Identification of A Putative T6SS Immunity Islet in Salmonella Typhi
Abstract
1. Introduction
2. Results
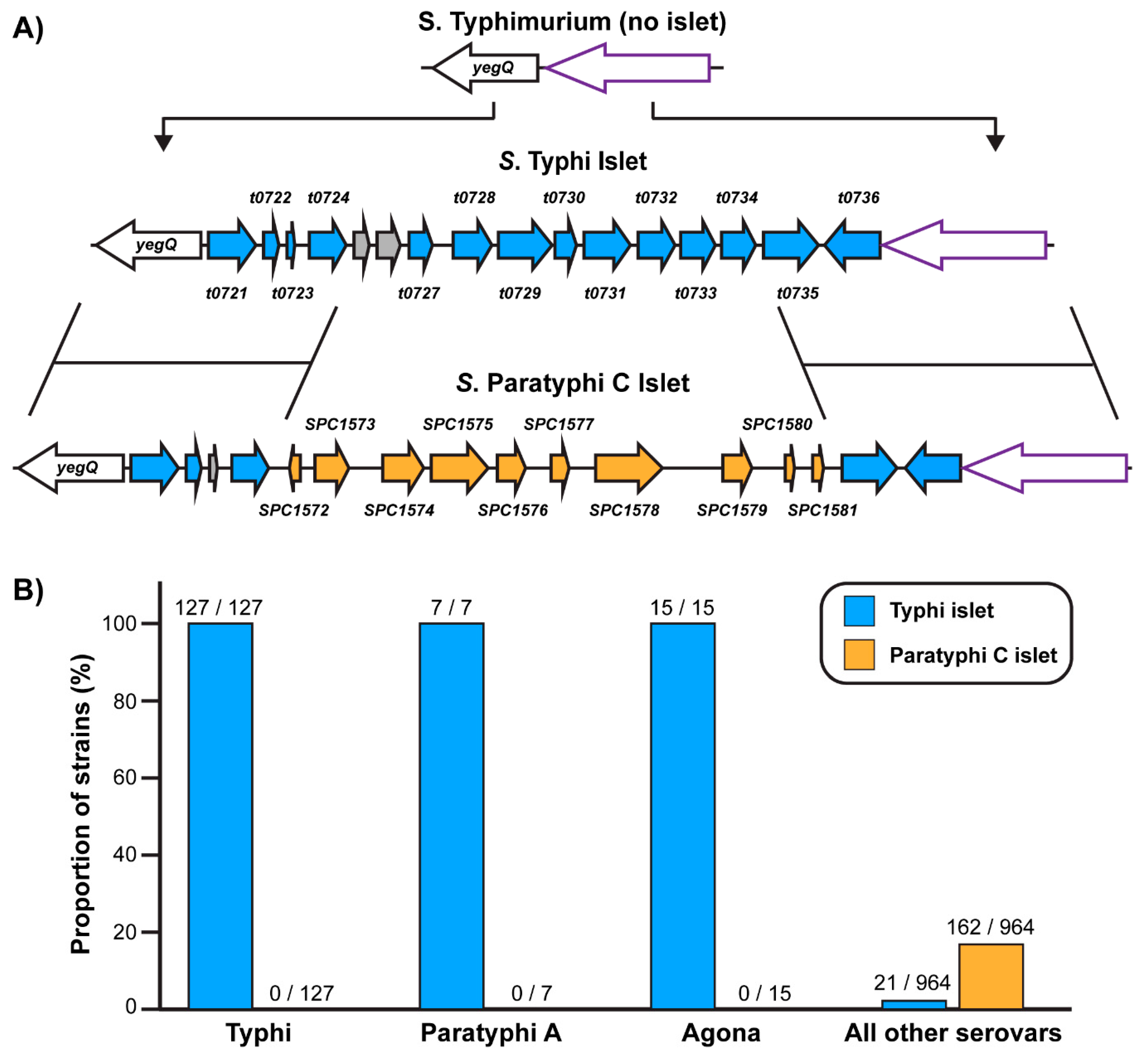
2.1. Identification and Phylogenetic Distribution of an Uncharacterized Genomic Islet Predominantly Encoded by Typhoidal Salmonellae
2.2. Conserved Domain Analysis Indicates the Uncharacterized Genomic Islet May be Involved in Interbacterial Antagonism
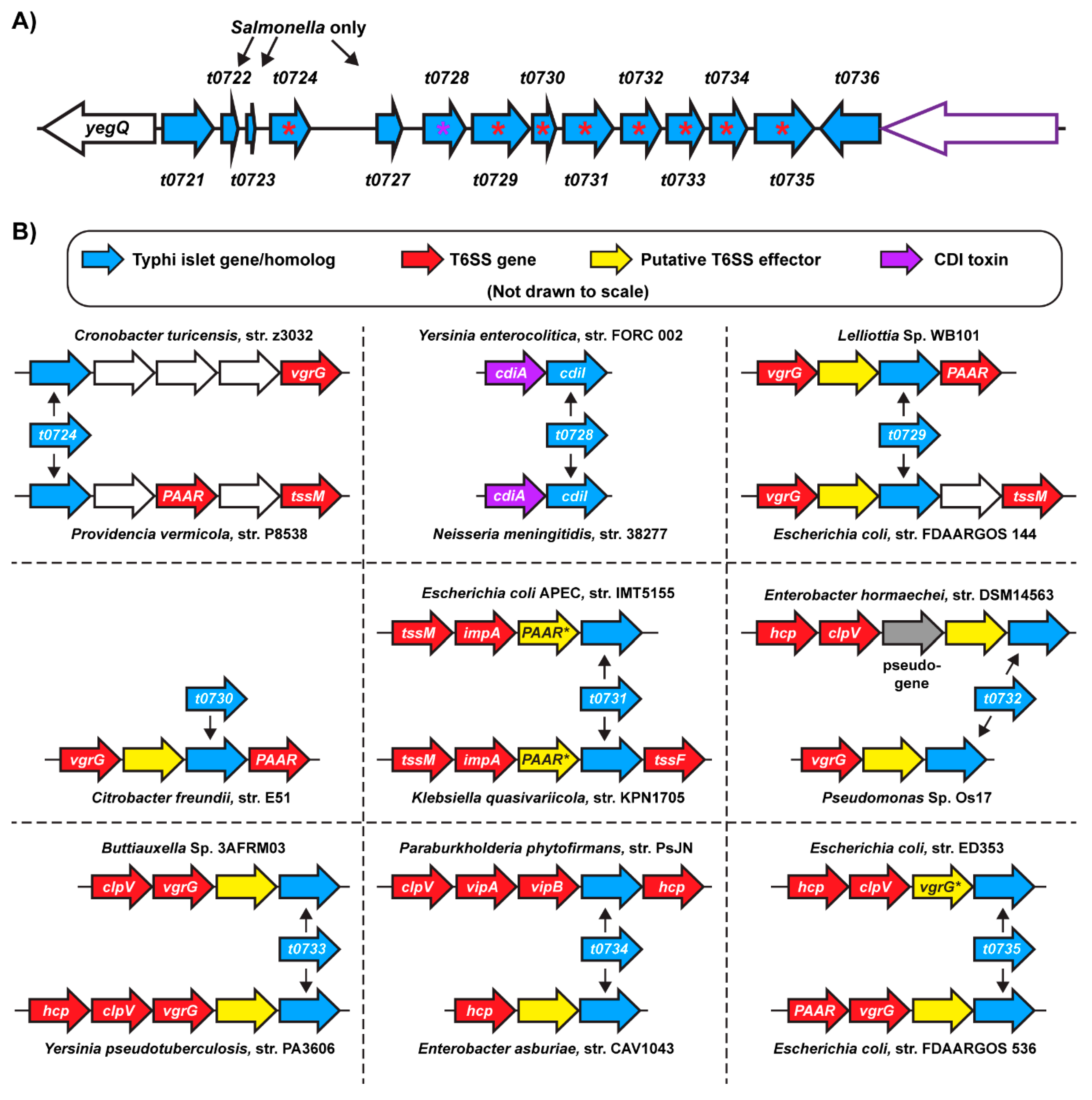
2.3. Homologs of S. Typhi Genomic Islet Genes are Found in T6SS/CDI Gene Clusters in Diverse Bacteria
2.4. Comparative Genomics and Hidden Markov Model (HMM) Homology Searches Indicate that the S. Typhi Genomic Islet is Primarily Comprised of Orphan Immunity Proteins
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Distribution Analyses
4.2. HMM Homology Searches and Conserved Domain Analyses
4.3. Genomic Context Analysis for S. Typhi Genomic Islet Homologs
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dougan, G.; Baker, S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev. Microbiol. 2014, 68, 317–336. [Google Scholar] [CrossRef] [PubMed]
- Parry, C.M.; Hien, T.T.; Dougan, G.; White, N.J.; Farrar, J.J. Typhoid Fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and acute infection of the gall bladder by salmonella Typhi: Understanding the carrier state. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef]
- Gopinath, S.; Carden, S.; Monack, D. Shedding light on salmonella carriers. Trends Microbiol. 2012, 20, 320–327. [Google Scholar] [CrossRef]
- The, C.S.; Chua, K.H.; Thong, K.L. Paratyphoid fever: Splicing the global analyses. Int. J. Med. Sci. 2014, 11, 732–741. [Google Scholar]
- McClelland, M.; Sanderson, K.E.; Clifton, S.W.; Latreille, P.; Porwollik, S.; Sabo, A.; Meyer, R.; Bieri, T.; Ozersky, P.; McLellan, M.; et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted Serovars of salmonella Enterica that cause typhoid. Nat. Genet. 2004, 36, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Achtman, M.; Parkhill, J.; Thomson, N.R.; Falush, D. A bimodal pattern of relatedness between the salmonella Paratyphi A and Typhi genomes: Convergence or divergence by homologous recombination. Genome Res. 2007, 17, 61–68. [Google Scholar] [CrossRef]
- Selander, R.K.; Beltran, P.; Smith, N.H.; Barker, R.M.; Crichton, P.B.; Old, D.C.; Musser, J.M.; Whittam, T.S. Genetic Population Structure, Clonal Phylogeny, and Pathogenicity of Salmonella Paratyphi B. Infect. Immun. 1990, 58, 1891–1901. [Google Scholar] [CrossRef]
