Contributions of Mass Spectrometry-Based Proteomics to Understanding Salmonella-Host Interactions
Abstract
1. Introduction
2. Large-Scale Analyses of Salmonella and Host Proteomes during Infection
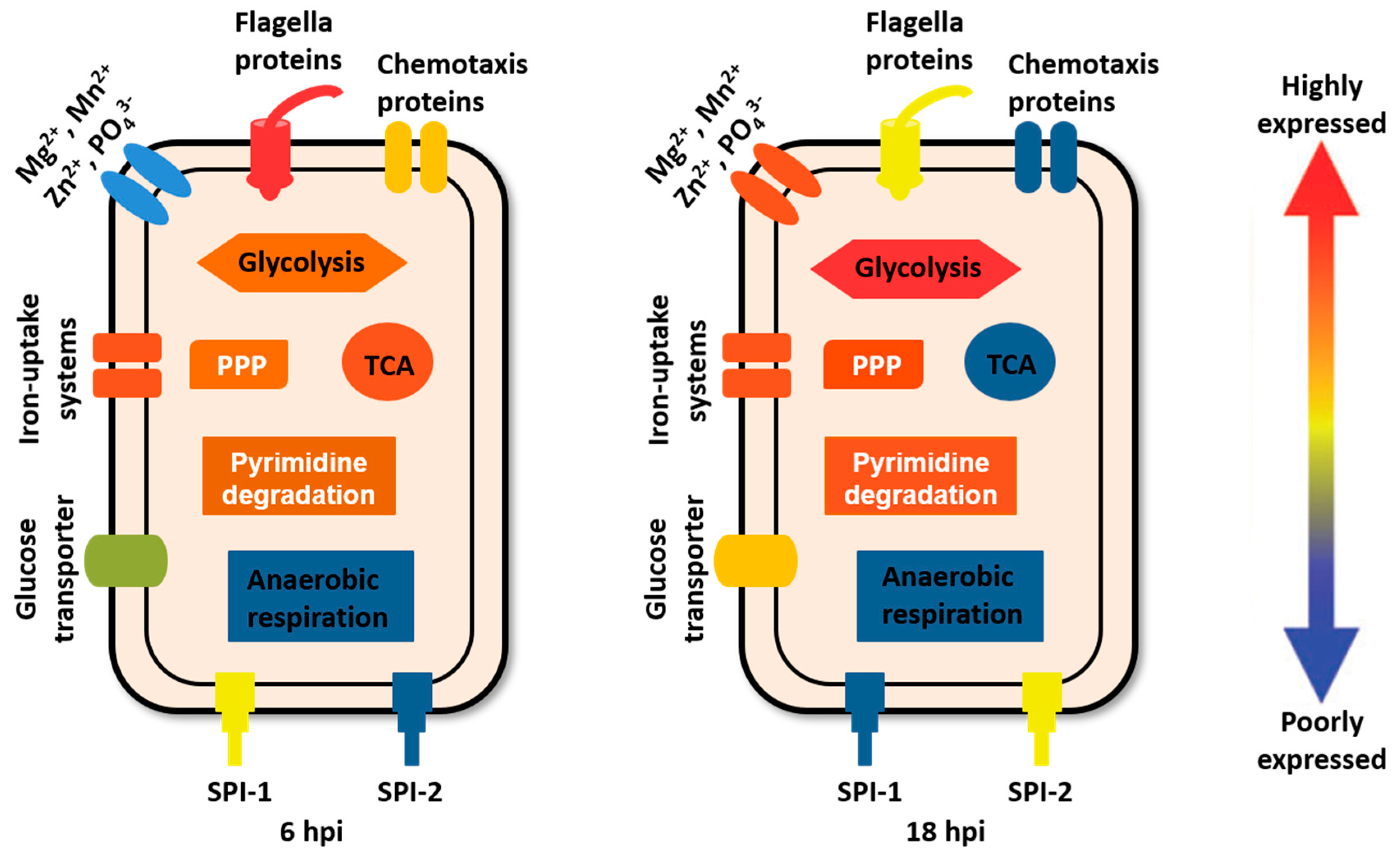
2.1. Research on Salmonella Proteome during Infection
2.2. Research on Host Proteome during Infection-Expression Profiling
2.3. Research on Host Proteome during Infection-PTM Profiling
3. Proteomic Tools Assist in the Study of Bacterial Virulence Factors
4. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Crump, J.A.; Sjolund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef]
- Richards, A.L.; Merrill, A.E.; Coon, J.J. Proteome sequencing goes deep. Curr. Opin. Chem. Biol. 2015, 24, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Greco, T.M.; Cristea, I.M. Proteomics tracing the footsteps of infectious disease. Mol. Cell. Proteom. 2017, 16, S5–S14. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Federspiel, J.D.; Sheng, X.; Cristea, I.M. Proteomics and integrative omic approaches for understanding host-pathogen interactions and infectious diseases. Mol. Syst. Biol. 2017, 13, 922. [Google Scholar] [CrossRef]
- Jenner, R.G.; Young, R.A. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 2005, 3, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Volker, U. Proteome analysis of host-pathogen interactions: Investigation of pathogen responses to the host cell environment. Proteomics 2011, 11, 3203–3211. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, M.; Yu, K.; Zeng, X.; Liu, X. Mass spectrometry-based proteomic approaches to study pathogenic bacteria-host interactions. Protein Cell 2015, 6, 265–274. [Google Scholar] [CrossRef]
- Semanjski, M.; Macek, B. Shotgun proteomics of bacterial pathogens: Advances, challenges and clinical implications. Expert Rev. Proteom. 2016, 13, 139–156. [Google Scholar] [CrossRef]
- Becker, D.; Selbach, M.; Rollenhagen, C.; Ballmaier, M.; Meyer, T.F.; Mann, M.; Bumann, D. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 2006, 440, 303–307. [Google Scholar] [CrossRef]
- Shi, L.; Adkins, J.N.; Coleman, J.R.; Schepmoes, A.A.; Dohnkova, A.; Mottaz, H.M.; Norbeck, A.D.; Purvine, S.O.; Manes, N.P.; Smallwood, H.S.; et al. Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: Identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J. Biol. Chem. 2006, 281, 29131–29140. [Google Scholar] [CrossRef]
- Steeb, B.; Claudi, B.; Burton, N.A.; Tienz, P.; Schmidt, A.; Farhan, H.; Maze, A.; Bumann, D. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 2013, 9, e1003301. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, B.; Novik, V.; Galan, J.E. Quantitative proteomics of intracellular Campylobacter jejuni reveals metabolic reprogramming. PLoS Pathog. 2012, 8, e1002562. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Q.; Hu, M.; Yu, K.; Fu, J.; Zhou, F.; Liu, X. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect. Immun. 2015, 83, 2897–2906. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, K.; Zhou, F.; Ding, T.; Yang, Y.; Hu, M.; Liu, X. Quantitative proteomics charts the landscape of Salmonella carbon metabolism within host epithelial cells. J. Proteome Res. 2017, 16, 788–797. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Qi, L.; Ding, T.; Wang, Z.; Fu, J.; Hu, M.; Li, M.; Song, J.; Liu, X. Temporal regulation of a Salmonella Typhimurium virulence factor by the transcriptional regulator YdcR. Mol. Cell. Proteom. 