.; AlIbrahim, M.; Kaddoura, M.; Hijazi, G.; Yassine, H.M.; Zaraket, H. Seasonality of Respiratory Viral Infections: Will COVID-19 Follow Suit? Front. Public Health 2020, 8, 567184. [Google Scholar] [CrossRef]
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, 7, 83–101. [Google Scholar] [CrossRef]
- Prasse, C.; Schlüsener, M.P.; Schulz, R.; Ternes, T.A. Antiviral Drugs in Wastewater and Surface Waters: A New Pharmaceutical Class of Environmental Relevance? Environ. Sci. Technol. 2010, 44, 1728–1735. [Google Scholar] [CrossRef]
- Eryildiz, B.; Yavuzturk Gul, B.; Koyuncu, I. A Sustainable Approach for the Removal Methods and Analytical Determination Methods of Antiviral Drugs from Water/Wastewater: A Review. J. Water Process Eng. 2022, 49, 103036. [Google Scholar] [CrossRef]
- Samal, K.; Mahapatra, S.; Hibzur Ali, M. Pharmaceutical Wastewater as Emerging Contaminants (EC): Treatment Technologies, Impact on Environment and Human Health. Energy Nexus 2022, 6, 100076. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse Transcriptase Droplet Digital PCR Shows High Resilience to PCR Inhibitors from Plant, Soil and Water Samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Jerome, K.R. Digital PCR-An Emerging Technology with Broad Applications in Microbiology. Clin. Chem. 2020, 66, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Zhernov, Y.V.; Kremb, S.; Helfer, M.; Schindler, M.; Harir, M.; Mueller, C.; Hertkorn, N.; Avvakumova, N.P.; Konstantinov, A.I.; Brack-Werner, R.; et al. Supramolecular Combinations of Humic Polyanions as Potent Microbicides with Polymodal Anti-HIV-Activities. New J. Chem. 2017, 41, 212–224. [Google Scholar] [CrossRef]
- Socol, D.C. Clinical Review of Humic Acid as an Antiviral: Leadup to Translational Applications in Clinical Humeomics. Front. Pharmacol. 2022, 13, 1018904. [Google Scholar] [CrossRef]
- Wang, Y.; Morimoto, S.; Ogawa, N.; Oomori, T.; Fujii, T. An Improved Method to Extract RNA from Soil with Efficient Removal of Humic Acids. J. Appl. Microbiol. 2009, 107, 1168–1177. [Google Scholar] [CrossRef]
- Sutlovic, D.; Gamulin, S.; Definis-Gojanovic, M.; Gugic, D.; Andjelinovic, S. Interaction of Humic Acids with Human DNA: Proposed Mechanisms and Kinetics. Electrophoresis 2008, 29, 1467–1472. [Google Scholar] [CrossRef]
- Chatterjee, A.; Zhang, K.; Parker, K.M. Binding of Dissolved Organic Matter to RNA and Protection from Nuclease-Mediated Degradation. Environ. Sci. Technol. 2023, 57, 16086–16096. [Google Scholar] [CrossRef]
- Al-Soud, W.A.; Jönsson, L.J.; Râdström, P. Identification and Characterization of Immunoglobulin G in Blood as a Major Inhibitor of Diagnostic PCR. J. Clin. Microbiol. 2000, 38, 345–350. [Google Scholar] [CrossRef]
- European Commission; Joint Research Centre; Global Water Research Coalition. The International Cookbook for Wastewater Practitioners. Vol. 1, SARS-CoV-2; Publications Office: Luxembourg, 2024. [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. Increasing the Utility of Wastewater-Based Disease Surveillance for Public Health Action: A Phase 2 Report; National Academies Press: Washington, DC, USA, 2024; p. 27516. ISBN 978-0-309-71620-8. [Google Scholar]
- Espinosa-Gongora, C.; Berg, C.; Rehn, M.; Varg, J.E.; Dillner, L.; Latorre-Margalef, N.; Székely, A.J.; Andersson, E.; Movert, E. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage Through Integrated Genomic Surveillance of Wastewater and COVID-19 Cases in Sweden, Weeks 31 to 38 2023. Eurosurveillance 2023, 28, 2300595. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linzner, N.; Bartel, A.; Schumacher, V.; Grau, J.H.; Wyler, E.; Preuß, H.; Garske, S.; Bitzegeio, J.; Kirst, E.B.; Liere, K.; et al. Effective Inhibitor Removal from Wastewater Samples Increases Sensitivity of RT-dPCR and Sequencing Analyses and Enhances the Stability of Wastewater-Based Surveillance. Microorganisms 2024, 12, 2475. https://doi.org/10.3390/microorganisms12122475
Linzner N, Bartel A, Schumacher V, Grau JH, Wyler E, Preuß H, Garske S, Bitzegeio J, Kirst EB, Liere K, et al. Effective Inhibitor Removal from Wastewater Samples Increases Sensitivity of RT-dPCR and Sequencing Analyses and Enhances the Stability of Wastewater-Based Surveillance. Microorganisms. 2024; 12(12):2475. https://doi.org/10.3390/microorganisms12122475
Chicago/Turabian StyleLinzner, Nico, Alexander Bartel, Vera Schumacher, José Horacio Grau, Emanuel Wyler, Henrike Preuß, Sonja Garske, Julia Bitzegeio, Elisabeth Barbara Kirst, Karsten Liere, and et al. 2024. "Effective Inhibitor Removal from Wastewater Samples Increases Sensitivity of RT-dPCR and Sequencing Analyses and Enhances the Stability of Wastewater-Based Surveillance" Microorganisms 12, no. 12: 2475. https://doi.org/10.3390/microorganisms12122475
APA StyleLinzner, N., Bartel, A., Schumacher, V., Grau, J. H., Wyler, E., Preuß, H., Garske, S., Bitzegeio, J., Kirst, E. B., Liere, K., Hoppe, S., Borodina, T. A., Altmüller, J., Landthaler, M., Meixner, M., Sagebiel, D., & Böckelmann, U. (2024). Effective Inhibitor Removal from Wastewater Samples Increases Sensitivity of RT-dPCR and Sequencing Analyses and Enhances the Stability of Wastewater-Based Surveillance. Microorganisms, 12(12), 2475. https://doi.org/10.3390/microorganisms12122475






