Prospection of Psychrotrophic Filamentous Fungi Isolated from the High Andean Paramo Region of Northern Ecuador: Enzymatic Activity and Molecular Identification
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Fungal Strain Identification
2.2.1. DNA Extraction
2.2.2. Polymerase Chain Reaction
2.2.3. DNA Sequencing
2.3. Preparation of Crude Enzyme Extract
2.4. Enzyme Activity Assay
2.4.1. Enzyme Determination with Natural Substrates
2.4.2. Enzyme Determination with Synthetic Substrates
2.5. Statistical Analysis
3. Results and Discussion
3.1. Sample Site Conditions and Soil Characteristics
3.2. Screening for Cultivable Fungi
3.3. Identification of Fungi
3.4. Enzymatic Characterization
3.4.1. Enzymatic Production of Cladosporium michoacanense
3.4.2. Enzymatic production of Cladosporium sp. (C. cladosporioides Species Complex)
3.4.3. Enzymatic Production of Didymella sp.
3.4.4. Comparison of Relative Enzyme Activity between Acid and Alkaline Phosphatases
3.4.5. Temperature-Dependent Enzyme Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiménez-Rivillas, C.; García, J.J.; Quijano-Abril, M.A.; Daza, J.M.; Morrone, J.J. A new biogeographical regionalisation of the Páramo biogeographic province. Aust. Syst. Bot. 2018, 31, 296–310. [Google Scholar] [CrossRef]
- Buytaert, W.; Deckers, J.; Wyseure, G. Regional variability of volcanic ash soils in south Ecuador: The relation with parent material, climate and land use. Catena 2007, 70, 143–154. [Google Scholar] [CrossRef]
- Bader, M.Y.; van Geloof, I.; Rietkerk, M. High solar radiation hinders tree regeneration above the alpine treeline in northern Ecuador. Plant Ecol. 2007, 191, 33–45. [Google Scholar] [CrossRef]
- Paucar, B.; Carpio, M.; Alvarado Ochoa, S.P.; Valverde, F.; Parra, R. Análisis de Solubilizadores de Fósforo en Los Suelos Andisoles de Sierra Norte y Centro de Ecuador. Simposio “El Suelo y la Nutrición de Cultivos en el Ecuador”. 2015. Available online: http://repositorio.iniap.gob.ec/handle/41000/2501 (accessed on 9 September 2021).
- Hassan, N.; Rafiq, M.; Hayat, M.; Shah, A.A.; Hasan, F. Psychrophilic and psychrotrophic fungi: A comprehensive review. Rev. Environ. Sci. Biotechnol. 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Rafiq, M.; Hassan, N.; Rehman, M.; Hasan, F. Adaptation mechanisms and applications of psychrophilic fungi. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S.M., Grube, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 157–174. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Barato, M.B.; Nobre, F.S.; Polezel, D.A.; de Oliveira, T.B.; dos Santos, J.A.; Rodrigues, A.; Sette, L.D. Production of cold-adapted enzymes by filamentous fungi from King George Island, Antarctica. Polar Biol. 2018, 41, 2511–2521. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, X.; Wu, W.; Hao, Y.; Su, Y.; Cai, L.; Xiang, M.; Liu, X. Psychrophilic fungi from the world’s roof. Pers. Mol. Phylogeny Evol. Fungi 2015, 34, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.J.; Garzon, L.N.; Valencia, J.B.; Madrinan, S. On the causes of rapid diversification in the paramos: Isolation by ecology and genomic divergence in espeletia. Front. Plant Sci. 2018, 9, 1700. [Google Scholar] [CrossRef] [PubMed]
- Avellaneda-Torres, L.M.; Pulido, C.P.G.; Rojas, E.T. Assessment of cellulolytic microorganisms in soils of Nevados Park, Colombia. Braz. J. Microbiol. 2014, 45, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Landinez-Torres, A.Y.; Becerra Abril, J.L.; Tosi, S.; Nicola, L. Soil microfungi of the colombian natural regions. Int. J. Environ. Res. Public Health 2020, 17, 8311. [Google Scholar] [CrossRef] [PubMed]
- Gualdrón-Arenas, C.; Suárez-Navarro, A.L.; Valencia-Zapata, H. Hongos del suelo aislados de zonas de vegetacion natural del paramo de chisaca, colombia. Caldasia 1997, 19, 235–245. [Google Scholar]
- Pinos León, A.J. Exploring the Microbiome Composition of the Rhizosphere Associated with the Wild Andean Blueberry (Vaccinium Floribundum, Kunth) in the Highlands of Ecuador. Master’s Thesis, Universidad San Francisco de Quito, Quito, Ecuador, 2020. Available online: https://repositorio.usfq.edu.ec/bitstream/23000/9113/1/141011.pdf (accessed on 5 December 2021).