- Liu, W.Q.; Feng, Y.; Wang, Y.; Zou, Q.H.; Chen, F.; Guo, J.T.; Peng, Y.H.; Jin, Y.; Li, Y.G.; Hu, S.N.; et al. Salmonella Paratyphi C: Genetic Divergence From Salmonella Choleraesuis and Pathogenic Convergence With Salmonella Typhi. PLoS ONE 2009, 4, e4510. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The global burden of Nontyphoidal salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Grassl, G.A.; Finlay, B.B. Pathogenesis of enteric salmonella infections. Curr. Opin. Gastroenterol. 2008, 24, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Yurist-Doutsch, S.; Arrieta, M.C.; Vogt, S.L.; Finlay, B.B. Gastrointestinal microbiota-mediated control of enteric pathogens. Annu. Rev. Genet. 2014, 48, 361–382. [Google Scholar] [CrossRef]
- Stecher, B.; Hardt, W.D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef]
- Anderson, C.J.; Kendall, M.M. Salmonella Enterica Serovar Typhimurium strategies for host adaptation. Front. Microbiol. 2017, 12, 1983. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella Enterica Serovar Typhimurium exploits inflammation to compete with the intestinal Microbiota. PLoS Biol. 2007, 5, e244. [Google Scholar] [CrossRef]
- Behnsen, J.; Jellbauer, S.; Wong, C.P.; Edwards, R.A.; George, M.D.; Ouyang, W.; Raffatellu, M. The cytokine Il-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 2014, 40, 262–273. [Google Scholar] [CrossRef]
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut inflammation provides a respiratory electron acceptor for salmonella. Nature 2010, 467, 426–429. [Google Scholar] [CrossRef]
- Thiennimitr, P.; Winter, S.E.; Winter, M.G.; Xavier, M.N.; Tolstikov, V.; Huseby, D.L.; Sterzenbach, T.; Tsolis, R.M.; Roth, J.R.; Bäumler, A.J. Intestinal Inflammation Allows Salmonella to Use Ethanolamine to Compete With the Microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 17480–17485. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Chávez, F.; Winter, S.E.; Lopez, C.A.; Xavier, M.N.; Winter, M.G.; Nuccio, S.P.; Russell, J.M.; Laughlin, R.C.; Lawhon, S.D.; Sterzenbach, T.; et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013, 9, e1003267. [Google Scholar] [CrossRef] [PubMed]
- Brunet, Y.R.; Khodr, A.; Logger, L.; Aussel, L.; Mignot, T.; Rimsky, S.; Cascales, E. H-NS silencing of the salmonella pathogenicity island 6-Encoded Type VI secretion system limits salmonella Enterica Serovar Typhimurium Interbacterial killing. Infect. Immun. 2015, 83, 2738–2750. [Google Scholar] [CrossRef]
