Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance of Myxobacterial Cultures
2.2. Standardized HPLC–MS Conditions for Analysis of Secondary Metabolism of Crude Extracts
2.3. Isolation of 1–3 Via Liquid–Liquid Extraction, Flash Chromatography and Semi-Preparative HPLC
2.4. NMR Based Structure Elucidation and Chiroptical Measurement
2.5. Bioactivity Profiling
2.6. In Vitro Succinylation Reactions of 2 and 3
2.7. Applied Software, DNA Sequence Analysis, and Bioinformatics Methods
3. Results
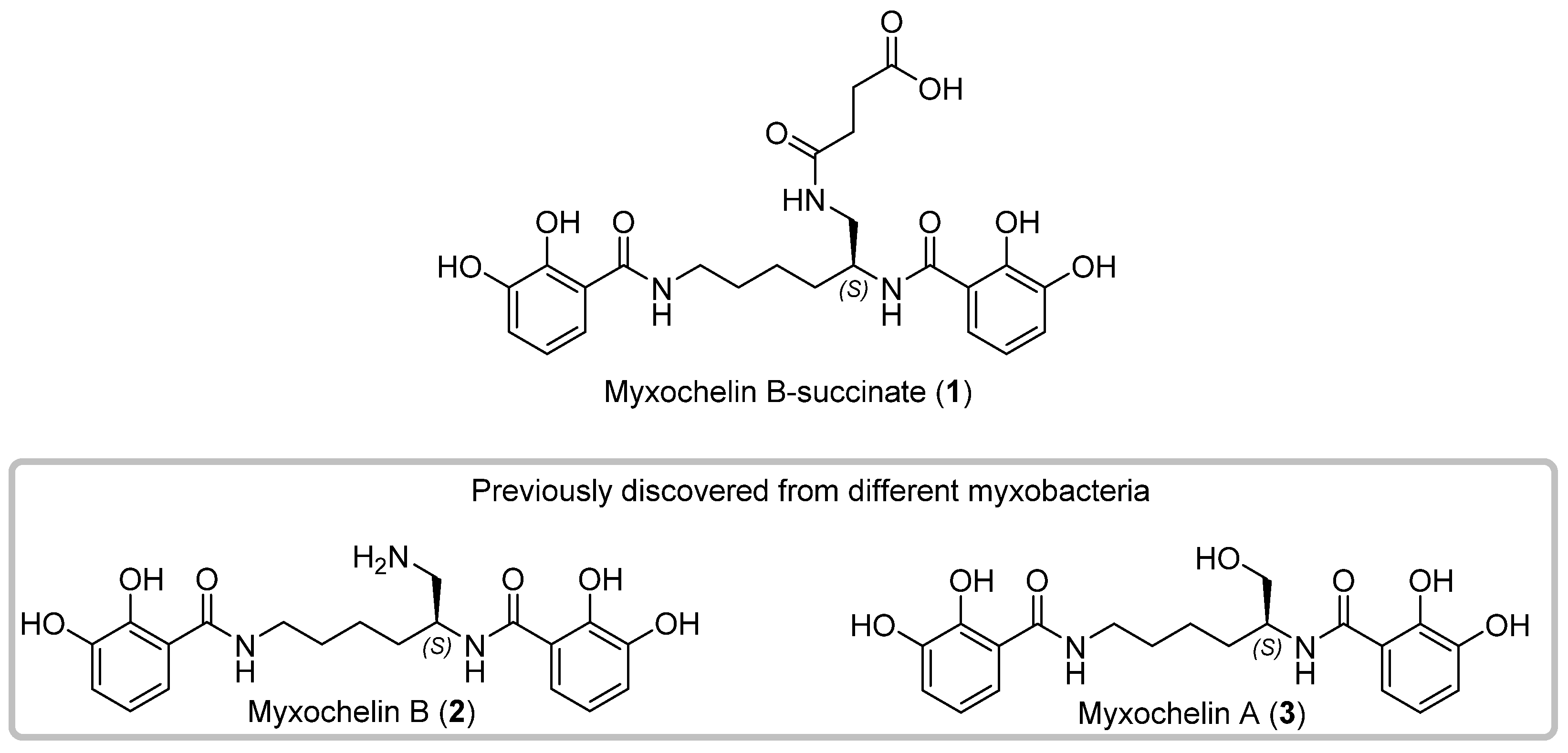
3.1. Discovery, Isolation and Structural Elucidation of 1
3.2. Bioactivity of 1–3
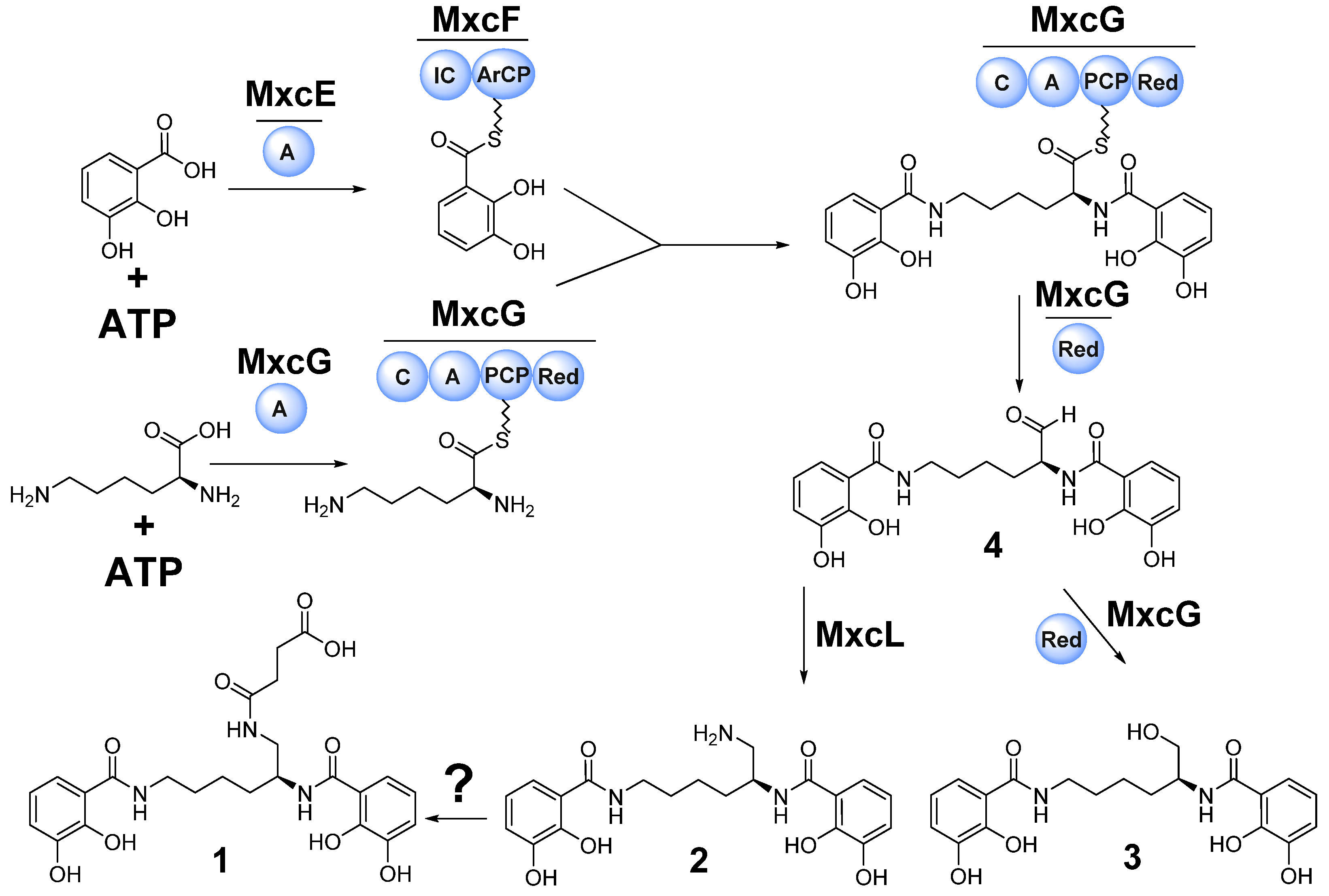
3.3. Biosynthesis of 1–3
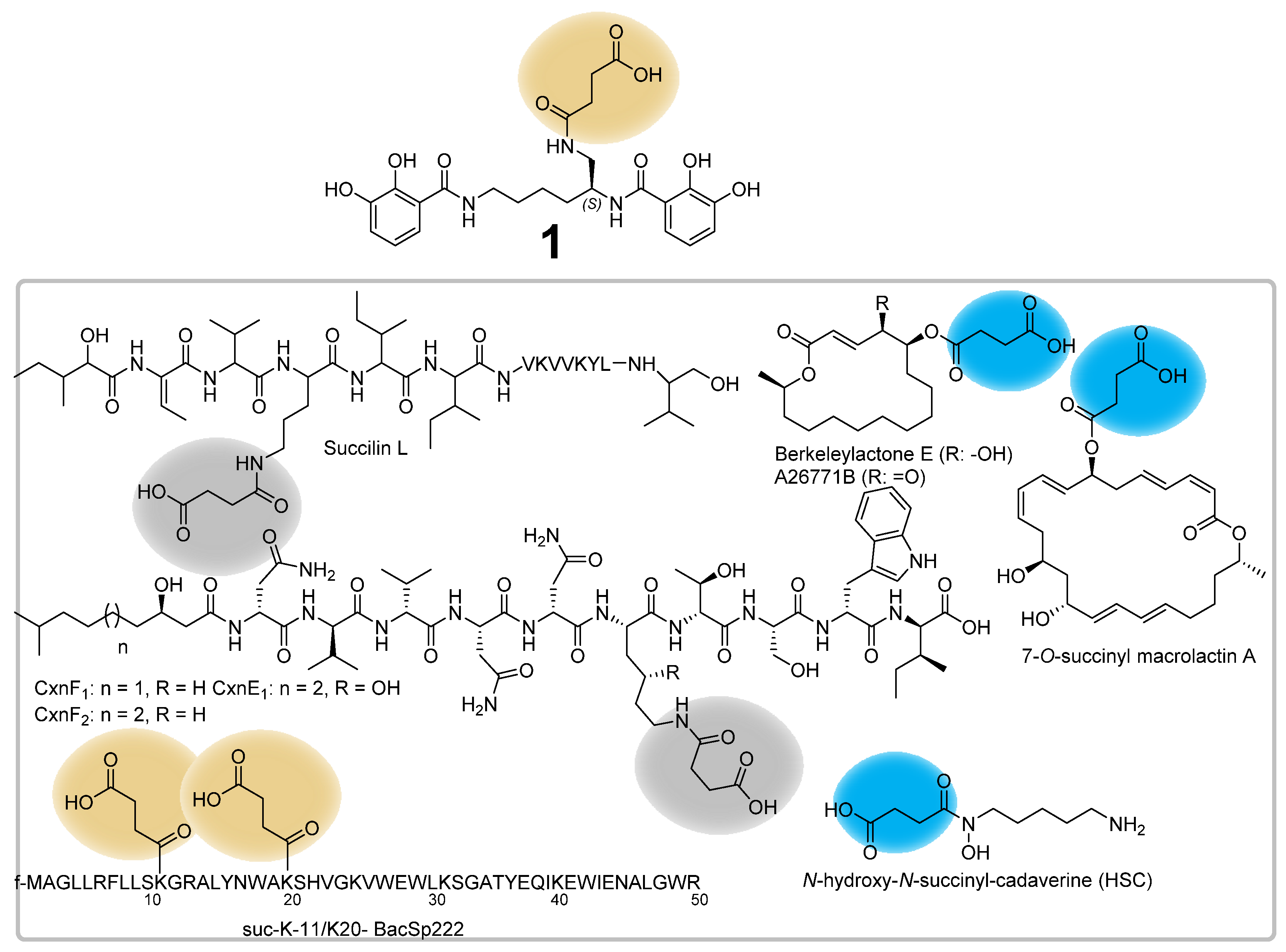
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crosa, J.H.; Mey, A.R.; Payne, S.M. (Eds.) Iron Transport in Bacteria; ASM Press: Washington, DC, USA, 2004; ISBN 9781683672050. [Google Scholar]
- Templeton, D. Molecular and Cellular Iron Transport; CRC Press: Boca Raton, FL, USA, 2002; ISBN 9780824744175. [Google Scholar]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Sandy, M.; Butler, A. Microbial iron acquisition: Marine and terrestrial siderophores. Chem. Rev. 2009, 109, 4580–4595. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.M.; Challis, G.L. Recent advances in siderophore biosynthesis. Curr. Opin. Chem. Biol. 2009, 13, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Barona-Gomez, F.; Wong, U.; Giannakopulos, A.E.; Derrick, P.J.; Challis, G.L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. [Google Scholar] [CrossRef]
- Barona-Gomez, F.; Lautru, S.; Francou, F.X.; Leblond, P.; Pernodet, J.L.; Challis, G.L. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 2006, 152, 3355–3366. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Rigouin, C.; Godet, J. An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ. Microbiol. 2020, 22, 1447–1466. [Google Scholar] [CrossRef]