2017, 16, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Torres, A.; Jones-Carson, J.; Baumler, A.J.; Falkow, S.; Valdivia, R.; Brown, W.; Le, M.; Berggren, R.; Parks, W.T.; Fang, F.C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 1999, 401, 804–808. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Fu, J.; Zhang, B.; Cheng, S.; Wu, M.; Wang, Z.; Jiang, J.; Chang, C.; Liu, X. Salmonella proteomic profiling during infection distinguishes the intracellular environment of host cells. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Noster, J.; Chao, T.C.; Sander, N.; Schulte, M.; Reuter, T.; Hansmeier, N.; Hensel, M. Proteomics of intracellular Salmonella enterica reveals roles of Salmonella pathogenicity island 2 in metabolism and antioxidant defense. PLoS Pathog. 2019, 15, e1007741. [Google Scholar] [CrossRef]
- Shi, L.; Chowdhury, S.M.; Smallwood, H.S.; Yoon, H.; Mottaz-Brewer, H.M.; Norbeck, A.D.; McDermott, J.E.; Clauss, T.R.; Heffron, F.; Smith, R.D.; et al. Proteomic investigation of the time course responses of RAW 264.7 macrophages to infection with Salmonella enterica. Infect. Immun. 2009, 77, 3227–3233. [Google Scholar] [CrossRef]
- Qi, L.; Hu, M.; Fu, J.; Liu, Y.; Wu, M.; Yu, K.; Liu, X. Quantitative proteomic analysis of host epithelial cells infected by Salmonella enterica serovar Typhimurium. Proteomics 2017, 17. [Google Scholar] [CrossRef]
- Salcedo, S.P.; Holden, D.W. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003, 22, 5003–5014. [Google Scholar] [CrossRef] [PubMed]
- Vogels, M.W.; van Balkom, B.W.; Heck, A.J.; de Haan, C.A.; Rottier, P.J.; Batenburg, J.J.; Kaloyanova, D.V.; Helms, J.B. Quantitative proteomic identification of host factors involved in the Salmonella typhimurium infection cycle. Proteomics 2011, 11, 4477–4491. [Google Scholar] [CrossRef] [PubMed]
- Vorwerk, S.; Krieger, V.; Deiwick, J.; Hensel, M.; Hansmeier, N. Proteomes of host cell membranes modified by intracellular activities of Salmonella enterica. Mol. Cell. Proteom. 2015, 14, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Duchateau, M.; Fredlund, J.; Weiner, A.; Mallet, A.; Schmitt, C.; Matondo, M.; Hourdel, V.; Chamot-Rooke, J.; Enninga, J. The COPII complex and lysosomal VAMP7 determine intracellular Salmonella localization and growth. Cell. Microbiol. 2015, 17, 1699–1720. [Google Scholar] [CrossRef] [PubMed]
- Hui, W.W.; Hercik, K.; Belsare, S.; Alugubelly, N.; Clapp, B.; Rinaldi, C.; Edelmann, M.J. Salmonella enterica Serovar Typhimurium alters the extracellular proteome of macrophages and leads to the production of proinflammatory exosomes. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Selkrig, J.; Li, N.; Hausmann, A.; Mangan, M.S.J.; Zietek, M.; Mateus, A.; Bobonis, J.; Sueki, A.; Imamura, H.; El Debs, B.; et al. Spatiotemporal proteomics uncovers cathepsin-dependent macrophage cell death during Salmonella infection. Nat. Microbiol. 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Kao, D.S.; Gu, B.; Bomjan, R.; Srivastava, M.; Lu, H.; Zhou, D.; Tao, W.A. Tracking pathogen infections by time-resolved chemical proteomics. Angew. Chem. Int. Ed. Engl. 2020, 59, 2235–2240. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Jensen, O.N.; Larsen, M.R. Analytical strategies for phosphoproteomics. Proteomics 2009, 9, 1451–1468. [Google Scholar] [CrossRef]
