Photoinactivation of Phage Phi6 as a SARS-CoV-2 Model in Wastewater: Evidence of Efficacy and Safety
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Bacterial Strain and Growth Conditions
2.3. Phage ϕ6 Preparation and Enrichment
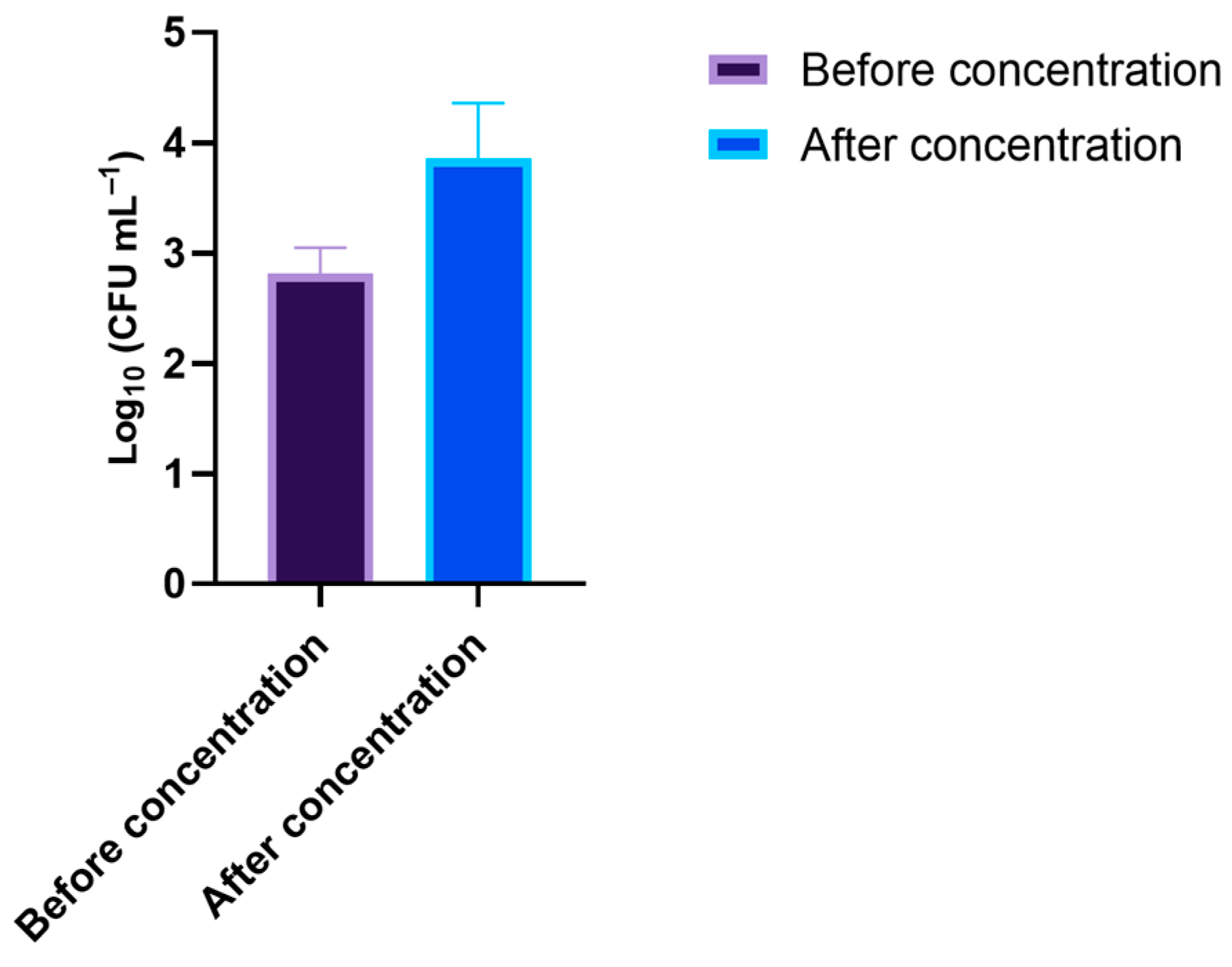
2.4. Wastewater Sample Collection
2.5. Phage φ6 Survival Assessment under Different Environmental Conditions
2.5.1. Temperature Experiments
2.5.2. pH Experiments
2.5.3. Salinity Experiments
2.5.4. UV-B Irradiation Experiments
2.5.5. Solar Radiation Experiments
2.6. Photodynamic Inactivation (PDI) Treatments
2.7. Effect of the PDI-Treated Effluent on Native Marine Water Microorganisms
- (i).
- non-irradiated control of the native marine microorganism concentrate (DC marine water);
- (ii).
- irradiated (50 mW cm−2) control of the native marine microorganism concentrate (LC marine water);
- (iii).
- non-irradiated controls of filtered WW added to the native marine microorganism concentrate in the ratios of 1:2, 1:10, 1:100, and 1:1000 (WW: native marine microorganism concentrate), DC-diluted;
- (iv).
- Irradiated (50 mW cm−2) controls of filtered WW added to the native marine microorganism concentrate in the ratios of 1:2, 1:10, 1:100, and 1:1000 (WW: native marine microorganism concentrate), LC-diluted;
- (v).
- non-irradiated samples with PDI-treated filtered WW added to native marine microorganism concentrate in the ratios of 1:2, 1:10, 1:100, and 1:1000 (WW: native marine microorganism concentrate), S (dark);
- (vi).
- irradiated samples (50 mW cm−2) with PDI-treated filtered WW added to native marine microorganism concentrate in the ratios of 1:2, 1:10, 1:100, and 1:1000 (WW: native marine microorganism concentrate), S (light).