- Sana, T.G.; Flaugnatti, N.; Lugo, K.A.; Lam, L.H.; Jacobson, A.; Baylot, V.; Durand, E.; Journet, L.; Cascales, E.; Monack, D.M. Salmonella Typhimurium Utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. USA 2016, 113, E5044–E5051. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Brunet, Y.R.; Cascales, E.; Mougous, J.D. Structure and regulation of the Type VI Secretion system. Annu. Rev. Microbiol. 2012, 66, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial Nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Wang, M.; Luo, Z.; Du, H.; Xu, S.; Ni, B.; Zhang, H.; Sheng, X.; Xu, H.; Huang, X. Molecular characterization of a functional type VI secretion system in salmonella Enterica Serovar Typhi. Curr. Microbiol. 2011, 63, 22–31. [Google Scholar] [CrossRef]
- Zhang, Y.; Brady, A.; Jones, C.; Song, Y.; Darton, T.C.; Jones, C.; Blohmke, C.J.; Pollard, A.J.; Magder, L.S.; Fasano, A.; et al. Compositional and functional differences in the human gut microbiome correlate with clinical outcome following infection with wild-type salmonella Enterica Serovar Typhi. MBio 2018, 9. [Google Scholar] [CrossRef]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of salmonella Enterica Serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Dougan, G.; James, K.D.; Thomson, N.R.; Pickard, D.; Wain, J.; Churcher, C.; Mungall, K.L.; Bentley, S.D.; Holden, M.T.; et al. Complete genome sequence of a multiple drug resistant Salmonella Enterica Serovar Typhi CT18. Nature 2001, 413, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Fowler, C.C.; Chang, S.J.; Gao, X.; Geiger, T.; Stack, G.; Galán, J.E. Emerging insights into the biology of typhoid toxin. Curr. Opin. Microbiol. 2017, 35, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Spanò, S.; Ugalde, J.E.; Galán, J.E. Delivery of a salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 2008, 3, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Gao, X.; Galán, J.E. Structure and function of the salmonella Typhi Chimaeric A(2)B(5) Typhoid Toxin. Nature 2013, 499, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Fowler, C.C.; Stack, G.; Jiao, X.; Lara-Tejero, M.; Galán, J.E. Alternate subunit assembly diversifies the function of a bacterial toxin. Nat. Commun. 2019, 10, 3684. [Google Scholar] [CrossRef]
- Pickard, D.; Wain, J.; Baker, S.; Line, A.; Chohan, S.; Fookes, M.; Barron, A.; Gaora, P.O.; Chabalgoity, J.A.; Thanky, N.; et al. Composition, acquisition, and distribution of the vi exopolysaccharide-encoding salmonella Enterica pathogenicity island SPI-7. J. Bacteriol. 2003, 185, 5055–5065. [Google Scholar] [CrossRef]