- Munoz-Dorado, J.; Marcos-Torres, F.J.; Garcia-Bravo, E.; Moraleda-Munoz, A.; Perez, J. Myxobacteria: Moving, Killing, Feeding, and Surviving Together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef]
- Herrmann, J.; Fayad, A.A.; Müller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef]
- Kunze, B.; Trowitzsch-Kienast, W.; Höfle, G.; Reichenbach, H. Nannochelins A, B and C, new iron-chelating compounds from Nannocystis exedens (myxobacteria). Production, isolation, physico-chemical and biological properties. J. Antibiot. 1992, 45, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Nadmid, S.; Plaza, A.; Lauro, G.; Garcia, R.; Bifulco, G.; Müller, R. Hyalachelins A-C, unusual siderophores isolated from the terrestrial myxobacterium Hyalangium minutum. Org. Lett. 2014, 16, 4130–4133. [Google Scholar] [CrossRef] [PubMed]
- Kunze, B.; Bedorf, N.; Kohl, W.; Höfle, G.; Reichenbach, H. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J. Antibiot. 1989, 42, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, H.D.; Hartmann, V.; Pistorius, D.; Reissbrodt, R.; Trowitzsch-Kienast, W. Myxochelins B, C, D, E and F: A new structural principle for powerful siderophores imitating nature. Eur. J. Org. Chem. 1998, 541–551. [Google Scholar] [CrossRef]
- Frank, N.A.; Széles, M.; Akone, S.H.; Rasheed, S.; Hüttel, S.; Frewert, S.; Hamed, M.M.; Herrmann, J.; Schuler, S.M.M.; Hirsch, A.K.H.; et al. Expanding the Myxochelin Natural Product Family by Nicotinic Acid Containing Congeners. Molecules 2021, 26, 4929. [Google Scholar] [CrossRef]
- Gaitatzis, N.; Kunze, B.; Müller, R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 2001, 98, 11136–11141. [Google Scholar] [CrossRef]