2.8. Statistical Analyses
3. Results
3.1. Assessment of the Effect of Environmental Factors on Phage φ6 Viability
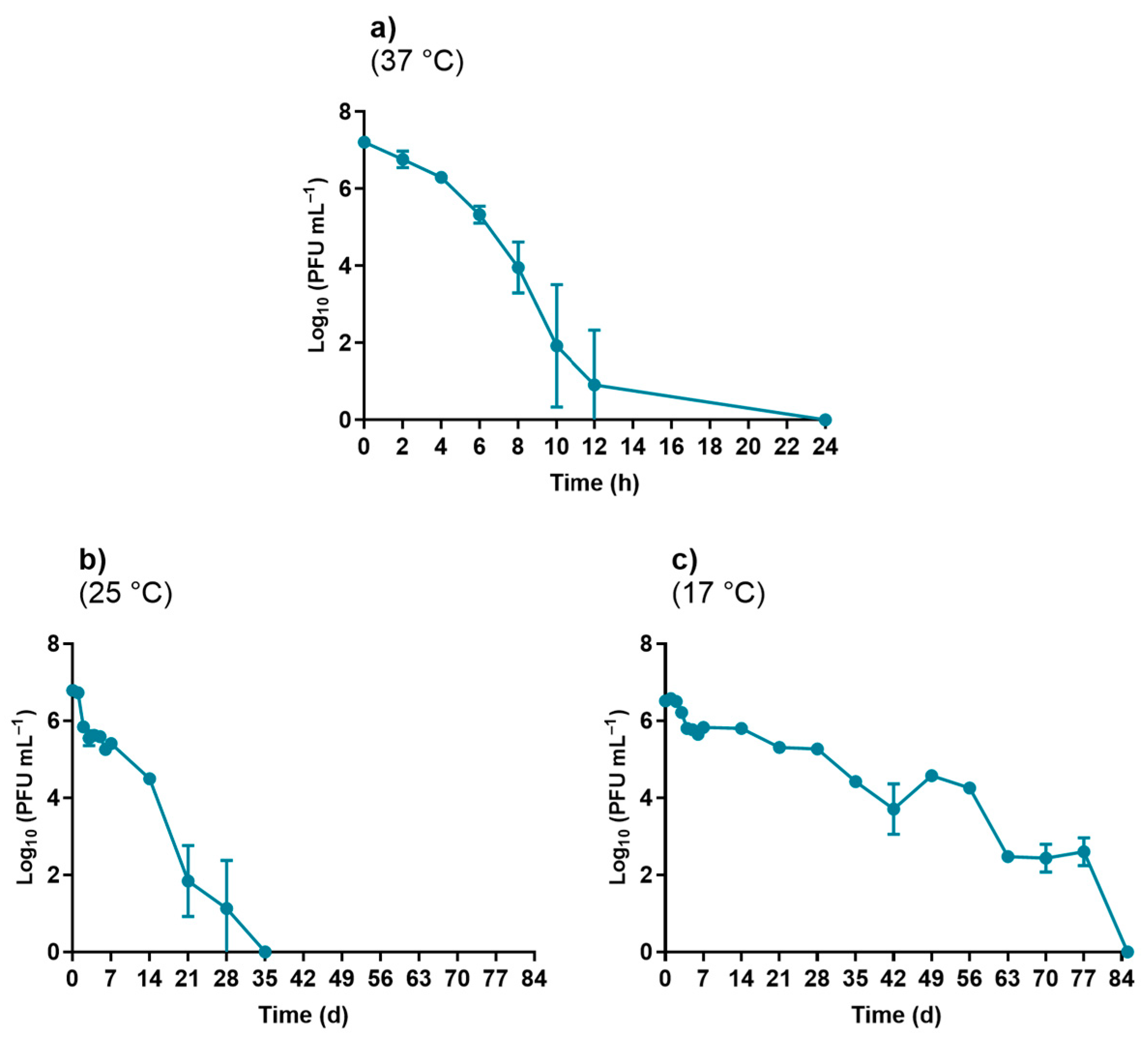
3.1.1. Temperature Experiments
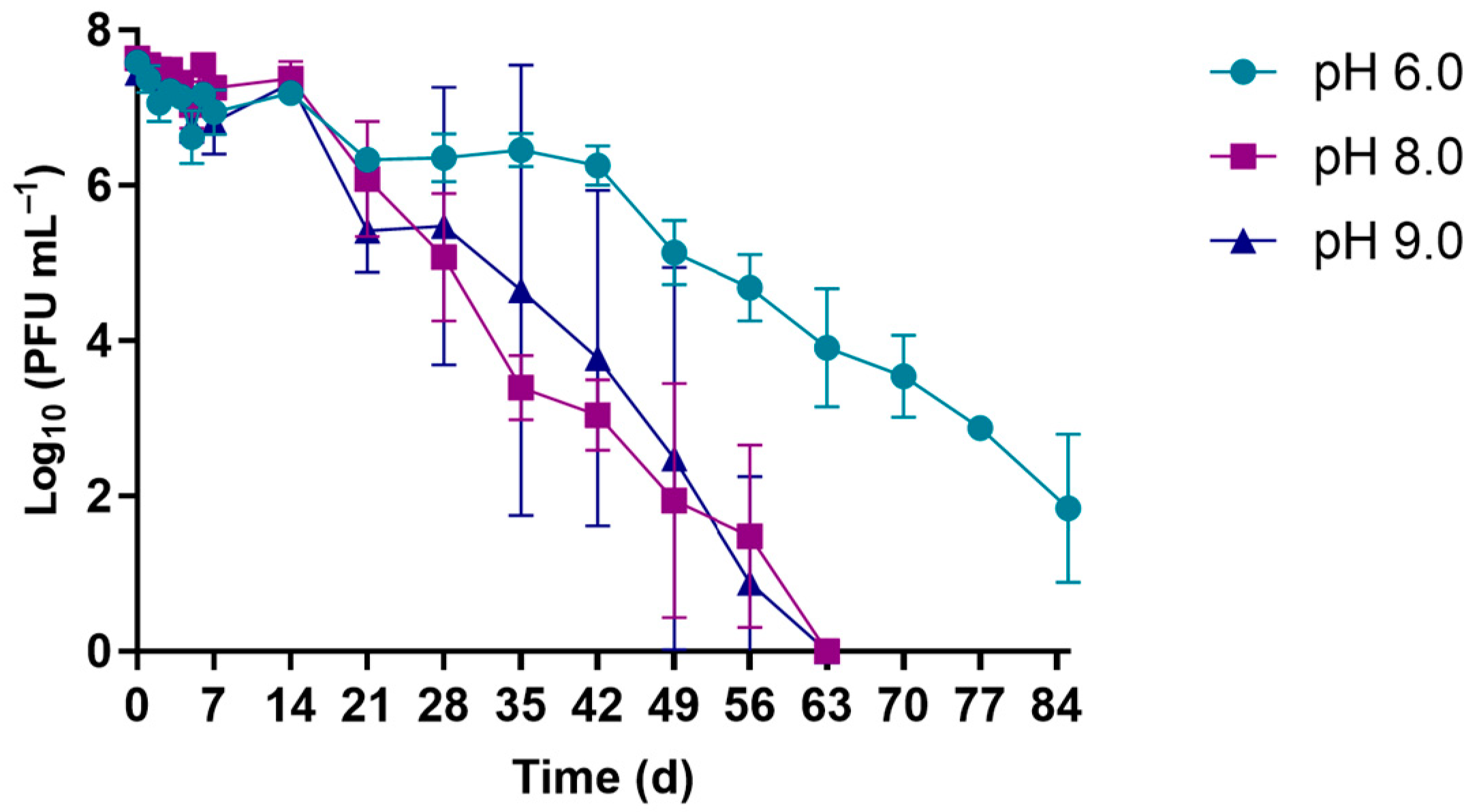
3.1.2. pH Experiments
3.1.3. Salinity Experiments
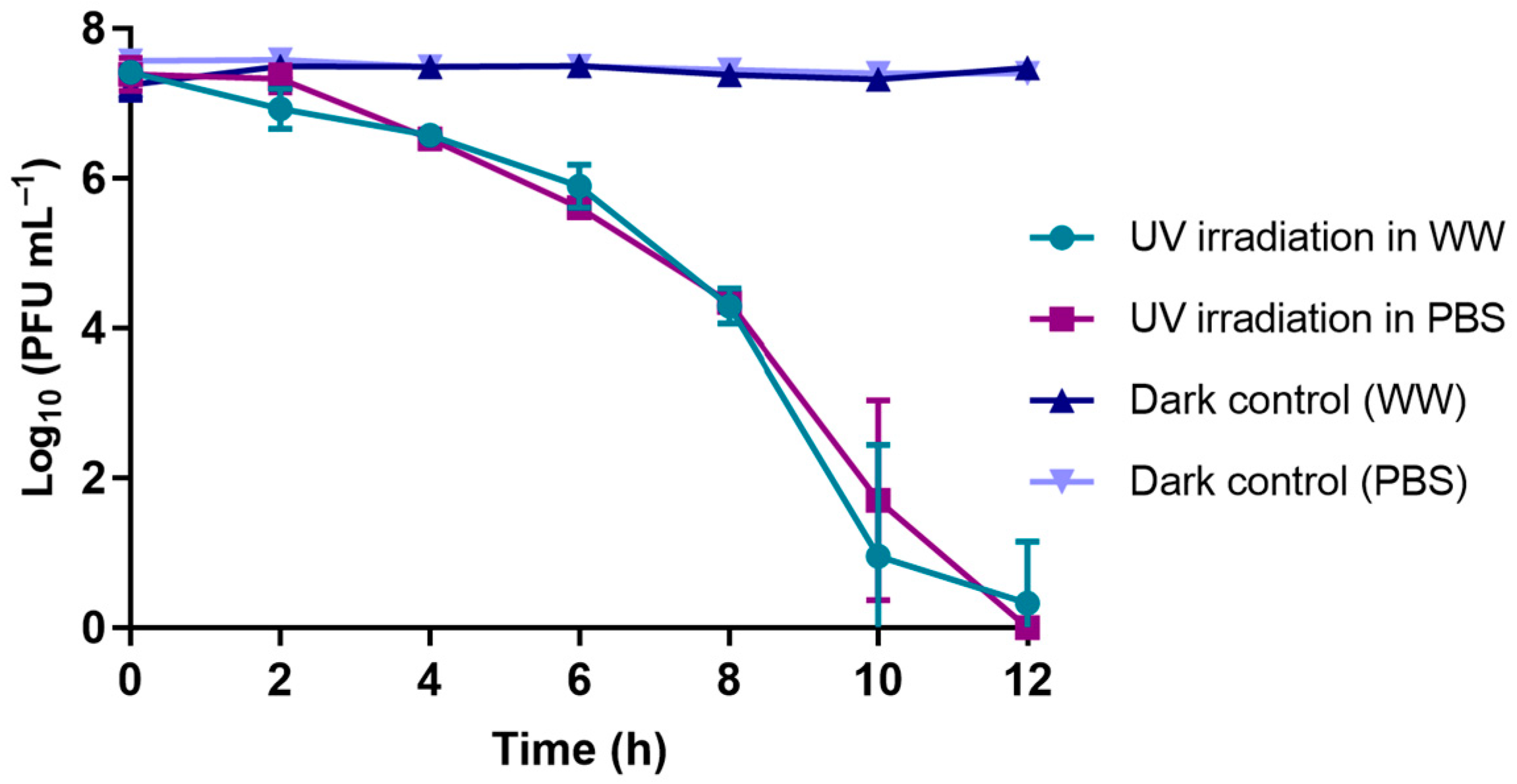
3.1.4. UV-B Exposure Experiments
3.1.5. Solar Radiation Experiments
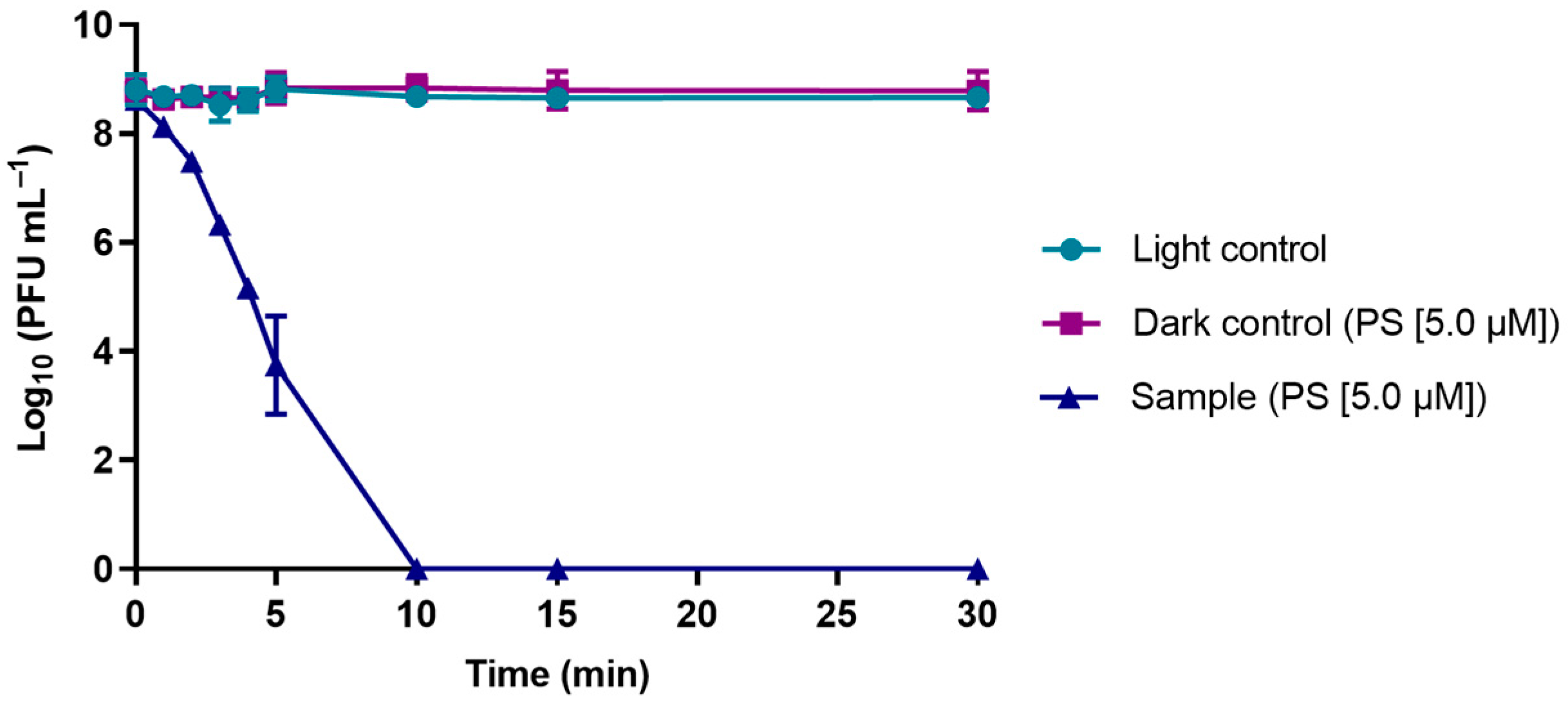
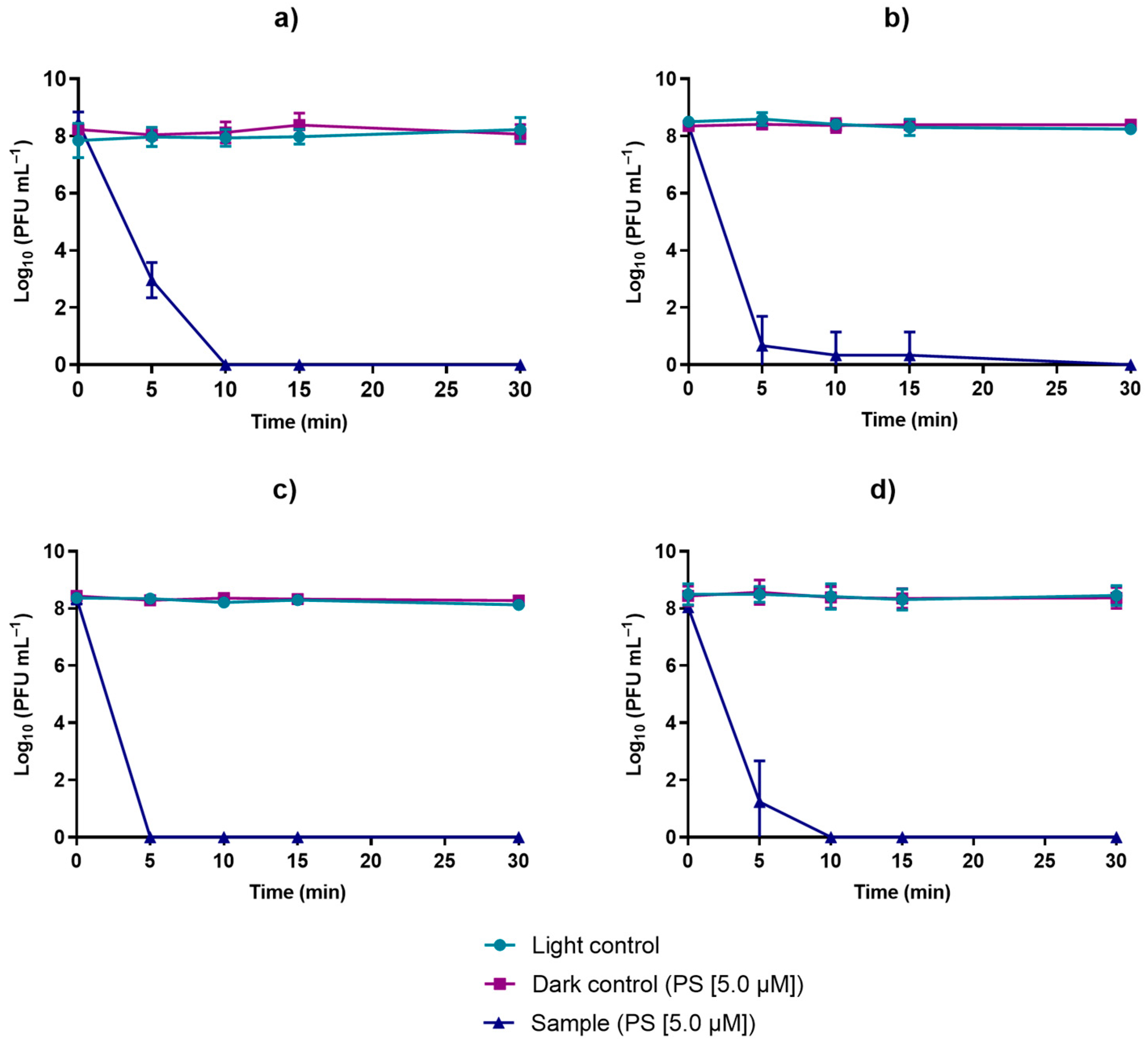
3.2. Evaluation of the Viability of Phage φ6 after PDI in the Presence of TetraPy(+)Me
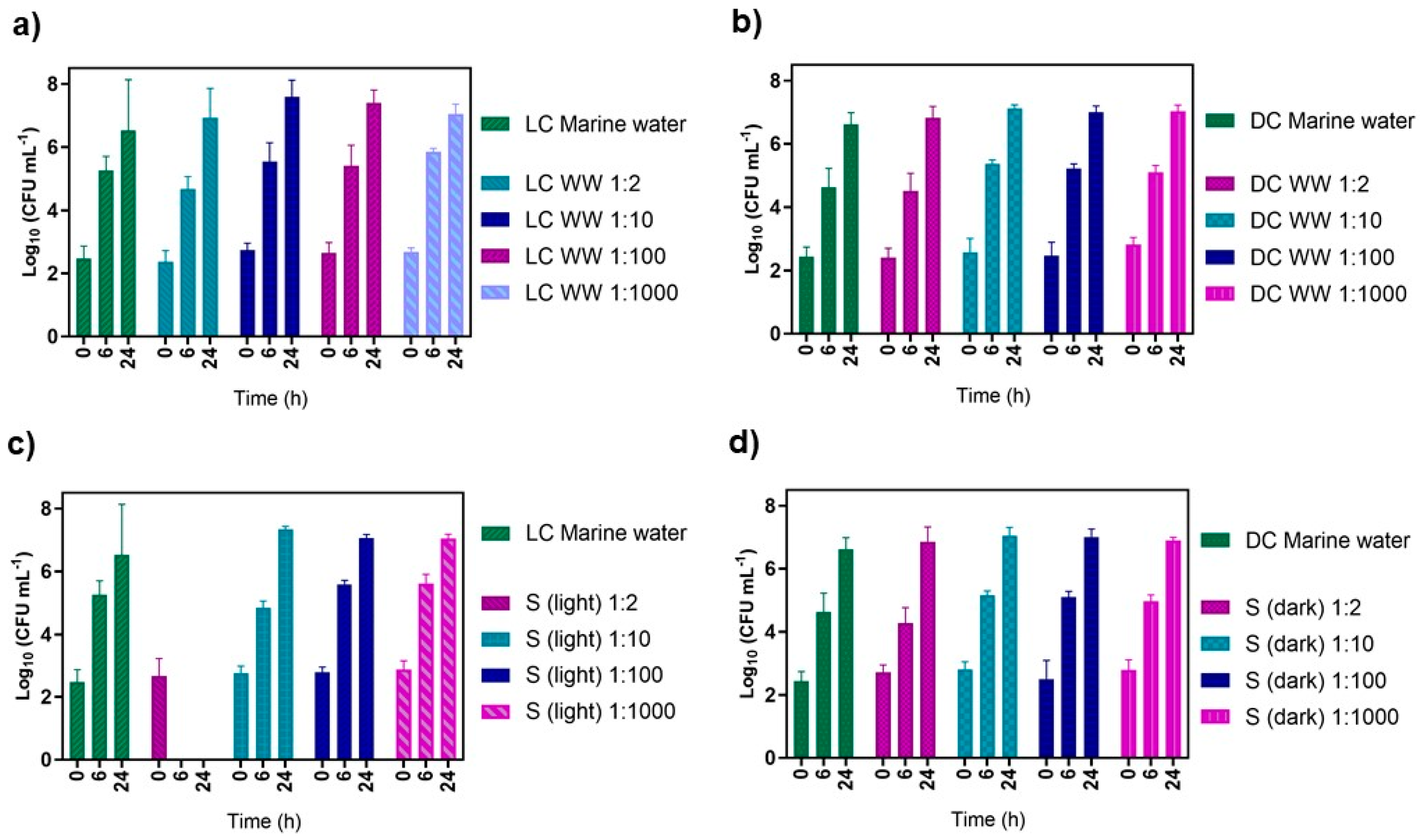
3.3. Effect of the PDI-Treated Effluent on Cultivable Native Marine Water Microorganisms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neher, R.A.; Dyrdak, R.; Druelle, V.; Hodcroft, E.B.; Albert, J. Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. Swiss Med. Wkly. 2020, 150, w20224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, S. Family Coronaviridae. In Viruses; Academic Press: Cambridge, MA, USA, 2017; pp. 140–158. [Google Scholar]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Duran, S.S.F.; Lim, W.Y.S.; Tan, C.K.I.; Cheong, W.C.D.; Suwardi, A.; Loh, X.J. SARS-CoV-2 in wastewater: From detection to evaluation. Mater. Today Adv. 2022, 13, 100211. [Google Scholar] [CrossRef] [PubMed]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef] [Green Version]
- Sobsey, M.D. Absence of virological and epidemiological evidence that SARS-CoV-2 poses COVID-19 risks from environmental fecal waste, wastewater and water exposures. J. Water Health 2022, 20, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, S.; Rente, D.; Cunha, M.V.; Marques, T.A.; Cardoso, E.; Vilaça, J.; Coelho, N.; Brôco, N.; Carvalho, M.; Santos, R. Discrimination and surveillance of infectious severe acute respiratory syndrome Coronavirus 2 in wastewater using cell culture and RT-qPCR. Sci. Total Environ. 2022, 815, 152914. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Weber, F.A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751. [Google Scholar] [CrossRef]
- Usman, M.; Farooq, M.; Hanna, K. Existence of SARS-CoV-2 in Wastewater: Implications for Its Environmental Transmission in Developing Communities. Environ. Sci. Technol. 2020, 54, 7758–7759. [Google Scholar] [CrossRef] [PubMed]
- Bartolomeu, M.; Reis, S.; Fontes, M.; Neves, M.G.P.M.S.; Faustino, M.A.F.; Almeida, A. Photodynamic Action against Wastewater Microorganisms and Chemical Pollutants: An Effective Approach with Low Environmental Impact. Water 2017, 9, 630. [Google Scholar] [CrossRef] [Green Version]
