A Farm-to-Fork Quantitative Microbial Exposure Assessment of β-Lactam-Resistant Escherichia coli among U.S. Beef Consumers
Abstract
:1. Introduction
2. Materials and Methods
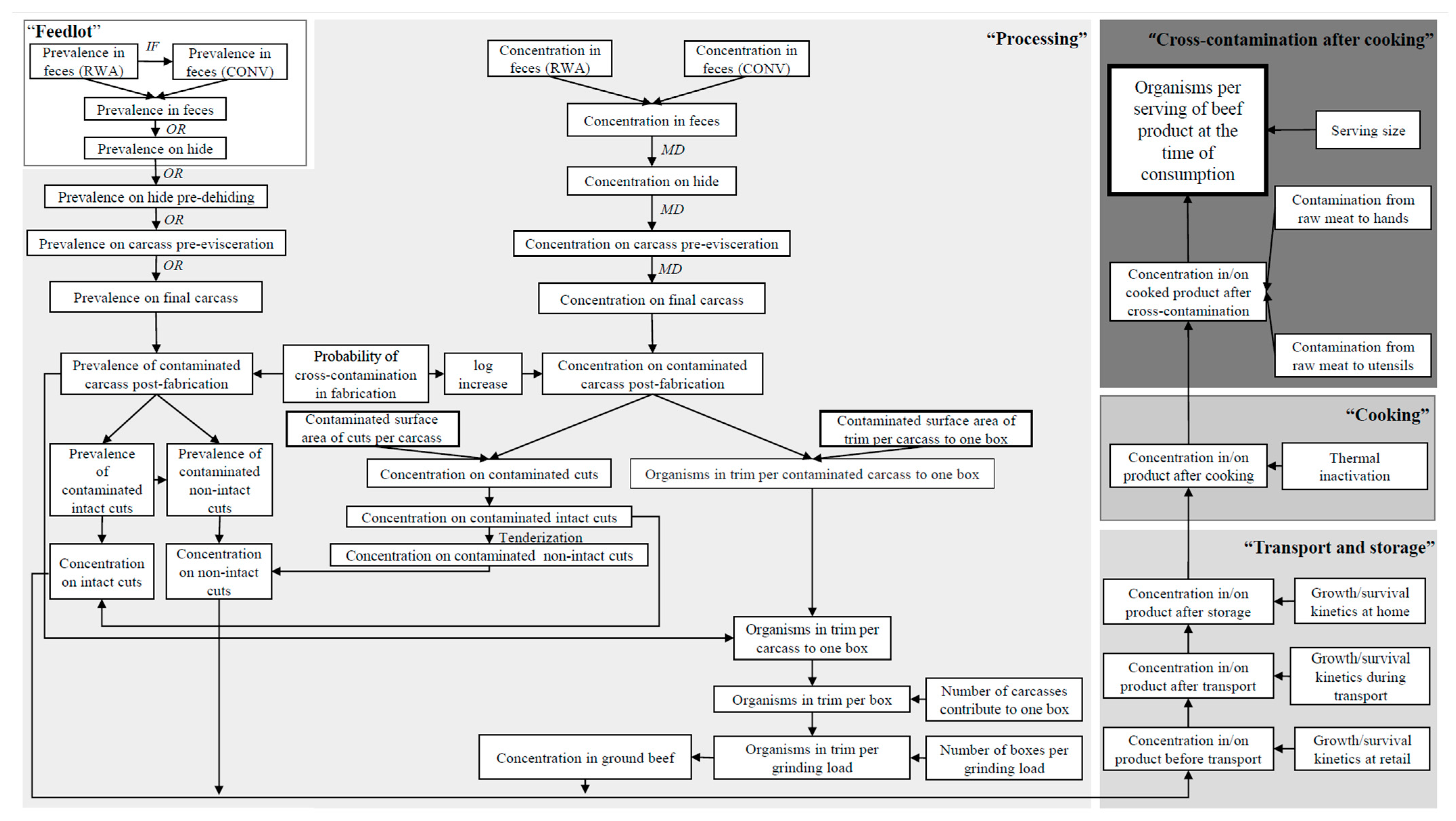
2.1. Model Overview
2.2. Exposure Assessment
2.2.1. Feedlot
Prevalence of BR-EC in Feces at the Feedlot
Prevalence of BR-EC on Hides at the Feedlot
2.2.2. Processing
Composition Variables of Beef Production
BR-EC Contamination on Hides before Dehiding
Primary Processing of Beef Carcasses
Secondary Processing of Beef Products