- Kuddus, M.; Roohi Arif, J.; Ramteke, P. An overview of cold-active microbial α-amylase: Adaptation strategies and biotechnological potentials. Biotechnology 2011, 10, 246–258. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes. Front Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech 2021, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.B.; de Lucas, R.C.; de Almeida Scarcella, A.S.; Pasin, T.M.; Contato, A.G.; Polizeli, M.L.T.M. Cold-active lytic enzymes and their applicability in the biocontrol of postharvest fungal pathogens. J. Agric. Food Chem. 2020, 68, 6461–6463. [Google Scholar] [CrossRef]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.-P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D.; et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Niñerola, A.; Ferrer-Rullan, R.; Vidal-Suñé, A. Climate change mitigation: Application of management production philosophies for energy saving in industrial processes. Sustainability 2020, 12, 717. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Pasin, T.M.; Scarcella, A.S.A.; de Oliveira, T.B.; Lucas, R.C.; Cereia, M.; Betini, J.H.A.; Polizeli, M.L.T.M. Paper industry wastes as carbon sources for Aspergillus species cultivation and production of an enzymatic cocktail for biotechnological applications. Ind. Biotechnol. 2020, 16, 56–60. [Google Scholar] [CrossRef]
- Duncan, S.M.; Farrell, R.L.; Thwaites, J.M.; Held, B.W.; Arenz, B.E.; Jurgens, J.A.; Blanchette, R.A. Endoglucanase-producing fungi isolated from Cape Evans historic expedition hut on Ross Island, Antarctica. Environ. Microbiol. 2006, 8, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Miri, S.; Naghdi, M.; Rouissi, T.; Kaur Brar, S.; Martel, R. Recent biotechnological advances in petroleum hydrocarbons degradation under cold climate conditions: A review. Crit. Rev. Environ. Sci. Technol. 2019, 49, 553–586. [Google Scholar] [CrossRef]
- Jiang, G.; Chen, P.; Bao, Y.; Wang, X.; Yang, T.; Mei, X.; Banerjee, S.; Wei, Z.; Xu, Y.; Shen, Q. Isolation of a novel psychrotrophic fungus for efficient low-temperature composting. Bioresour. Technol. 2021, 331, 125049. [Google Scholar] [CrossRef]
- Adhikari, P.; Jain, R.; Sharma, A.; Pandey, A. Plant Growth Promotion at Low Temperature by Phosphate-Solubilizing Pseudomonas spp. Isolated from High-Altitude Himalayan Soil. Microb. Ecol. 2021. [Google Scholar] [CrossRef]
- Geoportal Ecuador—Infraestructura de Datos Espaciales. Available online: http://www.geoportaligm.gob.ec/portal/ (accessed on 19 October 2021).
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Academic Press, Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Contato, A.G.; de Oliveira, T.B.; Aranha, G.M.; de Freitas, E.N.; Vici, A.C.; Nogueira, K.M.V.; de Lucas, R.C.; de Almeida Scarcella, A.S.; Buckeridge, M.S.; Silva, R.N.; et al. Prospection of fungal lignocellulolytic enzymes produced from jatoba (Hymenaea courbaril) and tamarind (Tamarindus indica) seeds: Scaling for bioreactor and saccharification profile of sugarcane bagasse. Microorganisms 2021, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Garen, A.; Siddiqi, O. Suppression of mutations in the alkaline phosphatase structural cistron of E. coli. Proc. Natl. Acad. Sci. USA 1962, 48, 1121–1127. [Google Scholar] [CrossRef]
- Kersters-Hilderson, H.; Claeyssens, M.; Van Doorslaer, E.; Saman, E.; De Bruyne, C.K. β-D-xylosidase from Baccilus pumilus. In Methods in Enzymology; Complex Carbohydrates Part D; Academic Press: Cambridge, MA, USA, 1982; Volume 83, pp. 631–639. Available online: https://www.sciencedirect.com/science/article/pii/0076687982830620 (accessed on 19 August 2021).