- Vernikos, G.S.; Thomson, N.R.; Parkhill, J. Genetic flux over time in the salmonella lineage. Genome Biol. 2007, 8, R100. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Morse, R.P.; Nikolakakis, K.C.; Willett, J.L.; Gerrick, E.; Low, D.A.; Hayes, C.S.; Goulding, C.W. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc. Natl. Acad. Sci. USA 2012, 109, 21480–21485. [Google Scholar] [CrossRef]
- Ruhe, Z.C.; Low, D.A.; Hayes, C.S. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013, 21, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimons, T.C.; Lewis, J.M.; Wright, A.; Kleifeld, O.; Schittenhelm, R.B.; Powell, D.; Harper, M.; Boyce, J.D. Identification of novel Acinetobacter Baumannii Type VI secretion system antibacterial effector and immunity pairs. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef]
- Cianfanelli, F.R.; Alcoforado Diniz, J.; Guo, M.; De Cesare, V.; Trost, M.; Coulthurst, S.J. VgrG and PAAR proteins define distinct versions of a functional Type VI secretion system. PLoS Pathog. 2016, 12, e1005735. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Hood, R.D.; Bui, N.K.; LeRoux, M.; Vollmer, W.; Mougous, J.D. Type VI secretion delivers Bacteriolytic effectors to target cells. Nature 2011, 475, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, W.; Liu, Q.; Gao, Y.; Gao, Y.; Wang, Y.; Wang, D.Z.; Li, Z.; Wang, T. Structural Insights on the bacteriolytic and self-protection mechanism of muramidase effector Tse3 in Pseudomonas aeruginosa. J. Biol. Chem. 2013, 288, 30607–30613. [Google Scholar] [CrossRef]
- Wang, T.; Ding, J.; Zhang, Y.; Wang, D.C.; Liu, W. Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1889–1900. [Google Scholar] [CrossRef]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Gao, Z.Q.; Wang, W.J.; Liu, G.F.; Xu, J.H.; Su, X.D.; Dong, Y.H. Structure of the Type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J. Biol. Chem. 2013, 288, 5928–5939. [Google Scholar] [CrossRef]
- Benz, J.; Reinstein, J.; Meinhart, A. Structural insights into the effector—Immunity system Tae4/Tai4 from salmonella Typhimurium. PLoS ONE 2013, 8, e67362. [Google Scholar] [CrossRef]
- Tang, B.L.; Yang, J.; Chen, X.L.; Wang, P.; Zhao, H.L.; Su, H.N.; Li, C.Y.; Yu, Y.; Zhong, S.; Wang, L. A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nat. Commun. 2020, 11, 285. [Google Scholar] [CrossRef]
- Weber, B.S.; Hennon, S.W.; Wright, M.S.; Scott, N.E.; de Berardinis, V.; Foster, L.J.; Ayala, J.A.; Adams, M.D.; Feldman, M.F. Genetic dissection of the type VI secretion system in acinetobacter and identification of a novel peptidoglycan hydrolase, Tagx, required for its biogenesis. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- de Campos, S.B.; Lardi, M.; Gandolfi, A.; Eberl, L.; Pessi, G. Mutations in Two Paraburkholderia phymatum Type VI secretion systems cause reduced fitness in Interbacterial competition. Front. Microbiol. 2017, 8, 2473. [Google Scholar] [CrossRef]