2.2.3. Transport and Storage
Transport from the Processing Plant to Retail
Retail Storage
Transport from Retail to Home
Home Storage
2.2.4. Cooking
2.2.5. Cross-Contamination after Cooking
2.3. Exposure Estimates of the Baseline Model
2.4. What-If Scenario Analysis
2.5. Sensitivity Analysis
3. Results and Discussion
3.1. Baseline Model Estimates and Validation
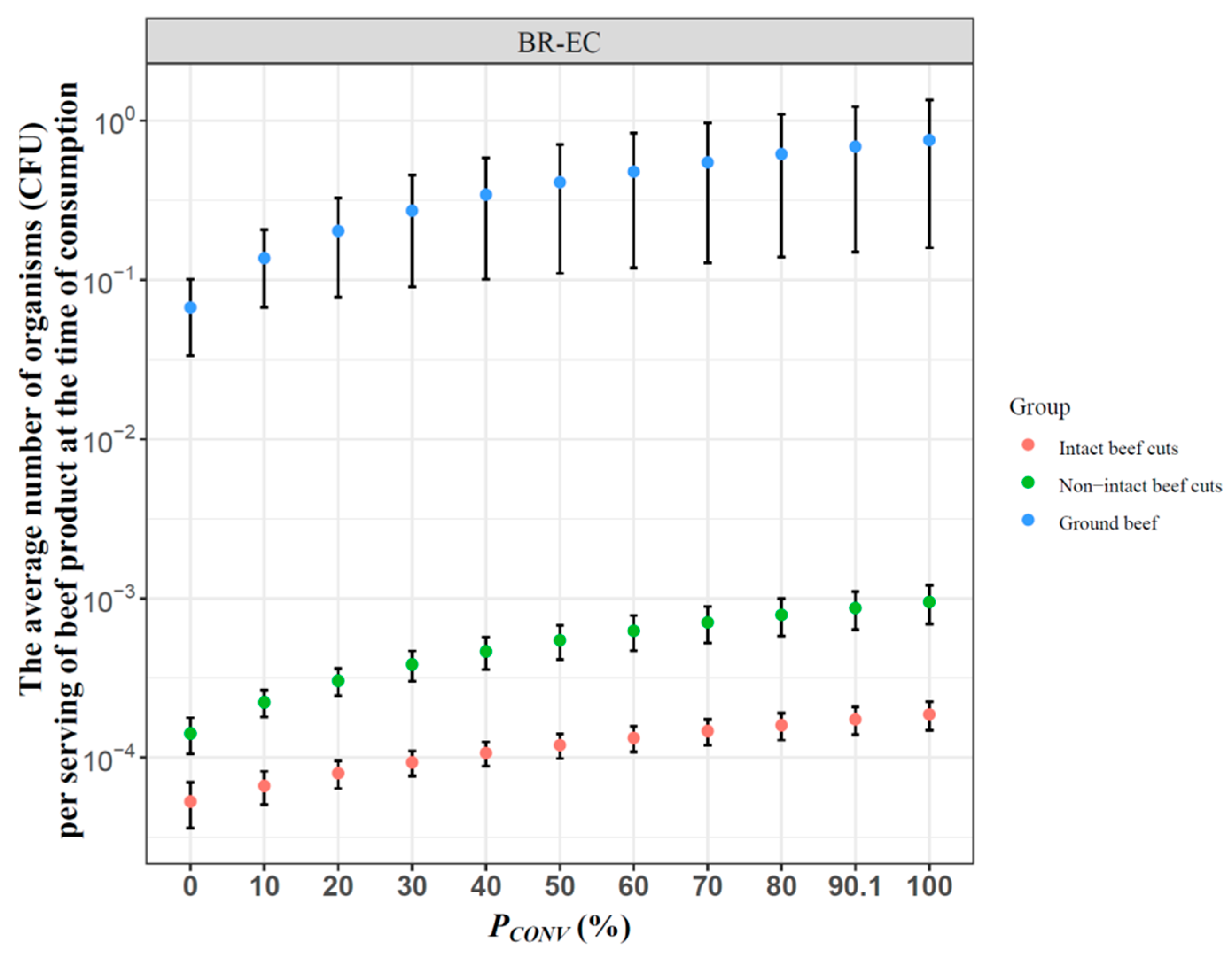
3.2. Effect of Antibiotic Use on Human Exposure to BR-EC via Consumption of Beef Meat
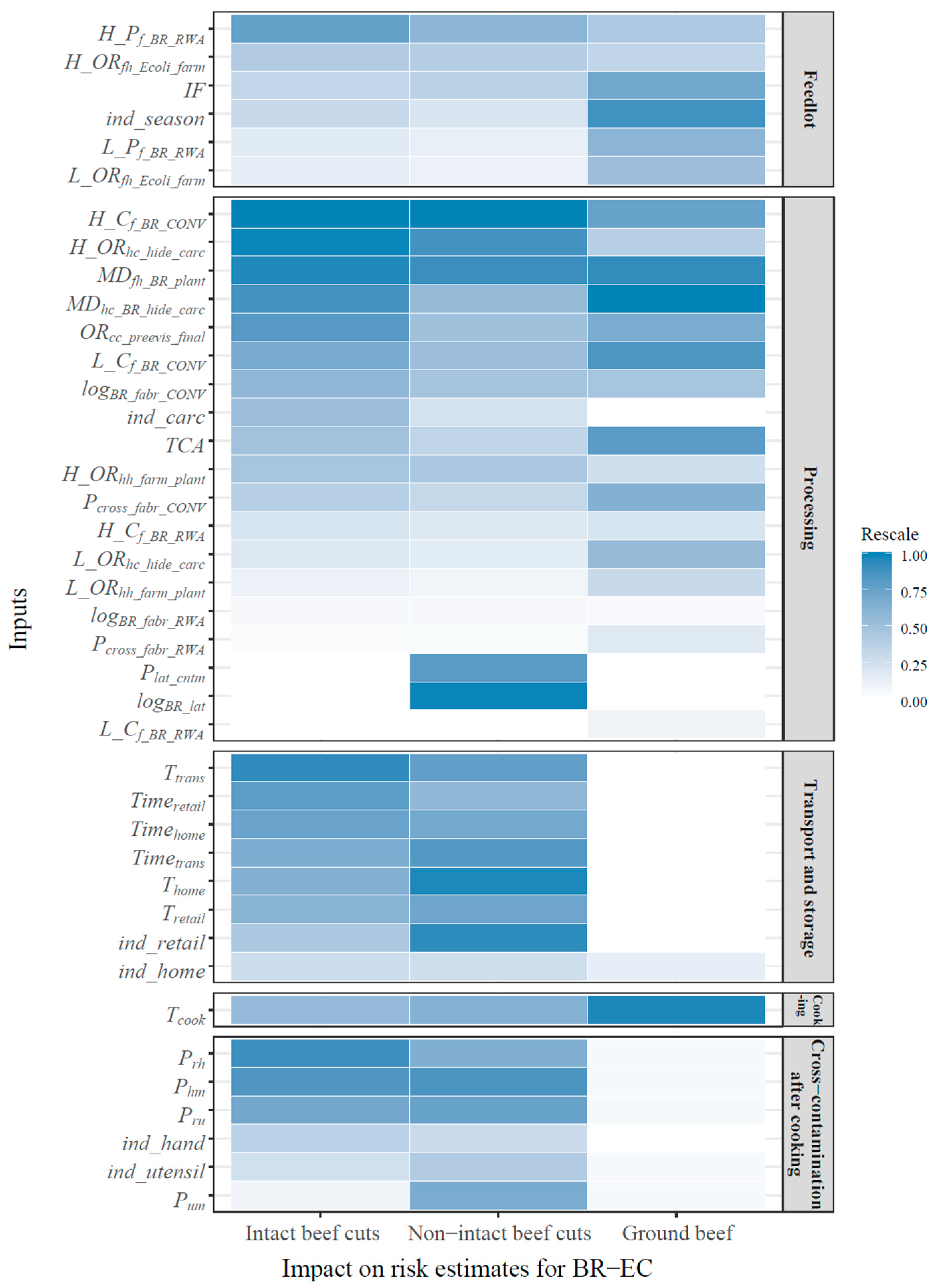
3.3. Significant Factors Controlling Human Exposure to BR-EC
3.4. General Discussion of the Model’s Development
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durso, L.M.; Cook, K.L. Impacts of antibiotic use in agriculture: What are the benefits and risks? Curr. Opin. Microbiol. 2014, 19, 37–44. [Google Scholar] [CrossRef]
- FDA. U.S. National Antimicrobial Resistance Monitoring System (NARMS) Retail Meat Annual Report. 2011. Available online: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM334834.pdf (accessed on 5 May 2018).