- Hribljan, J.A.; Suarez, E.; Heckman, K.A.; Lilleskov, E.A.; Chimner, R.A. Peatland carbon stocks and accumulation rates in the Ecuadorian paramo. Wetl. Ecol. Manag. 2016, 24, 113–127. [Google Scholar] [CrossRef]
- Roa García, C.; Brown, S.; Krzic, M.; Lavkulich, L.; Roa-García, M.C. Relationship of soil water retention characteristics and soil properties: A case study from the Colombian Andes. Can. J. Soil Sci. 2021, 101, 147–156. [Google Scholar] [CrossRef]
- Zúñiga-Silgado, D.; Rivera-Leyva, J.C.; Coleman, J.J.; Sánchez-Reyez, A.; Valencia-Díaz, S.; Serrano, M.; de-Bashan, L.E.; Folch-Mallol, J.L. Soil type affects organic acid production and phosphorus solubilization efficiency mediated by several native fungal strains from Mexico. Microorganisms 2020, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.C.; Grand, S.; Verrecchia, E.P. Calcium-mediated stabilisation of soil organic carbon. Biogeochemistry 2018, 137, 27–49. [Google Scholar] [CrossRef]
- Opfergelt, S.; Georg, R.B.; Delvaux, B.; Cabidoche, Y.-M.; Burton, K.W.; Halliday, A.N. Mechanisms of magnesium isotope fractionation in volcanic soil weathering sequences, Guadeloupe. Earth Planet Sci. Lett. 2012, 341, 176–185. [Google Scholar] [CrossRef]
- Rahimi, H.; Pazira, E.; Tajik, F. Effect of soil organic matter, electrical conductivity and sodium adsorption ratio on tensile strength of aggregates. Soil Tillage Res. 2000, 54, 145–153. [Google Scholar] [CrossRef]
- Ishiguro, M.; Makino, T. Sulfate adsorption on a volcanic ash soil (allophanic Andisol) under low pH conditions. Colloids Surf.-Physicochem. Eng. Asp. 2011, 384, 121–125. [Google Scholar] [CrossRef][Green Version]
- Hazelton, P.; Murphy, B. Interpreting Soil Test Results: What Do All the Numbers Mean? Csiro Publishing: Clayton, Australia, 2016; p. 201. [Google Scholar]
- Motsara, M.R.; Roy, R.N. Guide to Laboratory Establishment for Plant Nutrient Analysis. FAO Fertilizer and Plant Nutrition Bulletin; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008; p. 219. [Google Scholar]
- Sembiring, M. Bacterial and fungi phosphate solubilization effect to increase nutrient uptake and potatoes (Solanum tuberosum L.) production on Andisol Sinabung area. J. Agron. 2017, 16, 131–137. [Google Scholar] [CrossRef][Green Version]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef]
- Del-Cid, A.; Ubilla, P.; Ravanal, M.-C.; Medina, E.; Vaca, I.; Levicán, G.; Eyzaguirre, J.; Chávez, R. Cold-active xylanase produced by fungi associated with Antarctic marine sponges. Appl. Biochem. Biotechnol. 2014, 172, 524–532. [Google Scholar] [CrossRef]
- Wang, X.; Wu, X.; Jiang, S.; Yin, Q.; Li, D.; Wang, Y.; Wang, D.; Chen, Z. Whole genome sequence and gene annotation resource for Didymella bellidis associated with tea leaf spot. Plant Dis. 2021, 105, 1168–1170. [Google Scholar] [CrossRef]
- Zhang, J.; Bruton, B.D.; Biles, C.L. Cell wall-degrading enzymes of Didymella bryoniae in relation to fungal growth and virulence in cantaloupe fruit. Eur. J. Plant Pathol. 2014, 139, 749–761. [Google Scholar] [CrossRef]
- Rafiq, M.; Nadeem, S.; Hassan, N.; Hayat, M.; Sajjad, W.; Zada, S.; Sajjad, W.; Hasan, F. Fungal recovery and characterization from Hindu Kush mountain range, Tirich Mir glacier, and their potential for biotechnological applications. J. Basic Microbiol. 2020, 60, 444–457. [Google Scholar] [CrossRef]
- Hassan, N.; Hasan, F.; Nadeem, S.; Hayat, M.; All, P.; Khan, M.; Sajjad, W.; Zada, S.; Rafiq, M. Community analysis and characterization of fungi from Batura Glacier, Karakoram Mountain Range, Pakistan. Appl. Ecol. Environ. Res. 2018, 16, 5323–5341. [Google Scholar] [CrossRef]
- Pacelli, C.; Bryan, R.A.; Onofri, S.; Selbmann, L.; Shuryak, I.; Dadachova, E. Melanin is effective in protecting fast and slow growing fungi from various types of ionizing radiation. Environ. Microbiol. 2017, 19, 1612–1624. [Google Scholar] [CrossRef] [PubMed]
- Dadachova, E.; Casadevall, A. Ionizing radiation: How fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 2008, 11, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, R. Effect of transport phenomena of Cladosporium cladosporioides on decolorization and chemical oxygen demand of distillery spent wash. Int. J. Environ. Sci. Technol. 2015, 12, 1581–1590. [Google Scholar] [CrossRef][Green Version]
- Mohan Kumar, N.S.; Ramasamy, R.; Manonmani, H.K. Production and optimization of l-asparaginase from Cladosporium sp. using agricultural residues in solid state fermentation. Ind. Crops Prod. 2013, 43, 150–158. [Google Scholar] [CrossRef]
- Quintanilla, D.; Hagemann, T.; Hansen, K.; Gernaey, K.V. Fungal morphology in industrial enzyme production-modelling and monitoring. In Filaments in Bioprocesses; Krull, R., Bley, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 29–54. Available online: https://www.webofscience.com/wos/woscc/full-record/WOS:000365169300003 (accessed on 19 October 2021).
