A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites, Sampling and Environmental Descriptors
2.2. DNA Extraction
2.3. qPCR Standard Curves
2.4. PCR and Next-Generation Sequencing
2.5. Bioinformatic Pipeline
2.6. Statistics
2.7. Flow Cytometry Analysis
3. Results
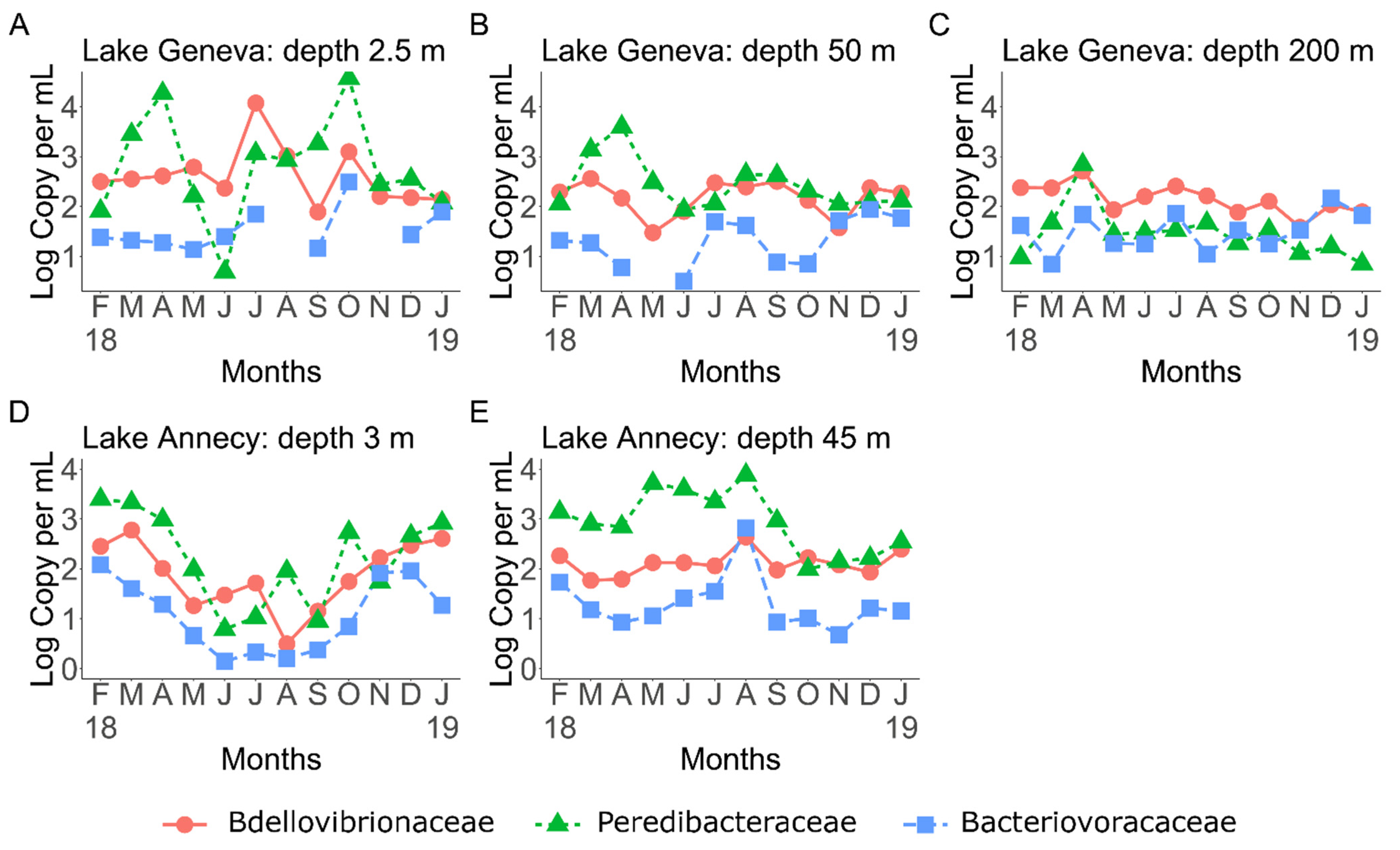
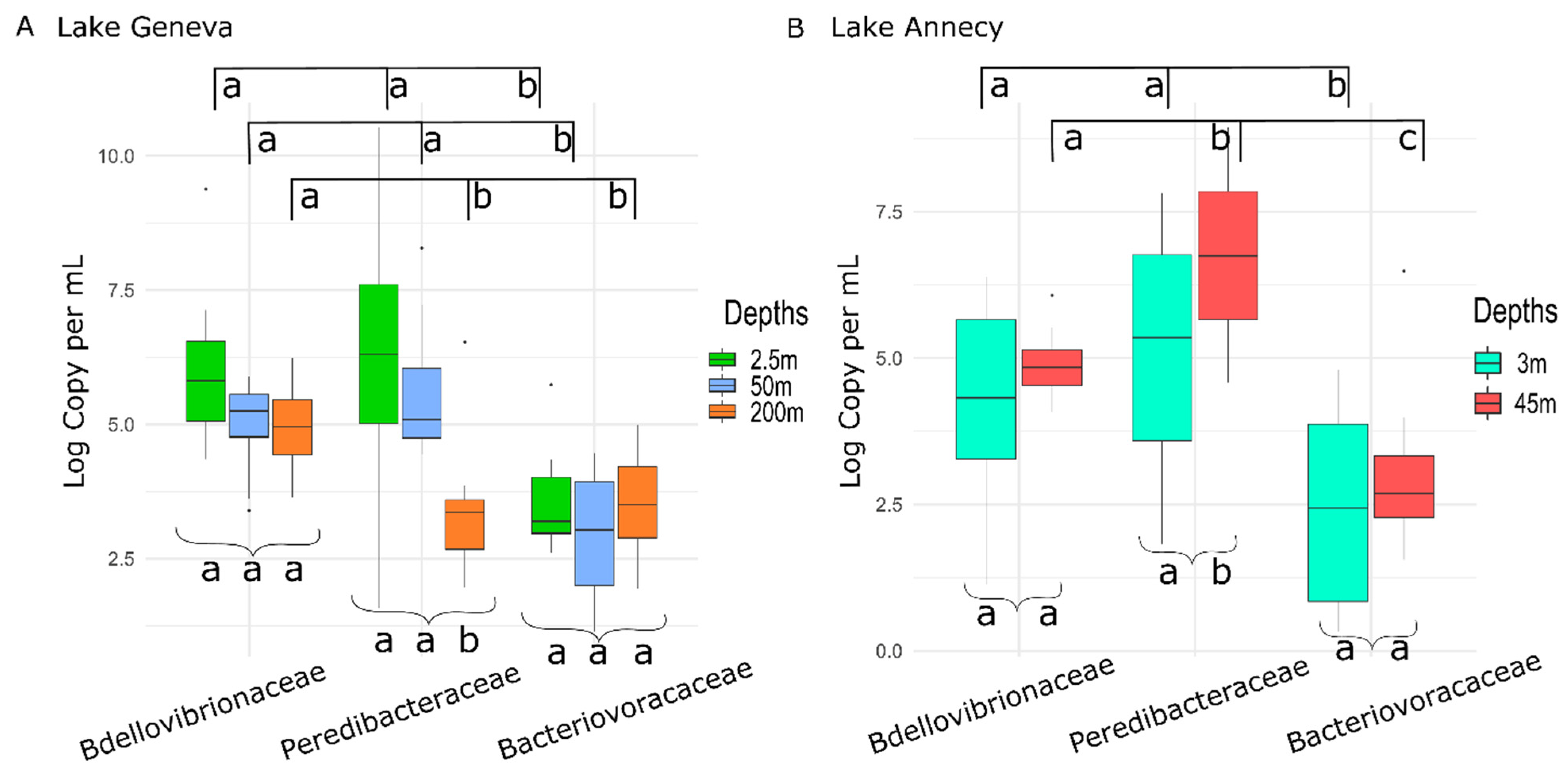
3.1. BALOs Abundance, Distribution and Dynamics
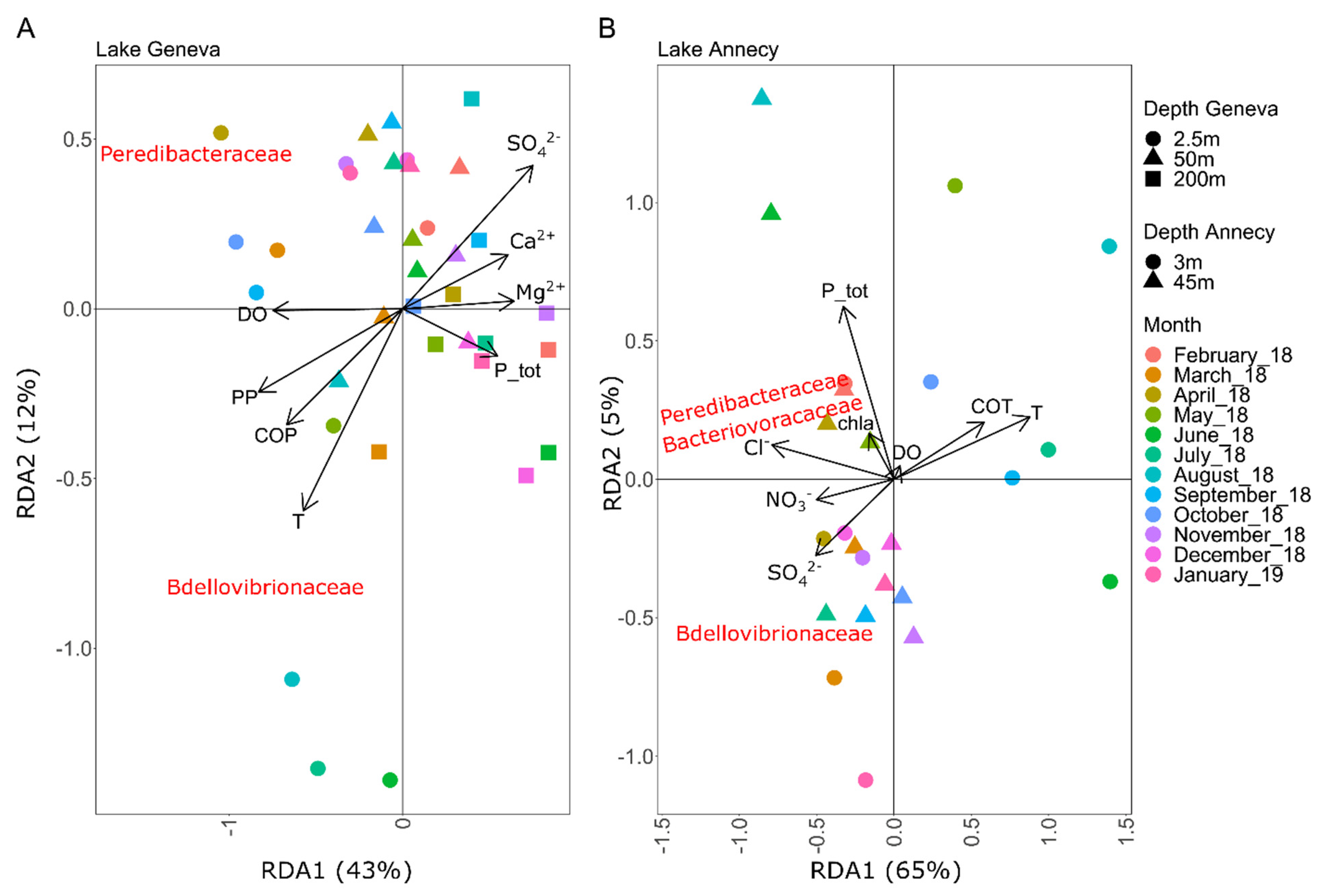
3.2. Relationships between BALOs Abundance and Environmental Variables
3.3. OTUs Diversity and Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jurkevitch, E.; Davidov, Y. Phylogenetic Diversity and Evolution of Predatory Prokaryotes. In Predatory Prokaryotes. Microbiology Monographs; Jurkevitch, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar] [CrossRef]
- Thiery, S.; Kaimer, C. The Predation Strategy of Myxococcus xanthus. Front. Microbiol. 2020, 11, 2. [Google Scholar] [CrossRef]
- Rotem, O.; Pasternak, Z.; Jurkevitch, E. The Genus Bdellovibrio and Like Organisms. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Iebba, V.; Totino, V.; Santangelo, F.; Gagliardi, A.; Ciotoli, L.; Virga, A.; Ambrosi, C.; Pompili, M.; De Biase, R.V.; Selan, L.; et al. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus cystic fibrosis isolates. Front. Microbiol. 