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial Drug Resistance in Escherichia coli from Humans and Food Animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Herman, L.; Haesebrouck, F.; Butaye, P. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010, 34, 295–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA. Animal & Veterinary: Cephalosporin Order of Prohibition Questions and Answers. Available online: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/ucm421538.htm (accessed on 7 May 2018).
- Jiang, X.; Yu, T.; Wu, N.; Meng, H.; Shi, L. Detection of qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli isolated from cooked meat products in Henan, China. Int. J. Food Microbiol. 2014, 187, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.W.; Agga, G.E.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Shackelford, S.D.; Wang, R.; Wheeler, T.L.; Arthur, T.M. Occurrence of antimicrobial-resistant Escherichia coli and Salmonella enterica in the beef cattle production and processing continuum. Appl. Environ. Microbiol. 2015, 81, 713–725. [Google Scholar] [CrossRef] [Green Version]
- Vikram, A.; Miller, E.; Arthur, T.M.; Bosilevac, J.M.; Wheeler, T.L.; Schmidt, J.W. Similar levels of antimicrobial resistance in U.S. food service ground beef products with and without a “Raised without Antibiotics” claim. J. Food Prot. 2018, 81, 2007–2018. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Rasheed, M.U.; Thajuddin, N.; Ahamed, P.; Teklemariam, Z.; Jamil, K. Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Rev. Inst. Med. Trop. São Paulo 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Dohoo, I.R.; Martin, S.W.; Stryhn, H. Methods in Epidemiologic Research; VER Inc.: Waldorf, MD, USA, 2012. [Google Scholar]
- USDA. Feedlot 2011 Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1000 or More Head. Available online: https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed11_dr_PartIV.pdf (accessed on 28 July 2018).
- Niyonzima, E.; Ongol, M.P.; Kimonyo, A.; Sindic, M. Risk factors and control measures for bacterial contamination in the bovine meat chain: A review on Salmonella and pathogenic E. coli. J. Food Res. 2015, 4, 98–121. [Google Scholar] [CrossRef]
- USDA-FSIS. Risk Assessment of the Public Health Impact of Escherichia coli O157:H7 in Ground Beef. Available online: https://www.fsis.usda.gov/wps/wcm/connect/1db13d79-1cd9-4e4d-b6ca-16ad89a085a1/00-023NReport.pdf?MOD=AJPERES (accessed on 28 July 2018).
- Vikram, A.; Rovira, P.; Agga, G.E.; Arthur, T.M.; Bosilevac, J.M.; Wheeler, T.L.; Morley, P.S.; Belk, K.E.; Schmidt, J.W. Impact of “raised without antibiotics” beef cattle production practices on occurrences of antimicrobial resistance. Appl. Environ. Microbiol. 2017, 83, e01682-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, B. Comparison of the efficacy of commercial antimicrobial interventions for reducing antibiotic resistant and susceptible beef-associated Salmonella and Escherichia coli strains. J. Consum. Prot. Food Saf. 2018, 13, 3–23. [Google Scholar] [CrossRef]
- Smith, B.A.; Fazil, A.; Lammerding, A.M. A risk assessment model for Escherichia coli O157:H7 in ground beef and beef cuts in Canada: Evaluating the effects of interventions. Food Control 2013, 29, 364–381. [Google Scholar] [CrossRef] [Green Version]
- Woerner, D.R.; Ransom, J.R.; Sofos, J.N.; Dewell, G.A.; Smith, G.C.; Salman, M.D.; Belk, K.E. Determining the prevalence of Escherichia coli O157 in cattle and beef from the feedlot to the cooler. J. Food Prot. 2006, 69, 2824–2827. [Google Scholar] [CrossRef] [PubMed]
- USDA-FSIS. Comparative Risk Assessment for Intact (Non-Tenderized) and Non-Intact (Tenderized) Beef: Technical Report. Available online: https://www.fsis.usda.gov/shared/PDF/Beef_Risk_Assess_Report_Mar2002.pdf (accessed on 7 August 2018).