- Rogers, L.D.; Brown, N.F.; Fang, Y.; Pelech, S.; Foster, L.J. Phosphoproteomic analysis of Salmonella-infected cells identifies key kinase regulators and SopB-dependent host phosphorylation events. Sci. Signal. 2011, 4, rs9. [Google Scholar] [CrossRef]
- Imami, K.; Bhavsar, A.P.; Yu, H.; Brown, N.F.; Rogers, L.D.; Finlay, B.B.; Foster, L.J. Global impact of Salmonella pathogenicity island 2-secreted effectors on the host phosphoproteome. Mol. Cell. Proteom. 2013, 12, 1632–1643. [Google Scholar] [CrossRef]
- Ashida, H.; Kim, M.; Sasakawa, C. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat. Rev. Microbiol. 2014, 12, 399–413. [Google Scholar] [CrossRef]
- Tanner, K.; Brzovic, P.; Rohde, J.R. The bacterial pathogen-ubiquitin interface: Lessons learned from Shigella. Cell. Microbiol. 2015, 17, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Fiskin, E.; Bionda, T.; Dikic, I.; Behrends, C. Global analysis of host and bacterial ubiquitinome in response to Salmonella Typhimurium infection. Mol. Cell 2016, 62, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Mohapatra, G.; Ahmad, S.M.; Rana, S.; Jain, S.; Khalsa, J.K.; Srikanth, C.V. Salmonella engages host micrornas to modulate SUMOylation: A new arsenal for intracellular survival. Mol. Cell. Biol. 2015, 35, 2932–2946. [Google Scholar] [CrossRef]
- Mohapatra, G.; Gaur, P.; Mujagond, P.; Singh, M.; Rana, S.; Pratap, S.; Kaur, N.; Verma, S.; Krishnan, V.; Singh, N.; et al. A SUMOylation-dependent switch of RAB7 governs intracellular life and pathogenesis of Salmonella Typhimurium. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Newson, J.P.M.; Scott, N.E.; Yeuk Wah Chung, I.; Wong Fok Lung, T.; Giogha, C.; Gan, J.; Wang, N.; Strugnell, R.A.; Brown, N.F.; Cygler, M.; et al. Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways. Mol. Cell. Proteom. 2019, 18, 1138–1156. [Google Scholar] [CrossRef]
- Niemann, G.S.; Brown, R.N.; Gustin, J.K.; Stufkens, A.; Shaikh-Kidwai, A.S.; Li, J.; McDermott, J.E.; Brewer, H.M.; Schepmoes, A.; Smith, R.D.; et al. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 2011, 79, 33–43. [Google Scholar] [CrossRef]
- Auweter, S.D.; Bhavsar, A.P.; de Hoog, C.L.; Li, Y.; Chan, Y.A.; van der Heijden, J.; Lowden, M.J.; Coombes, B.K.; Rogers, L.D.; Stoynov, N.; et al. Quantitative mass spectrometry catalogues Salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J. Biol. Chem. 2011, 286, 24023–24035. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, L.; Liu, Q.; Qi, L.; Yu, K.; Wang, Z.; Wu, M.; Liu, Y.; Fu, J.; Hu, M.; et al. Identification of a novel Salmonella type III effector by quantitative secretome profiling. Mol. Cell. Proteom. 2017, 16, 2219–2228. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, P.; Cheng, S.; Lu, Q.; Nowak, K.; Hopp, A.K.; Li, L.; Shi, X.; Zhou, Z.; Gao, W.; et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 2019, 178, 552–566 e520. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.D.; Kristensen, A.R.; Boyle, E.C.; Robinson, D.P.; Ly, R.T.; Finlay, B.B.; Foster, L.J. Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J. Proteom. 2008, 71, 97–108. [Google Scholar] [CrossRef]