2014, 5, 280. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, Y.; Yaacov, D.; Koval, S.; Jurkevitch, E. Samll eats big: Ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator-prey interactions. Agronomie 2003, 23, 433–439. [Google Scholar] [CrossRef]
- Johnke, J.; Fraune, S.; Bosch, T.C.G.; Hentschel, U.; Schulenburg, H. Bdellovibrio and Like Organisms Are Predictors of Microbiome Diversity in Distinct Host Groups. Microb. Ecol. 2020, 79, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Mun, W.; Kwon, H.; Im, H.; Choi, S.Y.; Monnappa, A.K.; Mitchell, R.J. Cyanide Production by Chromobacterium piscinae Shields It from Bdellovibrio bacteriovorus HD100 Predation. MBio 2017, 8, e01370-17. [Google Scholar] [CrossRef]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological Potential of Bdellovibrio and Like Organisms and Their Secreted Enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Willis, A.R.; Moore, C.; Mazon-Moya, M.; Krokowski, S.; Lambert, C.; Till, R.; Mostowy, S.; Sockett, R.E. Injections of Predatory Bacteria Work Alongside Host Immune Cells to Treat Shigella Infection in Zebrafish Larvae. Curr. Biol. 2016, 26, 3343–3351. [Google Scholar] [CrossRef]