- Huang, L.; Sheen, S. Quantitative analysis of vertical translocation and lateral cross-contamination of Escherichia coli O157:H7 during mechanical tenderization of beef. J. Food Saf. 2011, 31, 108–114. [Google Scholar] [CrossRef]
- Zhang, Y.; Schmidt, J.W.; Arthur, T.M.; Wheeler, T.L.; Wang, B. A comparative quantitative assessment of human exposure to various antimicrobial-resistant bacteria among US ground beef consumers. J. Food Prot. 2021, 84, 736–759. [Google Scholar] [CrossRef]
- Tamplin, M.L.; Paoli, G.; Marmer, B.S.; Phillips, J. Models of the behavior of Escherichia coli O157:H7 in raw sterile ground beef stored at 5 to 46 °C. Int. J. Food Microbiol. 2005, 100, 335–344. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Nastasijević, I.; Lakićević, B.; Petrović, Z. Cold Chain Management in Meat Storage, Distribution and Retail: A Review. In Proceedings of the 59th International Meat Industry Conference MEATCON2017, Zlatibor, Serbia, 1–4 October 2017; Available online: http://iopscience.iop.org/article/10.1088/1755-1315/85/1/012022/pdf (accessed on 7 August 2018).
- Jakubowski, T. Temperature monitoring in the transportation of meat products. J. Food Process. Technol. 2015, 6, 1000502. [Google Scholar] [CrossRef]
- EcoSure. 2007 U.S. Colding Temperature Data of Fresh Meat Products. Available online: http://foodrisk.org/resources/display/21 (accessed on 8 August 2018).
- Egan, A.F.; Eustace, I.J.; Shay, B.J. Meat packaging—Maintaining the quality and prolonging the storage life of chilled beef, pork and lamb. In Proceedings of the Industry Day: Part of the 34th International Congress of Meat Science and Technology, Brisbane, Australia, 29 August–2 September 1988; pp. 68–75. [Google Scholar]
- Godwin, S.; Coppings, R. Analysis of consumer food-handling practices from grocer to home including transport and storage of selected foods. J. Food Distrib. Res. 2005, 36, 55–62. [Google Scholar]
- Smadi, H.; Sargeant, J.M. Quantitative risk assessment of human salmonellosis in Canadian broiler chicken breast from retail to consumption. Risk Anal. 2013, 33, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Christensen, E. What’s the Right Amount of Meat to Serve per Person? Available online: https://www.thekitchn.com/whats-the-right-amount-of-meat-to-serve-per-person-112577 (accessed on 8 August 2018).
- USDA. Nutrition and Your Health: Dietary Guidelines for Americans. Available online: https://health.gov/dietaryguidelines/dga2000/dietgd.pdf (accessed on 8 August 2018).
- Vose, D. Risk Analysis: A Quantitative Guide, 3rd ed.; John Wiley & Sons: New York, NY, USA, 2008. [Google Scholar]
- Nekouei, O.; Checkley, S.; Waldner, C.; Smith, B.A.; Invik, J.; Carson, C.; Avery, B.; Sanchez, J.; Gow, S. Exposure to antimicrobial-resistant Escherichia coli through the consumption of ground beef in Western Canada. Int. J. Food Microbiol. 2018, 272, 41–48. [Google Scholar] [CrossRef] [PubMed]
- FAO; WHO. Microbiological Risk Assessment–Guidance for food; Food and Agriculture Organization: Rome, Italy, 2021; Volume 36. [Google Scholar]
- Evers, E.G.; Pielaat, A.; Smid, J.H.; van Duijkeren, E.; Vennemann, F.B.C.; Wijnands, L.M.; Chardon, J.E. Comparative exposure assessment of ESBL-producing Escherichia coli through meat consumption. PLoS ONE 2017, 12, e0169589. [Google Scholar] [CrossRef]
- Heiman, K.E.; Mody, R.K.; Johnson, S.D.; Griffin, P.M.; Gould, L.H. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 2015, 21, 1293–1301. [Google Scholar] [CrossRef]
- Barlow, R.S.; Gobius, K.S.; Desmarchelier, P.M. Shiga toxin-producing Escherichia coli in ground beef and lamb cuts: Results of a one-year study. Int. J. Food Microbiol. 2006, 111, 1–5. [Google Scholar] [CrossRef]
- Johns, D.F.; Bratcher, C.L.; Kerth, C.R.; McCaskey, T. Translocation of surface-inoculated Escherichia coli into whole muscle nonintact beef striploins following blade tenderization. J. Food Prot. 2011, 74, 1334–1337. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457. [Google Scholar] [CrossRef]