- Jana, B.; Salomon, D. Type VI secretion system: A modular toolkit for bacterial dominance. Future Microbiol. 2019, 14, 1451–1463. [Google Scholar] [CrossRef]
- Wood, T.E.; Howard, S.A.; Wettstadt, S.; Filloux, A. PAAR proteins act as the ’sorting hat’ of the type VI secretion system. Microbiology 2019, 165, 1203–1218. [Google Scholar] [CrossRef] [PubMed]
- Marcyjaniak, M.; Odintsov, S.G.; Sabala, I.; Bochtler, M. Peptidoglycan amidase MepA is a LAS metallopeptidase. J. Biol. Chem. 2004, 279, 43982–43989. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, S.S.; Hespanhol, J.T.; Nicastro, G.G.; Matsuyama, B.Y.; Mesnage, S.; Patel, A.; de Souza, R.F.; Guzzo, C.R.; Bayer-Santos, E. A Superfamily of T6SS Antibacterial Effectors Displaying L,D-carboxypeptidase activity towards peptidoglycan. bioRxiv 2020, 954545. [Google Scholar] [CrossRef]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.; Salinas, R.K.; et al. Bacterial killing via a Type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef]
- Zhang, D.; de Souza, R.F.; Anantharaman, V.; Iyer, L.M.; Aravind, L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012, 7, 18. [Google Scholar] [CrossRef]
- Wexler, A.G.; Bao, Y.; Whitney, J.C.; Bobay, L.M.; Xavier, J.B.; Schofield, W.B.; Barry, N.A.; Russell, A.B.; Tran, B.Q.; Goo, Y.A.; et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. USA 2016, 113, 3639–3644. [Google Scholar] [CrossRef]
- Kirchberger, P.C.; Unterweger, D.; Provenzano, D.; Pukatzki, S.; Boucher, Y. Sequential displacement of type vi secretion system effector genes leads to evolution of diverse immunity gene arrays in vibrio Cholerae. Sci. Rep. 2017, 7, 45133. [Google Scholar] [CrossRef] [PubMed]
- Ting, S.Y.; Bosch, D.E.; Mangiameli, S.M.; Radey, M.C.; Huang, S.; Park, Y.J.; Kelly, K.A.; Filip, S.K.; Goo, Y.A.; Eng, J.K.; et al. Bifunctional immunity proteins protect bacteria against FtsZ-Targeting ADP-Ribosylating toxins. Cell 2018, 175, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Ross, B.D.; Verster, A.J.; Radey, M.C.; Schmidtke, D.T.; Pope, C.E.; Hoffman, L.R.; Hajjar, A.M.; Peterson, S.B.; Borenstein, E.; Mougous, J.D. Human gut bacteria contain acquired Interbacterial defence systems. Nature 2019, 575, 224–228. [Google Scholar] [CrossRef]
- Sequence Manipulation Suite. Available online: https://www.bioinformatics.org/sms2/orf_find.html (accessed on 6 July 2020).