- Heller, L.; Mota, C.R.; Greco, D.B. COVID-19 faecal-oral transmission: Are we asking the right questions? Sci. Total Environ. 2020, 729, 138919. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Zhang, D.; Ling, H.; Huang, X.; Li, J.; Li, W.; Yi, C.; Zhang, T.; Jiang, Y.; He, Y.; Deng, S.; et al. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020, 741, 140445. [Google Scholar] [CrossRef]
- Lodder, W.; de Roda Husman, A.M. SARS-CoV-2 in wastewater: Potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020, 5, 533–534. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020, 25, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Xu, B.; Gamal El-Din, M. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020, 743. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.L.; Kase, J.A.; Harrison, L.M.; Balan, K.V.; Babu, U.; Chen, Y.; Macarisin, D.; Kwon, H.J.; Zheng, J.; Stevens, E.L.; et al. The persistence of bacterial pathogens in surface water and its impact on global food safety. Pathogens 2021, 10, 1391. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Monis, P.; Deere, D.; Ryan, U. Wastewater-based epidemiology—surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitol. Res. 2021, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Rasero, F.J.; Moya Ruano, L.A.; Rasero Del Real, P.; Cuberos Gómez, L.; Lorusso, N. Associations between SARS-CoV-2 RNA concentrations in wastewater and COVID-19 rates in days after sampling in small urban areas of Seville: A time series study. Sci. Total Environ. 2022, 806. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Zhang, H.; Wang, Z.; Chang, C.; Javed, A.; Ali, K.; Du, W.; Niazi, N.K.; Mao, K.; Yang, Z. Occurrence of various viruses and recent evidence of SARS-CoV-2 in wastewater systems. J. Hazard. Mater. 2021, 414. [Google Scholar] [CrossRef]
- Serra-Compte, A.; González, S.; Arnaldos, M.; Berlendis, S.; Courtois, S.; Loret, J.F.; Schlosser, O.; Yáñez, A.M.; Soria-Soria, E.; Fittipaldi, M.; et al. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021, 202. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.D.; Kulis, J.; Thomson, B.; Chapman, T.H.; Mawhinney, D.B. Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 2006, 366, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Calin, M.A.; Parasca, S.V. Light sources for photodynamic inactivation of bacteria. Lasers Med. Sci. 2008, 24, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Hamblin, M. Antimicrobial Photosensitizers: Drug Discovery Under the Spotlight. Curr. Med. Chem. 2015, 22, 2159–2185. [Google Scholar] [CrossRef]