- Nesme, J.; Cécillon, S.; Delmont, O.T.; Monier, J.-M.; Vogel, M.T.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef] [Green Version]
- Hernández, J.; González-Acuña, D. Anthropogenic antibiotic resistance genes mobilization to the polar regions. Infect. Ecol. Epidemiol. 2016, 6, 32112. [Google Scholar] [CrossRef] [Green Version]
- Alexander, T.W.; Inglis, G.D.; Yanke, L.J.; Topp, E.; Read, R.R.; Reuter, T.; McAllister, T.A. Farm-to-fork characterization of Escherichia coli associated with feedlot cattle with a known history of antimicrobial use. Int. J. Food Microbiol. 2010, 137, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Guarddon, M.; Miranda, J.M.; Rodríguez, J.A.; Vázquez, B.I.; Cepeda, A.; Franco, C.M. Quantitative detection of tetracycline-resistant microorganisms in conventional and organic beef, pork and chicken meat. CyTA J. Food 2014, 12, 383–388. [Google Scholar] [CrossRef]
- Muriana, P.M.; Eager, J.; Wellings, B.; Morgan, B.; Nelson, J.; Kushwaha, K. Evaluation of antimicrobial interventions against E. coli O157:H7 on the surface of raw beef to reduce bacterial translocation during blade tenderization. Foods 2019, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Signorini, M.; Costa, M.; Teitelbaum, D.; Restovich, V.; Brasesco, H.; García, D.; Superno, V.; Petroli, S.; Bruzzone, M.; Arduini, V.; et al. Evaluation of decontamination efficacy of commonly used antimicrobial interventions for beef carcasses against Shiga toxin-producing Escherichia coli. Meat Sci. 2018, 142, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.L.; Kalchayanand, N.; Bosilevac, J.M. Pre- and post-harvest interventions to reduce pathogen contamination in the U.S. beef industry. Meat Sci. 2014, 98, 372–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolhouse, M.E.J.; Ward, M.J. Sources of Antimicrobial Resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- Chapman, B.; Pintar, K.; Smith, B.A. Multi-exposure pathway model to compare Escherichia coli O157 risks and interventions. Risk Anal. 2018, 38, 392–409. [Google Scholar] [CrossRef]
- Swart, A.N.; van Leusden, F.; Nauta, M.J. A QMRA model for Salmonella in pork products during preparation and consumption. Risk Anal. 2016, 36, 516–530. [Google Scholar] [CrossRef]
- Nauta, M.J. Separation of uncertainty and variability in quantitative microbial risk assessment models. Int. J. Food Microbiol. 2000, 57, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Aiassa, E.; Higgins, J.P.T.; Frampton, G.K.; Greiner, M.; Afonso, A.; Amzal, B.; Deeks, J.; Dorne, J.L.; Glanville, J.; Lövei, G.L.; et al. Applicability and feasibility of systematic review for performing evidence-based risk assessment in food and feed safety. Crit. Rev. Food Sci. Nutr. 2015, 55, 1026–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkana, E.; Chorianopoulos, N.; Grounta, A.; Koutsoumanis, K.; Nychas, G.J.E. Effect of inoculum size, bacterial species, type of surfaces and contact time to the transfer of foodborne pathogens from inoculated to non-inoculated beef fillets via food processing surfaces. Food Microbiol. 2017, 62, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Agga, G.E.; Schmidt, J.W.; Arthur, T.M. Antimicrobial-resistant fecal bacteria from ceftiofur-treated and nonantimicrobial-treated comingled beef cows at a cow–calf operation. Microb. Drug Resist. 2016, 22, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.W.; Yanke, L.J.; Topp, E.; Olson, M.E.; Read, R.R.; Morck, D.W.; McAllister, T.A. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Appl. Environ. Microbiol. 2008, 74, 4405–4416. [Google Scholar] [CrossRef] [Green Version]
- Berge, A.C.B.; Moore, D.A.; Sischo, W.M. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl. Environ. Microbiol. 2006, 72, 3872–3878. [Google Scholar] [CrossRef] [Green Version]
- Checkley, S.L.; Campbell, J.R.; Chirino-Trejo, M.; Janzen, E.D.; Waldner, C.L. Associations between antimicrobial use and the prevalence of antimicrobial resistance in fecal Escherichia coli from feedlot cattle in western Canada. Can. Vet. J. 2010, 51, 853–861. [Google Scholar]