- Li, Y.; Weissman, K.J.; Müller, R. Myxochelin biosynthesis: Direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J. Am. Chem. Soc. 2008, 130, 7554–7555. [Google Scholar] [CrossRef]
- Silakowski, B.; Kunze, B.; Nordsiek, G.; Blöcker, H.; Höfle, G.; Müller, R. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur. J. Biochem. 2000, 267, 6476–6485. [Google Scholar] [CrossRef]
- Gaitatzis, N.; Kunze, B.; Müller, R. Novel insights into siderophore formation in myxobacteria. ChemBioChem 2005, 6, 365–374. [Google Scholar] [CrossRef]
- Krug, D.; Zurek, G.; Revermann, O.; Vos, M.; Velicer, G.J.; Müller, R. Discovering the Hidden Secondary Metabolome of Myxococcus xanthus: A Study of Intraspecific Diversity. Appl. Environ. Microbiol. 2008, 74, 3058–3068. [Google Scholar] [CrossRef]
- Kim, J.; Choi, J.N.; Kim, P.; Sok, D.E.; Nam, S.W.; Lee, C.H. LC-MS/MS Profiling-Based Secondary Metabolite Screening of Myxococcus xanthus. J. Microbiol. Biotechnol. 2009, 19, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Miyanaga, S.; Obata, T.; Onaka, H.; Fujita, T.; Saito, N.; Sakurai, H.; Saiki, I.; Furumai, T.; Igarashi, Y. Absolute configuration and antitumor activity of myxochelin A produced by Nonomuraea pusilla TP-A0861. J. Antibiot. 2006, 59, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Bitok, J.K.; Lemetre, C.; Ternei, M.A.; Brady, S.F. Identification of biosynthetic gene clusters from metagenomic libraries using PPTase complementation in a Streptomyces host. FEMS Microbiol. Lett. 2017, 364, fnx155. [Google Scholar] [CrossRef]