- Bonnett, R. Chemical Aspects of Photodynamic Therapy; Gordon and Breach Science Publishers: London, UK; Newark, NJ, USA, 2000; ISBN 90-5699-248-1. [Google Scholar]
- Costa, L.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Faustino, M.A.F.; Almeida, A. Susceptibility of non-enveloped DNA- and RNA-type viruses to photodynamic inactivation. Photochem. Photobiol. Sci. 2012, 11, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.F.; Langenberg, W.G.; Van Etten, J.L.; Vidaver, A.K. Ultrastructure of bacteriophage phi 6: Arrangement of the double stranded RNA and envelope. J. Gen. Virol. 1977, 35, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Lytle, C.D.; Budacz, A.P.; Keville, E.; Miller, S.A.; Prodouz, K.N. Differential Inactivation of Surrogate Viruses With Merocyanine 540. Photochem. Photobiol. 1991, 54, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, C.; Mu, Y.; Houston, H.; Martinez-Smith, M.; Noble-Wang, J.; Coulliette-Salmond, A.; Rose, L. Persistence of bacteriophage phi 6 on porous and nonporous surfaces and the potential for its use as an ebola virus or coronavirus surrogate. Appl. Environ. Microbiol. 2020, 86, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.I.; Boehm, A.B. Systematic Review and Meta-Analysis of the Persistence and Disinfection of Human Coronaviruses and Their Viral Surrogates in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 544–553. [Google Scholar] [CrossRef]
- Aquino De Carvalho, N.; Stachler, E.N.; Cimabue, N.; Bibby, K. Evaluation of Phi6 Persistence and Suitability as an Enveloped Virus Surrogate. Environ. Sci. Technol. 2017, 51, 8692–8700. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Chang, P.H.; Hartert, J.; Wigginton, K.R. Reactivity of Enveloped Virus Genome, Proteins, and Lipids with Free Chlorine and UV254. Environ. Sci. Technol. 2018, 52, 7698–7708. [Google Scholar] [CrossRef] [PubMed]
- Prussin, A.J.; Schwake, D.O.; Lin, K.; Gallagher, D.L.; Buttling, L.; Marr, L.C. Survival of the enveloped virus Phi6 in droplets as a function of relative humidity, absolute humidity, and temperature. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova, L.; Weaver, S. Evaluation of eluents for the recovery of an enveloped virus from hands by whole-hand sampling. J. Appl. Microbiol. Microbiol. 2015, 118, 1210–1216. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.W.; Dougherty, T.J. How does Photodynamic Therapy work? Photochem. Photobiol. 1992, 55, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Fedorenko, A.; Grinberg, M.; Orevi, T.; Kashtan, N. Survival of the enveloped bacteriophage Phi6 (a surrogate for SARS-CoV-2) in evaporated saliva microdroplets deposited on glass surfaces. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Linden, Y.S.; Gundy, P.M.; Gerba, C.P.; Sobsey, M.D.; Linden, K.G. Inactivation of Coronaviruses and Phage Phi6 from Irradiation across UVC Wavelengths. Environ. Sci. Technol. Lett. 2021, 8, 425–430. [Google Scholar] [CrossRef]
- Mäntynen, S.; Sundberg, L.R.; Poranen, M.M. Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family Cystoviridae. Arch. Virol. 2018, 163, 1117–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, C.M.B.; Alves, E.; Costa, L.; Tomé, J.P.C.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Almeida, A.; Cunha, Â.; et al. Functional cationic nanomagnet - Porphyrin hybrids for the photoinactivation of microorganisms. ACS Nano 2010, 4, 7133–7140. [Google Scholar] [CrossRef]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef] [Green Version]
- Gundy, P.M.; Gerba, C.P.; Pepper, I.L. Survival of Coronaviruses in Water and Wastewater. Food Environ. Virol. 2009, 1, 10–14. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, R.G.; Rose, J.B.; Hashsham, S.A.; Gerba, C.P.; Haase, C.N. Criteria for selection of surrogates used to study the fate and control of pathogens in the environment. Appl. Environ. Microbiol. 2012, 78, 1969–1977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages-review. Folia Microbiol. (Praha) 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchobanoglous, G.; Burton, F.L.; Stencel, H.D. Wastewater Engineering: Treatment and Reuse, 4th ed.; McGraw-Hill, B., Ed.; Metcalf & Eddy, Inc.: Boston, MA, USA, 2003; ISBN 0071122508. [Google Scholar]
- Jiang, L.Q.; Carter, B.R.; Feely, R.A.; Lauvset, S.K.; Olsen, A. Surface ocean pH and buffer capacity: Past, present and future. Sci. Reports 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mojica, K.D.A.; Brussaard, C.P.D. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumat, M.R.; Hong, P.-Y. Inactivation and Loss of Infectivity of Enterovirus 70 by Solar Irradiation. Water 2019, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Deller, S.; Mascher, F.; Platzer, S.; Reinthaler, F.F.; Marth, E. Effect of solar radiation on survival of indicator bacteria in bathing waters. Cent. Eur. J. Public Health 2006, 14, 133–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joux, F.; Jeffrey, W.H.; Lebaron, P.; Mitchell, D.L. Marine bacterial isolates display diverse responses to UV-B radiation. Appl. Environ. Microbiol. 