- Cao, H.; An, J.; Zheng, W.; He, S. Vibrio cholerae pathogen from the freshwater-cultured whiteleg shrimp Penaeus vannamei and control with Bdellovibrio bacteriovorus. J. Invertebr. Pathol. 2015, 130, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, C.; Sun, Q.; Liu, R.; Cai, J. Bdellovibrio and like organisms enhanced growth and survival of Penaeus monodon and altered bacterial community structures in its rearing water. Appl. Environ. Microbiol. 2014, 80, 6346–6354. [Google Scholar] [CrossRef]
- Youdkes, D.; Helman, Y.; Burdman, S.; Matan, O.; Jurkevitch, E. Potential Control of Potato Soft Rot Disease by the Obligate Predators Bdellovibrio and Like Organisms. Appl. Environ. Microbiol. 2020, 86, e02543-19. [Google Scholar] [CrossRef]
- Sivakala, K.K.; Jose, P.A.; Matan, O.; Zohar-Perez, C.; Nussinovitch, A.; Jurkevitch, E. In vivo predation and modification of the Mediterranean fruit fly Ceratitis capitata (Wiedemann) gut microbiome by the bacterial predator Bdellovibrio bacteriovorus. J. Appl. Microbiol. 2021, 131, 2971–2980. [Google Scholar] [CrossRef] [PubMed]
- Fratamico, P.M.; Whiting, R.C. Ability of Bdellovibrio bacteriovorus 109j to lyse gram-negative food-borne pathogenic and spoilage bacteria. J. Food Prot. 1995, 58, 160–164. [Google Scholar] [CrossRef]
- Fratamico, P.M.; Cooke, P.H. Isolation of Bdellovibrios that prey on Escherichia coli O157:H7 and Salmonella species and application for removal of prey from stainless steel surfaces. J. Food Saf. 1996, 16, 161–173. [Google Scholar] [CrossRef]