- Kiss, H.; Nett, M.; Domin, N.; Martin, K.; Maresca, J.A.; Copeland, A.; Lapidus, A.; Lucas, S.; Berry, K.W.; Glavina Del Rio, T.; et al. Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand. Genom. Sci. 2011, 5, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, E.C.; Stierhof, M.; Goralczyk, S.; Vabre, F.M.; Pellissier, L.; Hanssen, K.Ø.; La Cruz, M.d.; Díaz, C.; Witte, P.d.; Copmans, D.; et al. Pseudochelin A, a siderophore of Pseudoalteromonas piscicida S2040. Tetrahedron 2017, 73, 2633–2637. [Google Scholar] [CrossRef]
- Miyanaga, S.; Sakurai, H.; Saiki, I.; Onaka, H.; Igarashi, Y. Synthesis and evaluation of myxochelin analogues as antimetastatic agents. Bioorg. Med. Chem. 2009, 17, 2724–2732. [Google Scholar] [CrossRef]
- Korp, J.; König, S.; Schieferdecker, S.; Dahse, H.-M.; König, G.M.; Werz, O.; Nett, M. Harnessing Enzymatic Promiscuity in Myxochelin Biosynthesis for the Production of 5-Lipoxygenase Inhibitors. ChemBioChem 2015, 16, 2445–2450. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Wang, D.-G.; Niu, L.; Lin, Z.-M.; Wang, J.-J.; Gao, D.-F.; Sui, H.-Y.; Li, Y.-Z.; Wu, C. The Discovery and Biosynthesis of Nicotinic Myxochelins from an Archangium sp. SDU34. J. Nat. Prod. 2021, 84, 2744–2748. [Google Scholar] [CrossRef]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; van Dien, S.J.; Rabinowitz, J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Simon, E.J.; Shemin, D. The Preparation of S-Succinyl Coenzyme A. Adv. Ceram. Mater. 1953, 75, 2520. [Google Scholar] [CrossRef]
- Okoth, D.A.; Hug, J.J.; Mándi, A.; Kurtán, T.; Garcia, R.; Müller, R. Structure and biosynthesis of sorangipyranone—A new γ-dihydropyrone from the myxobacterial strain MSr12020. J. Ind. Microbiol. 2021, 48, kuab029. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 803. [Google Scholar] [CrossRef]
- Ahn, J.-W.; Lee, C.-O.; Baek, S.-H. Myxochelin A, a cytotoxic antibiotic from the myxobacterium Angiococcus disciformis. Orient. Pharm. Exp. Med. 2002, 2, 64–67. [Google Scholar] [CrossRef]