1999, 65, 3820–3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinton, L.W.; Davies-Colley, R.J.; Bell, R.G. Inactivation of enterococci and fecal coliforms from sewage and meatworks effluents in seawater chambers. Appl. Environ. Microbiol. 1994, 60, 2040–2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipe, O.M.S.; Santos, E.B.H.; Otero, M.; Gonçalves, E.A.C.; Neves, M.G.P.M.S. Photodegradation of metoprolol in the presence of aquatic fulvic acids. Kinetic studies, degradation pathways and role of singlet oxygen, OH radicals and fulvic acids triplet states. J. Hazard. Mater. 2020, 385, 121523. [Google Scholar] [CrossRef] [PubMed]
- Dimou, A.D.; Sakkas, V.A.; Albanis, T.A. Metolachlor photodegradation study in aqueous media under natural and simulated solar irradiation. J. Agric. Food Chem. 2005, 53, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Laurinavičius, S.; Käkelä, R.; Bamford, D.H.; Somerharju, P. The origin of phospholipids of the enveloped bacteriophage phi6. Virology 2004, 326, 182–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vatter, P.; Hoenes, K.; Hessling, M. Photoinactivation of the Coronavirus Surrogate phi6 by Visible Light. Photochem. Photobiol. 2020, 97, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Vatter, P.; Hoenes, K.; Hessling, M. Blue light inactivation of the enveloped RNA virus Phi6. BMC Res. Notes 2021, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.C.; Torres-Franco, A.F.; Lopes, B.C.; da Silva Santos, B.S.Á.; Costa, E.A.; Costa, M.S.; Reis, M.T.P.; Melo, M.C.; Polizzi, R.B.; Teixeira, M.M.; et al. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021, 195. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.T.; Fuhrman, J.A. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 1997, 63, 77–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, R.L.; Knowlton, D.R.; Winston, P.E. Mechanism of inactivation of enteric viruses in fresh water. Appl. Environ. Microbiol. 1986, 52, 450–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-D.; Unno, H. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 1996, 33, 243–250. [Google Scholar] [CrossRef]