- Abrha, B.; Gashe, B.A. Cellulase production and activity in a species of Cladosporium. World J. Microbiol. Biotechnol. 1992, 8, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Maya-Yescas, M.E.; Revah, S.; Le Borgne, S.; Valenzuela, J.; Palacios-González, E.; Terrés-Rojas, E.; Vigueras-Ramírez, G. Growth of Leucoagaricus gongylophorus Möller (Singer) and production of key enzymes in submerged and solid-state cultures with lignocellulosic substrates. Biotechnol. Lett. 2021, 43, 845–854. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Arst, H.N. Regulation of gene expression by ambient ph in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 426–446. [Google Scholar] [CrossRef]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular enzymes and bioactive compounds from Antarctic terrestrial fungi for bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Rafiq, M.; Hayat, M.; Nadeem, S.; Shah, A.A.; Hasan, F. Potential of psychrotrophic fungi isolated from siachen glacier, Pakistan, to produce antimicrobial metabolites. Appl. Ecol. Environ. Res. 2017, 15, 1157–1171. [Google Scholar] [CrossRef]
- Grujic, M.; Dojnov, B.; Potocnik, I.; Atanasova, L.; Duduk, B.; Srebotnik, E.; Druzhinina, I.S.; Kubicek, C.P.; Vujčić, Z. Superior cellulolytic activity of Trichoderma guizhouense on raw wheat straw. World J. Microbiol. Biotechnol. 2019, 35, 194. [Google Scholar] [CrossRef]
- Ghazali, M.F.S.M.; Zainudin, N.A.I.M.; Abd Aziz, N.A.; Mustafa, M. Screening of lignocellulolytic fungi for hydrolyzation of lignocellulosic materials in paddy straw for bioethanol production. Malays. J. Microbiol. 2019, 15, 379–386. [Google Scholar]
- Ji, L.; Yang, J.; Fan, H.; Yang, Y.; Li, B.; Yu, X.; Zhu, N.; Yuan, H. Synergy of crude enzyme cocktail from cold-adapted Cladosporium cladosporioides Ch2-2 with commercial xylanase achieving high sugars yield at low cost. Biotechnol. Biofuels 2014, 7, 130. [Google Scholar] [CrossRef]