- Corbin, J.L.; Bulen, W.A. The isolation and identification of 2,3-dihydroxybenzoic acid and 2-N,6-N-di-92,3-dihydroxybenzoyl)-L-lysine formed by iron-deficient Azotobacter vinelandii. Biochemistry 1969, 8, 757–762. [Google Scholar] [CrossRef]
- Sester, A.; Winand, L.; Pace, S.; Hiller, W.; Werz, O.; Nett, M. Myxochelin- and Pseudochelin-Derived Lipoxygenase Inhibitors from a Genetically Engineered Myxococcus xanthus Strain. J. Nat. Prod. 2019, 82, 2544–2549. [Google Scholar] [CrossRef]
- Sinha, S.; Doble, M.; Manju, S.L. 5-Lipoxygenase as a drug target: A review on trends in inhibitors structural design, SAR and mechanism based approach. Bioorg. Med. Chem. 2019, 27, 3745–3759. [Google Scholar] [CrossRef]
- Schieferdecker, S.; König, S.; Koeberle, A.; Dahse, H.-M.; Werz, O.; Nett, M. Myxochelins target human 5-lipoxygenase. J. Nat. Prod. 2015, 78, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Schieferdecker, S.; König, S.; Pace, S.; Werz, O.; Nett, M. Myxochelin-Inspired 5-Lipoxygenase Inhibitors: Synthesis and Biological Evaluation. ChemMedChem 2017, 12, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef] [PubMed]
- Śmiałek, J.; Nowakowski, M.; Bzowska, M.; Bocheńska, O.; Wlizło, A.; Kozik, A.; Dubin, G.; Mak, P. Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. Int. J. Mol. Sci. 2021, 22, 6256. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.B.; Breugst, M.; Mayr, H. Towards a general scale of nucleophilicity? Angew. Chem. Int. Ed. 2006, 45, 3869–3874. [Google Scholar] [CrossRef]
- Wagner, G.R.; Bhatt, D.P.; O’Connell, T.M.; Thompson, J.W.; Dubois, L.G.; Backos, D.S.; Yang, H.; Mitchell, G.A.; Ilkayeva, O.R.; Stevens, R.D.; et al. A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell Metab. 2017, 25, 823–837.e8. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Bochmann, S.M.; Spieß, T.; Kötter, P.; Entian, K.