- Wommack, K.E.; Hill, R.T.; Muller, T.A.; Colwell, R.R. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 1996, 62, 1336–1341. [Google Scholar] [CrossRef] [Green Version]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Lytle, C.D.; Sagripanti, J.-L. Predicted Inativation of Viruses of Relevance to Biodefense by Solar Radiation. J. Virol. 2005, 79, 14244–14252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, Y.J.; Costa, L.; Pereira, C.; Cunha, Â.; Calado, R.; Gomes, N.C.M.; Almeida, A. Influence of environmental variables in the efficiency of phage therapy in aquaculture. Microb. Biotechnol. 2014, 7, 401–413. [Google Scholar] [CrossRef]
- Teitelbaum, S.; Azevedo, L.H.; Bernaola-Paredes, W.E. Antimicrobial Photodynamic Therapy Used as First Choice to Treat Herpes Zoster Virus Infection in Younger Patient: A Case Report. Photobiomodulation Photomed. Laser Surg. 2020, 38, 232–236. [Google Scholar] [CrossRef]
- Wiehe, A.; O’brien, J.M.; Senge, M.O. Trends and targets in antiviral phototherapy. Photochem. Photobiol. Sci. 2019, 18, 2565–2612. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.; Esteves, A.C.; Correia, A.; Moreirinha, C.; Delgadillo, I.; Cunha, Â.; Neves, M.G.P.M.S.; Faustino, M.A.F.; Almeida, A. SDS-PAGE and IR spectroscopy to evaluate modifications in the viral protein profile induced by a cationic porphyrinic photosensitizer. J. Virol. Methods 2014, 209, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Bartolomeu, M.; Oliveira, C.; Pereira, C.; Neves, M.G.P.M.S.; Faustino, M.A.F.; Almeida, A. Antimicrobial photodynamic approach in the inactivation of viruses in wastewater: Influence of alternative adjuvants. Antibiotics 2021, 10, 767. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Costa, L.; Faustino, M.A.F.; Almeida, A. Photodynamic inactivation of multidrug-resistant bacteria in hospital wastewaters: Influence of residual antibiotics. Photochem. Photobiol. Sci. 2014, 13, 626–633. [Google Scholar] [CrossRef]
- Costa, L.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, Â.; Almeida, A. Photodynamic inactivation of mammalian viruses and bacteriophages. Viruses 2012, 4, 1034–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, L.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Faustino, M.A.F.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Evaluation of resistance development and viability recovery by a non-enveloped virus after repeated cycles of aPDT. Antiviral Res. 2011, 91, 278–282. [Google Scholar] [CrossRef]
- Kong, J.; Lu, Y.; Ren, Y.; Chen, Z.; Chen, M. The virus removal in UV irradiation, ozonation and chlorination. Water Cycle 2021, 2, 23–31. [Google Scholar] [CrossRef]