- Aslam, M.S.; Aishy, A.; Samra, Z.Q.; Gull, I.; Athar, M.A. Identification, Purification and characterization of a novel extracellular laccase from Cladosporium cladosporioides. Biotechnol. Biotechnol. Equip. 2012, 26, 3345–3350. [Google Scholar] [CrossRef]
- Halaburgi, V.M.; Sharma, S.; Sinha, M.; Singh, T.P.; Karegoudar, T.B. Purification and characterization of a thermostable laccase from the ascomycetes Cladosporium cladosporioides and its applications. Process Biochem. 2011, 46, 1146–1152. [Google Scholar] [CrossRef]
- Gawas-Sakhalkar, P.; Singh, S.M.; Naik, S.; Ravindra, R. High-temperature optima phosphatases from the cold-tolerant Arctic fungus Penicillium citrinum. Polar Res. 2012, 31, 11105. [Google Scholar] [CrossRef]
- Krishnan, A.; Convey, P.; Gonzalez-Rocha, G.; Alias, S.A. Production of extracellular hydrolase enzymes by fungi from King George Island. Polar Biol. 2016, 39, 65–76. [Google Scholar] [CrossRef]
- Fenice, M.; Selbmann, L.; Zucconi, L.; Onofri, S. Production of extracellular enzymes by Antarctic fungal strains. Polar Biol. 1997, 17, 275–280. [Google Scholar] [CrossRef]
- Wang, N.; Zang, J.; Ming, K.; Liu, Y.; Wu, Z.; Ding, H. Production of cold-adapted cellulase by Verticillium sp. isolated from Antarctic soils. Electron. J. Biotechnol. 2013, 16, 10. Available online: http://www.ejbiotechnology.info/index.php/ejbiotechnology/article/view/1302 (accessed on 9 December 2021). [CrossRef]
- Salunke, M.; Sondge, D.B.; Yadav, S.; Warkhade, R.; Rathod, S.; Kate, S. Alkaline phosphatase production by Enterobacter hormaechei isolated from birds fecal waste and its optimization. Int. J. Adv. Sci. Technol. 2020, 29, 9. [Google Scholar]
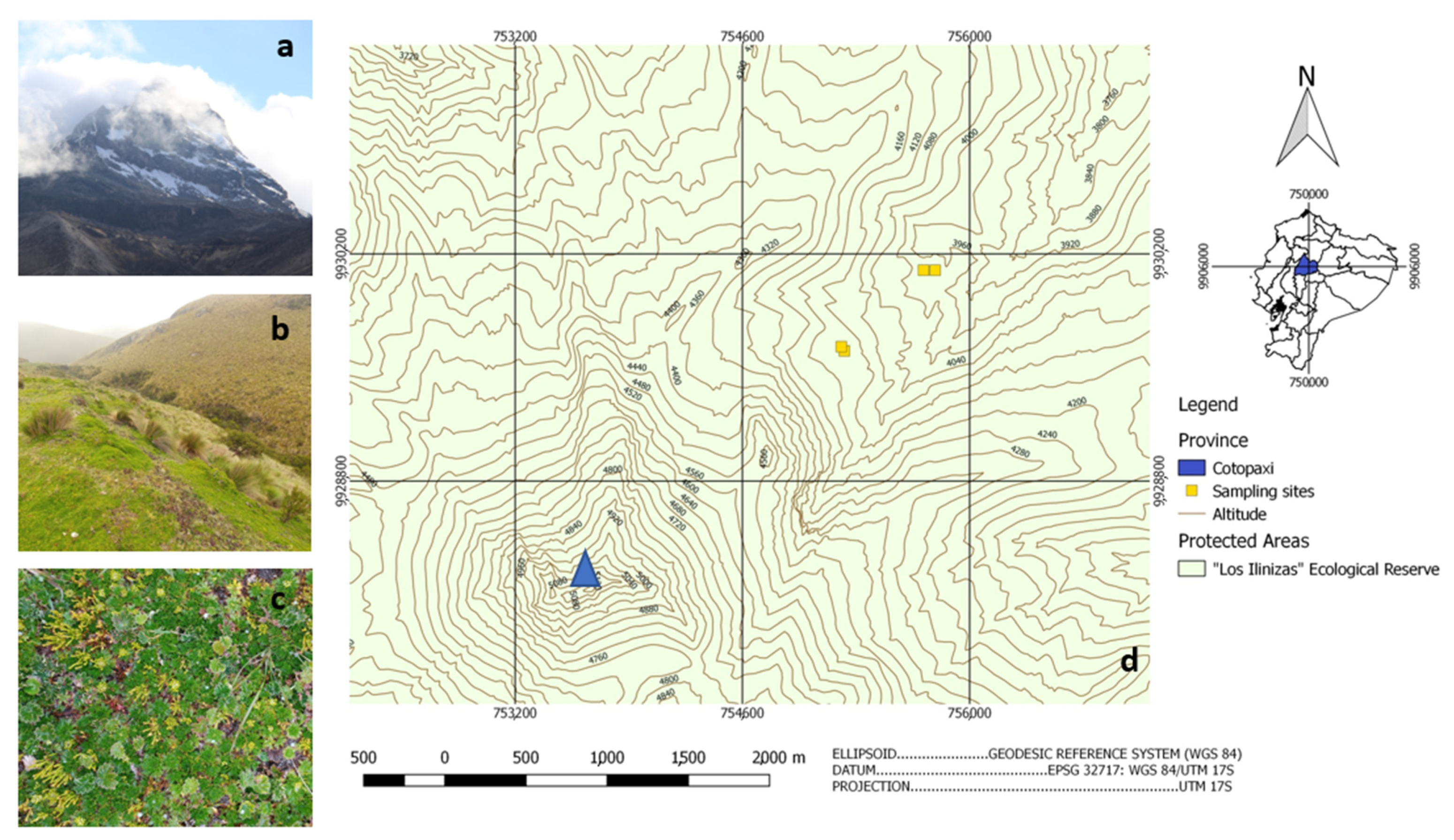

| Sample Site | Soil Type | Soil Texture | pH | COND (µS/cm2) | Nt % | OM % | P (ppm) | S (ppm) | K (meq/100 g) | Ca (meq/100 g) | Mg (meq/100 g) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Andisol | sandy loam, loamy | 6.15 | 21.5 | 0.31 | 8.8 | 9.01 | 8.4 | 0.54 | 9.78 | 1.71 |