- Koval, S.F.; Williams, H.N.; Colin Stine, O. Reclassification of Bacteriovorax marinus as Halobacteriovorax marinus gen. Nov., comb. nov. and Bacteriovorax litoralis as Halobacteriovorax litoralis comb. nov.; description of Halobacteriovoraceae fam. nov. in the class Deltaproteobacteria. Int. J. Syst. Evol. Microbiol. 2015, 65, 593–597. [Google Scholar] [CrossRef]
- Hahn, M.W.; Schmidt, J.; Koll, U.; Rohde, M.; Verbarg, S.; Pitt, A.; Nakai, R.; Naganuma, T.; Lang, E. Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales. Int. J. Syst. Evol. Microbiol. 2017, 67, 2555–2568. [Google Scholar] [CrossRef]
- Ezzedine, J.A.; Chardon, C.; Jacquet, S. New 16S rRNA primers to uncover Bdellovibrio and like organisms diversity and abundance. J. Microbiol. Methods 2020, 175, 105996. [Google Scholar] [CrossRef]
- Babut, M.; Mulhauser, G. La Lettre du Léman. Bulletin de la Commission Internationale pour la Protection des eaux du Léman 2018, 57, 1–8. [Google Scholar]
- Gerdeaux, D.; Druart, J.-C.; Laine, L.; Lazzarotto, J.; Perga, M.-E.; Suivi de la Qualité des Eaux du lac d’Annecy—Rapport 2006. Syndicat mixte du lac d’Annecy 2006. Available online: https://hal.archives-ouvertes.fr/ (accessed on 23 March 2020).