- Cho, S.; Bender, J.B.; Diez-Gonzalez, F.; Fossler, C.P.; Hedberg, C.W.; Kaneene, J.B.; Ruegg, P.L.; Warnick, L.D.; Wells, S.J. Prevalence and characterization of Escherichia coli O157 isolates from Minnesota dairy farms and county fairs. J. Food Prot. 2006, 69, 252–259. [Google Scholar] [CrossRef]
- Cho, S.; Diez-Gonzalez, F.; Fossler, C.P.; Wells, S.J.; Hedberg, C.W.; Kaneene, J.B.; Ruegg, P.L.; Warnick, L.D.; Bender, J.B. Prevalence of shiga toxin-encoding bacteria and shiga toxin-producing Escherichia coli isolates from dairy farms and county fairs. Vet. Microbiol. 2006, 118, 289–298. [Google Scholar] [CrossRef]
- Cho, S.; Fossler, C.P.; Diez-Gonzalez, F.; Wells, S.J.; Hedberg, C.W.; Kaneene, J.B.; Ruegg, P.L.; Warnick, L.D.; Bender, J.B. Antimicrobial susceptibility of shiga toxin-producing Escherichia coli isolated from organic dairy farms, conventional dairy farms, and county fairs in Minnesota. Foodborne Pathog. Dis. 2007, 4, 178–186. [Google Scholar] [CrossRef]
- Lowrance, T.C.; Loneragan, G.H.; Kunze, D.J.; Platt, T.M.; Ives, S.E.; Scott, H.M.; Norby, B.; Echeverry, A.; Brashears, M.M. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 2007, 68, 501–507. [Google Scholar] [CrossRef]
- Sato, K.; Bartlett, P.C.; Saeed, M.A. Antimicrobial susceptibility of Escherichia coli isolates from dairy farms using organic versus conventional production methods. J. Am. Vet. Med. Assoc. 2005, 226, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Munns, K.; Alexander, T.; Entz, T.; Mirzaagha, P.; Yanke, L.J.; Mulvey, M.; Topp, E.; McAllister, T. Diversity and distribution of commensal fecal Escherichia coli bacteria in beef cattle administered selected subtherapeutic antimicrobials in a feedlot setting. Appl. Environ. Microbiol. 2008, 74, 6178–6186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walk, S.T.; Mladonicky, J.M.; Middleton, J.A.; Heidt, A.J.; Cunningham, J.R.; Bartlett, P.; Sato, K.; Whittam, T.S. Influence of antibiotic selection on genetic composition of Escherichia coli populations from conventional and organic dairy farms. Appl. Environ. Microbiol. 2007, 73, 5982–5989. [Google Scholar] [CrossRef] [Green Version]
- Arthur, T.M.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Guerini, M.N.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 2007, 70, 280–286. [Google Scholar] [CrossRef] [Green Version]
- Dewell, G.A.; Simpson, C.A.; Dewell, R.D.; Hyatt, D.R.; Belk, K.E.; Scanga, J.A.; Morley, P.S.; Grandin, T.; Smith, G.C.; Dargatz, D.A.; et al. Impact of transportation and lairage on hide contamination with Escherichia coli O157 in finished beef cattle. J. Food Prot. 2008, 71, 1114–1118. [Google Scholar] [CrossRef]
- Fluckey, W.M.; Loneragan, G.H.; Warner, R.; Brashears, M.M. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 2007, 70, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Ransom, J.; Belk, K. Investigation of on-Farm Management Practices as Pre-Harvest Beef Microbiological Interventions. Available online: https://www.beefresearch.org/CMDocs/BeefResearch/Safety_Project_Summaries/FY02_Investigation_of_On_Farm_Management_Practices.pdf (accessed on 30 July 2018).
- Stephens, T.P.; Loneragan, G.H.; Karunasena, E.; Brashears, M.M. Reduction of Escherichia coli O157 and Salmonella in feces and on hides of feedlot cattle using various doses of a direct-fed microbial. J. Food Prot. 2007, 70, 2386–2391. [Google Scholar] [CrossRef]
- Wells, J.E.; Shackelford, S.D.; Berry, E.D.; Kalchayanand, N.; Guerini, M.N.; Varel, V.H.; Arthur, T.M.; Bosilevac, J.M.; Freetly, H.C.; Wheeler, T.L.; et al. Prevalence and level of Escherichia coli O157:H7 in feces and on hides of feedlot steers fed diets with or without wet distillers grains with solubles. J. Food Prot. 2009, 72, 1624–1633. [Google Scholar] [CrossRef]
- USDA. Livestock Slaughter 2017 Summary. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/r207tp32d/cn69m6457/pc289m639/LiveSlauSu-04-18-2018.pdf (accessed on 5 August 2018).