- Ramos, P.; Valente Neves, M. Monitorização de descargas de águas residuais no mar através de veículos submarinos autónomos (VSAs). Ing. del agua 2009, 16, 201–216. [Google Scholar] [CrossRef] [Green Version]
- Castro, K.A.D.F.; Moura, N.M.M.; Figueira, F.; Ferreira, R.I.; Simões, M.M.Q.; Cavaleiro, J.A.S.; Faustino, M.A.F.; Silvestre, A.J.D.; Freire, C.S.R.; Tomé, J.P.C.; et al. New Materials Based on Cationic Porphyrins Conjugated to Chitosan or Titanium Dioxide: Synthesis, Characterization and Antimicrobial Efficacy. Int. J. Mol. Sci. 2019, 20, 2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesquita, M.Q.; Menezes, J.C.J.M.D.S.; Pires, S.M.G.; Neves, M.G.P.M.S.; Simões, M.M.Q.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, A.; Daniel-da-Silva, A.L.; Almeida, A.; et al. Pyrrolidine-fused chlorin photosensitizer immobilized on solid supports for the photoinactivation of Gram negative bacteria. Dye. Pigment. 2014, 110, 123–133. [Google Scholar] [CrossRef]
- Alves, E.; Rodrigues, J.M.M.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Lin, Z.; Cunha, Â.; Nadais, M.H.; Tomé, J.P.C.; Almeida, A. A new insight on nanomagnet-porphyrin hybrids for photodynamic inactivation of microorganisms. Dye. Pigment. 2014, 110, 80–88. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, M.; Bartolomeu, M.; Vieira, C.; Gomes, A.T.P.C.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Almeida, A. Photoinactivation of Phage Phi6 as a SARS-CoV-2 Model in Wastewater: Evidence of Efficacy and Safety. Microorganisms 2022, 10, 659. https://doi.org/10.3390/microorganisms10030659
Gomes M, Bartolomeu M, Vieira C, Gomes ATPC, Faustino MAF, Neves MGPMS, Almeida A. Photoinactivation of Phage Phi6 as a SARS-CoV-2 Model in Wastewater: Evidence of Efficacy and Safety. Microorganisms. 2022; 10(3):659. https://doi.org/10.3390/microorganisms10030659
Chicago/Turabian StyleGomes, Marta, Maria Bartolomeu, Cátia Vieira, Ana T. P. C. Gomes, Maria Amparo F. Faustino, Maria Graça P. M. S. Neves, and Adelaide Almeida. 2022. "Photoinactivation of Phage Phi6 as a SARS-CoV-2 Model in Wastewater: Evidence of Efficacy and Safety" Microorganisms 10, no. 3: 659. https://doi.org/10.3390/microorganisms10030659
APA StyleGomes, M., Bartolomeu, M., Vieira, C., Gomes, A. T. P. C., Faustino, M. A. F., Neves, M. G. P. M. S., & Almeida, A. (2022). Photoinactivation of Phage Phi6 as a SARS-CoV-2 Model in Wastewater: Evidence of Efficacy and Safety. Microorganisms, 10(3), 659. https://doi.org/10.3390/microorganisms10030659