- Rimet, F.; Anneville, O.; Barbet, D.; Chardon, C.; Crépin, L.; Domaizon, I.; Dorioz, J.-M.; Espinat, L.; Frossard, V.; Guillard, J.; et al. The Observatory on LAkes (OLA) database: Sixty years of environmental data accessible to the public. J. Limnol. 2020, 79, 164–178. [Google Scholar] [CrossRef]
- Ezzedine, J.A.; Desdevises, Y.; Jacquet, S. Exploring archaeal and bacterial diversity and co-occurrence in Lake Geneva. Adv. Oceanogr. Limnol. 2020, 11, 2. Available online: https://www.pagepressjournals.org/index.php/aiol/article/view/aiol.2020.9099 (accessed on 23 March 2020).
- Chonova, T.; Keck, F.; Labanowski, J.; Montuelle, B.; Rimet, F.; Bouchez, A. Separate treatment of hospital and urban wastewaters: A real scale comparison of effluents and their effect on microbial communities. Sci. Total Environ. 2016, 542, 965–975. [Google Scholar] [CrossRef]
- Paix, B.; Ezzedine, J.A.; Jacquet, S. Diversity, dynamics and distribution of Bdellovibrio and like organisms in peri-alpine lakes. Appl. Environ. Microbiol. 2019, 85, e02494-18. [Google Scholar] [CrossRef]
- Ezzedine, J.A.; Jacas, L.; Desdevises, Y.; Jacquet, S. Bdellovibrio and Like Organisms in Lake Geneva: An Unseen Elephant in the Room? Front. Microbiol. 2020, 11, 98. [Google Scholar] [CrossRef]
- Van Essche, M.; Sliepen, I.; Loozen, G.; Van Eldere, J.; Quirynen, M.; Davidov, Y.; Jurkevitch, E.; Boon, N.; Teughels, W. Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ. Microbiol. Rep. 2009, 1, 228–233. [Google Scholar] [CrossRef]
- Whelan, J.A.; Russel, N.B.; Whelan, M.A. A method for the absolute quantification of cDNA using real time PCR. J. Immunol. 2003, 278, 261–269. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarmv2: Highly-scalable and high-resolution amplicon clustering. PeerJ 2015, 2015, e1420. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Glo, F.O.; Yarza, P. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2018. Available online: https://ggplot2.tidyverse.org/ (accessed on 23 March 2020).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package “vegan” Title Community Ecology Package. Community Ecol. Packag. 2019, 2, 1–297. [Google Scholar]
- Powell, J. Ecological Analysis of Communities; R Notes Community Ecology; Hawkesbury Institute for the Environment, University of Western Sydney: Richmond, Australia, 2018. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Visualization of a Correlation Matrix. Statistician 2017, 56, 316–324. [Google Scholar]
- Jacquet, S.; Dorigo, U.; Personnic, S. A Few Tests Prior to Flow Cytometry and Epifluorescence Analyses of Freshwater Bacterio- and Virioplankton Communities. Flow Cytom 2013, 1, 1–30. [Google Scholar]
- Vaulot, D.; Courties, C.; Partensky, F. A simple method to preserve oceanic phytoplankton for flow cytometric analyses. Cytometry 1989, 10, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Davidov, Y.; Jurkevitch, E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1439–1452. [Google Scholar] [CrossRef]
- Ezzedine, J.; Pavard, G.; Gardillon, M.; Jacquet, S. Bdellovibrio sp.: An Important Bacterial Predator in Lake Geneva? J. Microbiol. Biotechnol. 2020, 5, 10-23880. [Google Scholar] [CrossRef]
- Ezzedine, J.A.; Janicot, A.; Rasconi, S.; Domaizon, I.; Jacquet, S. Short-Term Dynamics of Bdellovibrio and Like Organisms in Lake Geneva in Response to a Simulated Climatic Extreme Event. Microb. Ecol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Zhang, S.; Chen, Z.; Li, C. Isolation and application of predatory Bdellovibrio-and-like organisms for municipal waste sludge biolysis and dewaterability enhancement. Front. Environ. Sci. Eng. 2017, 11, 10. [Google Scholar] [CrossRef]
- Davidov, Y.; Huchon, D.; Koval, S.F.; Jurkevitch, E. A new α-proteobacterial clade of Bdellovibrio-like predators: Implications for the mitochondrial endosymbiotic theory. Environ. Microbiol. 2006, 8, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Kandel, P.P.; Pasternak, Z.; van Rijn, J.; Nahum, O.; Jurkevitch, E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol. Ecol. 2014, 89, 149–161. [Google Scholar] [CrossRef]