-D. Synthesis and succinylation of subtilin-like lantibiotics are strongly influenced by glucose and transition state regulator AbrB. Appl. Environ. Microbiol. 2015, 81, 614–622. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Surgenor, R.R.; Khey, K.M.W.; Vederas, J.C. Total Synthesis and Stereochemical Assignment of the Antimicrobial Lipopeptide Cerexin A1. Org. Lett. 2015, 17, 5428–5431. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Vries, R.H.d.; Chakraborty, P.; Song, C.; Zhao, X.; Scheffers, D.-J.; Roelfes, G.; Kuipers, O.P. Novel modifications of nonribosomal peptides from Brevibacillus laterosporus MG64 and investigation of their mode of action. Appl. Environ. Microbiol. 2020, 86, e01981-20. [Google Scholar] [CrossRef]
- Rebets, Y.; Nadmid, S.; Paulus, C.; Dalem, C.; Hermann, J.; Hübner, H.; Rückert, C.; Kiemer, A.; Gmeiner, P.; Kalinowski, J.; et al. Perquinolines A-C: Unprecedented bacterial tetrahydroisoquinolines involving an intriguing biosynthesis. Angew. Chem. Int. Ed. 2019, 58, 12930–12934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, J.; Zhang, L.; Zhang, C.; Liu, B.; Hu, Y. Self-Resistance in the Biosynthesis of Fungal Macrolides Involving Cycles of Extracellular Oxidative Activation and Intracellular Reductive Inactivation. Angew. Chem. Int. Ed. 2021, 60, 6639–6645. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Dong, S.; Liu, Y.; Feng, Y.; Li, H.; Yun, C.-H.; Cui, Q.; Li, W. Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase. J. Am. Chem. Soc. 2020, 142, 16031–16038. [Google Scholar] [CrossRef] [PubMed]
- Ronan, J.L.; Kadi, N.; McMahon, S.A.; Naismith, J.H.; Alkhalaf, L.M.; Challis, G.L. Desferrioxamine biosynthesis: Diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Alleyn, M.; Breitzig, M.; Lockey, R.; Kolliputi, N. The dawn of succinylation: A posttranslational modification. Am. J. Physiol. Cell Physiol. 2018, 314, C228–C232. [Google Scholar] [CrossRef] [PubMed]