| 2 | 6.78 | 21.3 | 0.36 | 9.7 | 14 | 9.3 | 0.48 | 9.76 | 1.28 | ||
| 3 | Andisol | sandy loam | 5.93 | 15.51 | 0.25 | 7.7 | 6.04 | 4.7 | 0.42 | 7.49 | 1.01 |
| 4 | 5.87 | 20.8 | 0.27 | 9 | 12 | 4.1 | 0.37 | 4.87 | 0.73 |
| Agitation | Static | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | 24 h | 48 h | 72 h | 96 h | 120 h | 24 h | 48 h | 72 h | 96 h | 120 h |
| Endoglucanase | 932.2 ± 20.7 | 1514.4 ± 100.5 | 2503.7 ± 207.1 * | 394.5 ± 51.8 | 385.2 ± 170.0 | 887.4 ± 84.8 | 1061.0 ± 23.3 | 917.6 ± 17.1 | 791.4 ± 70.4 | 730.6 ± 26.9 |
| Exoglucanase | ND | ND | ND | 1.4 ± 0.7 | 1.4 ± 0.3 | ND | ND | ND | ND | 0.2 ± 0.1 |
| β-Glucosidase | ND | ND | ND | 1.7 ± 0.0 | ND | ND | ND | ND | ND | 0.2 ± 0.0 |
| Xylanase | 360.2 ± 105.7 | 969.4 ± 120.1 | 2430.1 ± 2.1 * | 274.6 ± 81.3 | 145.2 ± 34.7 | 737.8 ± 88.3 | 435.5 ± 111.7 | 412.2 ± 53.9 | 353.0 ± 119.5 | 384.3 ± 70.6 |
| β-Xylosidase | ND | ND | 0.7±0. | 1.1 ± 0.3 | 1.4 ± 0.3 * | 0.5 ± 0.1 | ND | ND | ND | ND |
| Acid phosphatase | 1.4 ± 0.6 | 1.7 ± 1.0 | 31.0 ± 0.3 | 5.9 | 34.9 ± 0.1 * | ND | ND | ND | ND | 0.7 ± 0.4 |
| Alkaline phosphatase | 0.3±0.1 | 1.4 ± 0.3 | 2.3 ± 0.4 * | 0.2 ± 0.2 | ND | 1.4 ± 0.3 | 0.6 ± 0.3 | ND | ND | ND |
| Agitation | Static | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | 24 h | 48 h | 72 h | 96 h | 120 h | 24 h | 48 h | 72 h | 96 h | 120 h |
| Endoglucanase | 393.2 ± 97.3 | 1274.0 ± 102.0 | 328.5 ± 68.6 | 14.0 ± 11.4 | 14.6 ± 11.9 | 761.0 ± 70.4 | 638.8 ± 140.4 | 945.3 ± 120.6 | 1076.4 ± 149.4 | 775.0 ± 156.2 |
| Exoglucanase | ND | ND | 3.6 ± 1.0 | 6.0 ± 0.3 | 12.0 ± 0.6 * | ND | ND | 0.7 ± 0.4 | 0.9 ± 0.6 | 1.0 ± 0.0 |
| β-Glucosidase | 0.2 ± 0.2 | 0.8 ± 0.2 | 5.7 ± 1.0 | 8.7 ± 2.9 | 11.1 ± 0.8 * | ND | ND | 0.7 ± 0.4 | ND | ND |
| Xylanase | 156.0 ± 8.3 | 861.0 ± 11.9 | 958.9 ± 15.5 | 481.0 ± 57.0 | ND | 522.1 ± 87.6 | 660.0 ± 56.1 | 1113.0 ± 865 | 937.3 ± 40.4 | 898.0 ± 62.1 |