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely reimplemented MPI bioinformatics Toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein data bank (PDB): The single global macromolecular structure archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar]
| Gene | Domains | E Value $ | Comment |
|---|---|---|---|
| t0736/SPC1582 (Both islets) | Tetratricopeptide repeat (TPR) domain (pfam13424) | 7.65 × 10−4 | TPR domains are protein interaction modules found in a broad array of functionally-diverse proteins |
| t0728 (Typhi islet only) | CDI inhibitor, EC869-like domain (cd13445) | 6.1 × 10−69 | A conserved domain of immunity proteins that protect against the activity of contact-dependent growth inhibition (CDI) system toxins related to E. coli EC869 |
| SPC1575 (Paratyphi C islet only) | Rhs-repeat core domain (TIGR03696) RhsA domain (COG3209) OCRE domain (cd16074) | 5.1 × 10−31 2.7 × 10−22 9.8 × 10−3 | Rhs repeats are found in toxic bacterial proteins, including certain insecticidal toxins and T6SS-delivered effector proteins OCRE domains have no defined function |
| SPC1576 (Paratyphi C islet only) | Tetratricopeptide repeat (TPR) domain (cl34042) | 1.6 × 10−3 | TPR domains are protein interaction modules found in a broad array of functionally-diverse proteins |
| SPC1577 (Paratyphi C islet only) | PAAR-like domain (cd14738) | 6.7 × 10−4 | A conserved domain found in the proline-alanine-alanine-arginine (PAAR) repeat superfamily. The domain forms the sharp conical extension of the T6SS VgrG spike |
| SPC1578 (Paratyphi C islet only) | PRK09687 domain HEAT repeat domain (COG1413) HEAT repeat domain (pfam13646) | 7.2 × 10−145 3.8 × 10−15 3.0 × 10−8 | PRK09687 is a conserved domain of unknown function HEAT repeats are found in a broad array of functionally-diverse proteins |
| SPC1581 (Paratyphi C islet only) | InsA C-terminal domain (cl37596) InsA domain (COG3677) | 8.4 × 10−17 4.3 × 10−6 | InsA domains are associated with IS1-family transposases |
| Gene | Homolog DS of CDI/T6SS Toxin $ | Homology to Immunity Protein # | Putative Function * |
|---|---|---|---|
| t0728 | Yes | Yes | Immunity protein for an EC869-like CDI toxin, which is a Zn2+-dependent DNase that degrades genomic DNA. |
| t0729 | Yes | Not detected | Immunity protein for a peptidoglycan-degrading T6SS effector protein |
| t0730 | Yes | Yes | Immunity protein for a Tae4-like peptidoglycan-degrading T6SS effector |
| t0731 | Yes | Not detected | Immunity protein for a T6SS effector of unknown function |
| t0732 | Yes | Yes | Immunity protein for a peptidoglycan-degrading T6SS effector protein |
| t0733 | Yes | Not detected | Immunity protein for a peptidoglycan-degrading T6SS effector protein |
| t0734 | Yes | Not detected | Immunity protein for a peptidoglycan-degrading T6SS effector protein |
| t0735 | Yes | Not detected | Immunity protein for a peptidoglycan-degrading T6SS effector protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barretto, L.A.F.; Fowler, C.C. Identification of A Putative T6SS Immunity Islet in Salmonella Typhi. Pathogens 2020, 9, 559. https://doi.org/10.3390/pathogens9070559
Barretto LAF, Fowler CC. Identification of A Putative T6SS Immunity Islet in Salmonella Typhi. Pathogens. 2020; 9(7):559. https://doi.org/10.3390/pathogens9070559
Chicago/Turabian StyleBarretto, Luke A. F., and Casey C. Fowler. 2020. "Identification of A Putative T6SS Immunity Islet in Salmonella Typhi" Pathogens 9, no. 7: 559. https://doi.org/10.3390/pathogens9070559
APA StyleBarretto, L. A. F., & Fowler, C. C. (2020). Identification of A Putative T6SS Immunity Islet in Salmonella Typhi. Pathogens, 9(7), 559. https://doi.org/10.3390/pathogens9070559