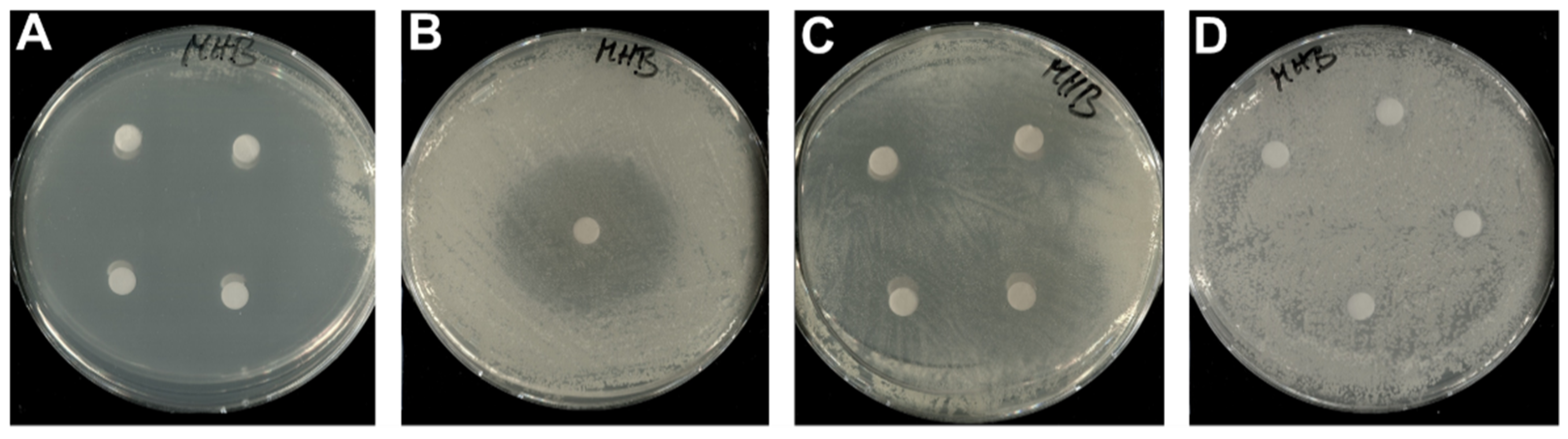

| Zone of Inhibition in mm | |||||||
|---|---|---|---|---|---|---|---|
| Microorganism | 1 | 2 | 3 | Kanamycin | Fusidic Acid | Cycloheximide | MeOH/DMSO |
| Escherichia coli HS 996 | 6 | 6 | 6 | 30 | NT | NT | 6 |
| E. coli BW251123 | 6 | 6 | 6 | 32 | NT | NT | 6 |
| Micrococcus luteus DSM 1790 | 40 | 40 | 6 | NT | 44 | NT | 6 |
| Bacillus subtilis DSM 10 | 6 | 10 | 6 | NT | 27 | NT | 6 |
| Mucor hiemalis DSM 2656 | 6 | 6 | 6 | NT | NT | 15 | 6 |
| Pichia anomala DSM 6766 | 6 | 6 | 6 | NT | NT | 52 | 6 |
| IC50 Values of 1–3 in μg/mL | ||||
|---|---|---|---|---|
| Cancer Cell Line | 1 | 2 | 3 | Doxorubicin |
| HCT-116 | 23.3 | 23.2 | 24.3 | 0.1 |
| KB-3-1 | 22.7 | 21.2 | 41.7 | 0.6 |
| U-2 OS | 31.5 | 29.1 | 34.6 | 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okoth, D.A.; Hug, J.J.; Garcia, R.; Müller, R. Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020. Microorganisms 2022, 10, 1959. https://doi.org/10.3390/microorganisms10101959
Okoth DA, Hug JJ, Garcia R, Müller R. Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020. Microorganisms. 2022; 10(10):1959. https://doi.org/10.3390/microorganisms10101959
Chicago/Turabian StyleOkoth, Dorothy A., Joachim J. Hug, Ronald Garcia, and Rolf Müller. 2022. "Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020" Microorganisms 10, no. 10: 1959. https://doi.org/10.3390/microorganisms10101959
APA StyleOkoth, D. A., Hug, J. J., Garcia, R., & Müller, R. (2022). Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial Strain MSr12020. Microorganisms, 10(10), 1959. https://doi.org/10.3390/microorganisms10101959