- AGWEB. Dairy Cattle Beef Up Beef Industry. Available online: https://www.agweb.com/article/dairy-cattle-beef-up-beef-industry-wyatt-bechtel/ (accessed on 30 July 2018).
- Nold, R. How Much Meat Can you Expect from a Fed Steer? Available online: http://igrow.org/livestock/beef/how-much-meat-can-you-expect-from-a-fed-steer/ (accessed on 5 August 2018).
- Reddish, R.L. Cutting the Pork Carcass. University of Florida IFAS Extension, 2003. Available online: https://www.yumpu.com/en/document/read/21968387/cutting-the-pork-carcass-edis-university-of-florida (accessed on 11 March 2022).
- Kalchayanand, N.; Brichta-Harhay, D.M.; Arthur, T.M.; Bosilevac, J.M.; Guerini, M.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M. Prevalence rates of Escherichia coli O157:H7 and Salmonella at different sampling sites on cattle hides at a feedlot and processing plant. J. Food Prot. 2009, 72, 1267–1271. [Google Scholar] [CrossRef] [Green Version]
- Bosilevac, J.M.; Arthur, T.M.; Bono, J.L.; Brichta-Harhay, D.M.; Kalchayanand, N.; King, D.A.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Prevalence and enumeration of Escherichia coli O157:H7 and Salmonella in U.S. abattoirs that process fewer than 1000 head of cattle per day. J. Food Prot. 2009, 72, 1272–1278. [Google Scholar] [CrossRef]
- Brichta-Harhay, D.M.; Arthur, T.M.; Bosilevac, J.M.; Guerini, M.N.; Kalchayanand, N.; Koohmaraie, M. Enumeration of Salmonella and Escherichia coli O157:H7 in ground beef, cattle carcass, hide and faecal samples using direct plating methods†. J. Appl. Microbiol. 2007, 103, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Rahman, S.M.E.; Purev, U.; Oh, D.-H. Modelling of Escherichia coli O157:H7 growth at various storage temperatures on beef treated with electrolyzed oxidizing water. J. Food Eng. 2010, 97, 497–503. [Google Scholar] [CrossRef]
- Bogard, A.K.; Fuller, C.C.; Radke, V.; Selman, C.A.; Smith, K.E. Ground beef handling and cooking practices in restaurants in eight states. J. Food Prot. 2013, 76, 2132–2140. [Google Scholar] [CrossRef] [PubMed]
- Brichta-Harhay, D.M.; GUERINI, M.N.; Arthur, T.M.; Bosilevac, J.M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Salmonella and Escherichia coli O157:H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: An evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl. Environ. Microbiol. 2008, 74, 6289–6297. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, W.V. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions; Version 6.0 (Updated July 2019); Available online: www.training.cochrane.org/handbook (accessed on 11 March 2022).
- Hedges, L.V. A random effects model for effect sizes. Psychol. Bull. 1983, 93, 388–395. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducing meta-analyses in R with the metafor package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Schmidt, J.W.; Arthur, T.M.; Wheeler, T.L.; Zhang, Q.; Wang, B. A Farm-to-Fork Quantitative Microbial Exposure Assessment of β-Lactam-Resistant Escherichia coli among U.S. Beef Consumers. Microorganisms 2022, 10, 661. https://doi.org/10.3390/microorganisms10030661
Zhang Y, Schmidt JW, Arthur TM, Wheeler TL, Zhang Q, Wang B. A Farm-to-Fork Quantitative Microbial Exposure Assessment of β-Lactam-Resistant Escherichia coli among U.S. Beef Consumers. Microorganisms. 2022; 10(3):661. https://doi.org/10.3390/microorganisms10030661
Chicago/Turabian StyleZhang, Yangjunna, John W. Schmidt, Terrance M. Arthur, Tommy L. Wheeler, Qi Zhang, and Bing Wang. 2022. "A Farm-to-Fork Quantitative Microbial Exposure Assessment of β-Lactam-Resistant Escherichia coli among U.S. Beef Consumers" Microorganisms 10, no. 3: 661. https://doi.org/10.3390/microorganisms10030661
APA StyleZhang, Y., Schmidt, J. W., Arthur, T. M., Wheeler, T. L., Zhang, Q., & Wang, B. (2022). A Farm-to-Fork Quantitative Microbial Exposure Assessment of β-Lactam-Resistant Escherichia coli among U.S. Beef Consumers. Microorganisms, 10(3), 661. https://doi.org/10.3390/microorganisms10030661