- Williams, H.N.; Piñeiro, S. Ecology of the predatory Bdellovibrio and like organisms. In Predatory Prokaryotes. Microbiology Monographs; Jurkevitch, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4, pp. 214–247. [Google Scholar] [CrossRef]
- Sutton, D.C.; Besant, P.J. Ecology and characteristics of bdellovibrios from three tropical marine habitats. Mar. Biol. 1994, 119, 313–320. [Google Scholar] [CrossRef]
- Jashnsaz, H.; Al Juboori, M.; Weistuch, C.; Miller, N.; Nguyen, T.; Meyerhoff, V.; McCoy, B.; Perkins, S.; Wallgren, R.; Ray, B.D.; et al. Hydrodynamic Hunters. Biophys. J. 2017, 112, 1282–1289. [Google Scholar] [CrossRef]
- Hespell, R.B.; Thomashow, M.F.; Rittenberg, S.C. Changes in cell composition and viability of Bdellovibrio bacteriovorus during starvation. Arch. Microbiol. 1974, 97, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Sathyamoorthy, R.; Kushmaro, Y.; Rotem, O.; Matan, O.; Kadouri, D.E.; Huppert, A.; Jurkevitch, E. To hunt or to rest: Prey depletion induces a novel starvation survival strategy in bacterial predators. ISME J. 2020, 15, 109–123. [Google Scholar] [CrossRef]
- Varon, M.; Zeigler, B.P. Bacterial predator-prey interaction at low prey density. Appl. Environ. Microbiol. 1978, 36, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Varon, M.; Fine, M.; Stein, A. The maintenance of Bdellovibrio at low prey density. Microb. Ecol. 1984, 10, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Schoeffield, A.J.; Williams, H.N.; Turng, B.F.; Falkler, W.A. A comparison of the survival of intraperiplasmic and attack phase bdellovibrios with reduced oxygen. Microb. Ecol. 1996, 32, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Roeßler, M.; Sewald, X.; Müller, V. Chloride dependence of growth in bacteria. FEMS Microbiol. Lett. 2003, 225, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.C.; Starr, M.P. Effects of calcium and magnesium ions and host viability on growth of bdellovibrios. Antonie Van Leeuwenhoek 1973, 39, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Dashiff, A.; Junka, R.A.; Libera, M.; Kadouri, D.E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 2011, 110, 431–444. [Google Scholar] [CrossRef]
- Seidler, R.J.; Starr, M.P. Isolation and characterization of host-independent Bdellovibrios. J. Bacteriol. 1969, 100, 769–785. [Google Scholar] [CrossRef]
- Morin, M.; Enjalbert, B.; Ropers, D.; Girbal, L.; Cocaign-Bousquet, M. Genomewide Stabilization of mRNA during a “Feast-to-Famine” Growth Transition in Escherichia coli. mSphere 2020, 5, e00276-20. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, C.; Spitzer, K.; Elster, J.; Faßhauer, F.; Brinkmann, N.; Lepka, D.; Diwan, V.; Friedl, T. Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol. Ecol. 2016, 92, fiw122. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol Biol Evol 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Creevey, C.J.; Pentony, M.M.; Naughton, T.J.; McInerney, J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006, 6, 29. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In 2nd International Symposium on Information Theory; Petrov, B.N., Csaki, F., Eds.; Akadémiai Kiadó: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezzedine, J.A.; Scheifler, M.; Desdevises, Y.; Jacquet, S. A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva. Microorganisms 2022, 10, 1960. https://doi.org/10.3390/microorganisms10101960
Ezzedine JA, Scheifler M, Desdevises Y, Jacquet S. A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva. Microorganisms. 2022; 10(10):1960. https://doi.org/10.3390/microorganisms10101960
Chicago/Turabian StyleEzzedine, Jade A., Mathilde Scheifler, Yves Desdevises, and Stéphan Jacquet. 2022. "A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva" Microorganisms 10, no. 10: 1960. https://doi.org/10.3390/microorganisms10101960
APA StyleEzzedine, J. A., Scheifler, M., Desdevises, Y., & Jacquet, S. (2022). A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva. Microorganisms, 10(10), 1960. https://doi.org/10.3390/microorganisms10101960