| β-Xylosidase | 1.2 ± 0.3 | 0.4 ± 0.1 | ND | ND | ND | 1.0 ± 0.1 | ND | ND | ND | ND |
| Acid phosphatase | 0.7 ± 0.4 | 1.9 ± 0.7 | 3.4 ± 0.7 | 6.8 ± 0.3 | 13.9 ± 0.5 * | ND | ND | 0.3 ± 0.2 | 1.0 ± 0.7 | 0.9 ± 0.1 |
| Alkaline phosphatase | ND | ND | 1.2 ± 0.8 | 1.9 ± 1.6 | 8.0 ± 1.5 * | ND | ND | ND | ND | 1.0 ± 0.3 |
| Agitation | Static | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | 24 h | 48 h | 72 h | 96 h | 120 h | 24 h | 48 h | 72 h | 96 h | 120 h |
| Endoglucanase | 1077.4 ± 6.7 | 1037.4 ± 99.0 | 354.5 ± 6.7 | 334.4 ± 10.9 | 119.0 ± 39.4 | 870.1 ± 99.8 | 1243.0 ± 108.1 | 1151.6 ± 189.5 | 1413.0 ± 165.7 * | 926.7 ± 15.0 |
| Exoglucanase | 0.4 ± 0.2 | 0.7 ± 0.2 | 1.0 ± 0.3 | 1.7 ± 0.4 | 1.7 ± 0.4 | 0.7 ± 0.2 | 1.7 ± 0.1 | 0.6 ± 0.2 | ND | ND |
| β-Glucosidase | ND | ND | ND | 1.6 ± 0.7 | 0.5 ± 0.1 | ND | ND | ND | ND | ND |
| Xylanase | 202.3 ± 15.0 | 233.0 ± 98.2 | 383.0 ± 103.2 | 289.2 ± 62.1 | ND | 873.2 ± 93.7 | 893.3 ± 125.9 | 906.0 ± 186.4 | 1623.5 ± 196.4 * | 624.7 ± 141.4 |
| β-Xylosidase | 1.0 ± 0.0 | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.1 ± 0.0 | ND | ND | ND | ND | ND | ND |
| Acid phosphatase | ND | 2.7 ± 0.6 | 5.7 ± 1.1 | 7.3 ± 1.1 | 7.5 ± 1.5 | ND | ND | ND | ND | 0.3 ± 0.1 |
| Alkaline phosphatase | 0.4 ± 0.3 | 0.4 ± 0.1 | 1.8 ± 0.1 | 2.1 ± 0.5 | ND | ND | ND | ND | ND | ND |
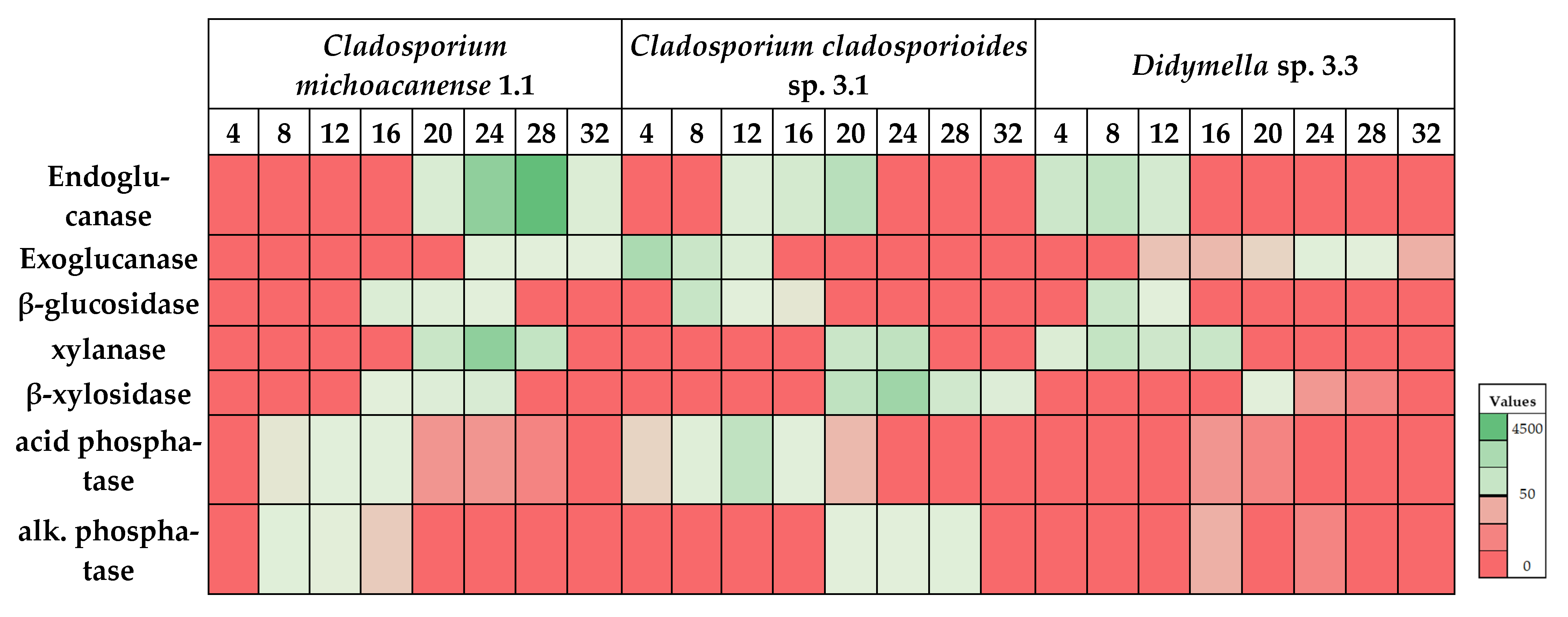
| Cladosporium Michoacanense 1.1 | Cladosporium Cladosporioides Complex 3.1 | Didymella sp. 3.3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Maximum Yield ± SD | T opt °C | IT (h) | Maximum Yield ± SD | T opt °C | IT (h) | Maximum Yield ± SD | T opt °C | IT (h) |
| Endoglucanase | 4563 ± 209 a | 28 | 72 | 1553 ± 330 b | 20 | 48 | 1247 ± 21 b | 8 | 72 |
| Exoglucanase | 107 ± 26 a | 24 | 96 | 2037 ± 254 b | 4 | 120 | 127 ± 5 a | 24 | 96 |
| β-Glucosidase | 303 ± 39 a | 16 | 96 | 1013 ± 151 b | 8 | 96 | 917 ± 12 b | 8 | 120 |
| Xylanase | 3036 ± 634 a | 24 | 72 | 1290 ± 122 ab | 24 | 72 | 1150 ± 121 b | 8 | 96 |
| β-Xylosidase | 430 ± 29 ab | 24 | 120 | 2457 ± 336 a | 24 | 120 | 71 ± 5 b | 20 | 48 |
| Acid phosphatase | 97 ± 12 ab | 12 | 120 | 1273 ± 360 a | 12 | 120 | 17 ± 5 b | 16 | 120 |
| Alkaline phosphatase | 127 ± 21 a | 8 | 72 | 137 ± 37 b | 28 | 120 | 26 ± 7 a | 16 | 96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brück, S.A.; Contato, A.G.; Gamboa-Trujillo, P.; de Oliveira, T.B.; Cereia, M.; de Moraes Polizeli, M.d.L.T. Prospection of Psychrotrophic Filamentous Fungi Isolated from the High Andean Paramo Region of Northern Ecuador: Enzymatic Activity and Molecular Identification. Microorganisms 2022, 10, 282. https://doi.org/10.3390/microorganisms10020282
Brück SA, Contato AG, Gamboa-Trujillo P, de Oliveira TB, Cereia M, de Moraes Polizeli MdLT. Prospection of Psychrotrophic Filamentous Fungi Isolated from the High Andean Paramo Region of Northern Ecuador: Enzymatic Activity and Molecular Identification. Microorganisms. 2022; 10(2):282. https://doi.org/10.3390/microorganisms10020282
Chicago/Turabian StyleBrück, Stefan Alexander, Alex Graça Contato, Paul Gamboa-Trujillo, Tássio Brito de Oliveira, Mariana Cereia, and Maria de Lourdes Teixeira de Moraes Polizeli. 2022. "Prospection of Psychrotrophic Filamentous Fungi Isolated from the High Andean Paramo Region of Northern Ecuador: Enzymatic Activity and Molecular Identification" Microorganisms 10, no. 2: 282. https://doi.org/10.3390/microorganisms10020282
APA StyleBrück, S. A., Contato, A. G., Gamboa-Trujillo, P., de Oliveira, T. B., Cereia, M., & de Moraes Polizeli, M. d. L. T. (2022). Prospection of Psychrotrophic Filamentous Fungi Isolated from the High Andean Paramo Region of Northern Ecuador: Enzymatic Activity and Molecular Identification. Microorganisms, 10(2), 282. https://doi.org/10.3390/microorganisms10020282