- Humphreys, D.; Davidson, A.; Hume, P.J.; Koronakis, V. Salmonella virulence effector SopE and host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 2012, 11, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Pilar, A.V.C.; Reid-Yu, S.A.; Cooper, C.A.; Mulder, D.T.; Coombes, B.K. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLOS Pathog. 2012, 8, e1002773. [Google Scholar] [CrossRef]
- Panagi, I.; Jennings, E.; Zeng, J.; Günster, R.A.; Stones, C.D.; Mak, H.; Jin, E.; Stapels, D.A.C.; Subari, N.Z.; Pham, T.H.M.; et al. Salmonella effector SteE converts the mammalian serine/threonine kinase GSK3 into a tyrosine kinase to direct macrophage polarization. Cell Host Microbe 2020, 27, 41–53.e46. [Google Scholar] [CrossRef]
- McEwan, D.G.; Richter, B.; Claudi, B.; Wigge, C.; Wild, P.; Farhan, H.; McGourty, K.; Coxon, F.P.; Franz-Wachtel, M.; Perdu, B.; et al. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe 2015, 17, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Kamanova, J.; Sun, H.; Lara-Tejero, M.; Galán, J.E. The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 2016, 12, e1005552. [Google Scholar] [CrossRef] [PubMed]
- Fiskin, E.; Bhogaraju, S.; Herhaus, L.; Kalayil, S.; Hahn, M.; Dikic, I. Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA. Nat. Commun. 2017, 8, 14004. [Google Scholar] [CrossRef]
- Sontag, R.L.; Nakayasu, E.S.; Brown, R.N.; Niemann, G.S.; Sydor, M.A.; Sanchez, O.; Ansong, C.; Lu, S.-Y.; Choi, H.; Valleau, D.; et al. Identification of novel host interactors of effectors secreted by Salmonella and Citrobacter. mSystems 2016, 1, e00032-00015. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- D’Costa, V.M.; Coyaud, E.; Boddy, K.C.; Laurent, E.M.N.; St-Germain, J.; Li, T.; Grinstein, S.; Raught, B.; Brumell, J.H. BioID screen of Salmonella type 3 secreted effectors reveals host factors involved in vacuole positioning and stability during infection. Nat. Microbiol. 2019, 4, 2511–2522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, W.; Zhang, R.; Xu, J.; Wang, R.; Wang, L.; Zhao, X.; Li, J. First acetyl-proteome profiling of Salmonella Typhimurium revealed involvement of lysine acetylation in drug resistance. Vet. Microbiol. 2018, 226, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Santin, Y.G.; Doan, T.; Lebrun, R.; Espinosa, L.; Journet, L.; Cascales, E. In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat. Microbiol. 2018, 3, 1304–1313. [Google Scholar] [CrossRef]
- Olson, M.G.; Widner, R.E.; Jorgenson, L.M.; Lawrence, A.; Lagundzin, D.; Woods, N.T.; Ouellette, S.P.; Rucks, E.A. Proximity labeling to map host-pathogen interactions at the membrane of a bacterium-containing vacuole in Chlamydia trachomatis-infected human cells. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef]
- Dickinson, M.S.; Anderson, L.N.; Webb-Robertson, B.M.; Hansen, J.R.; Smith, R.D.; Wright, A.T.; Hybiske, K. Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites. PLoS Pathog. 2019, 15, e1007698. [Google Scholar] [CrossRef]
- Wang, Z.; McCloskey, A.; Cheng, S.; Wu, M.; Xue, C.; Yu, Z.; Fu, J.; Liu, Y.; Luo, Z.Q.; Liu, X. Regulation of the small GTPase Rab1 function by a bacterial glucosyltransferase. Cell Discov. 2018, 4, 53. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Liu, B.; Zhou, Y.; Zhang, X.; Zou, Q.; Liu, X. Contributions of Mass Spectrometry-Based Proteomics to Understanding Salmonella-Host Interactions. Pathogens 2020, 9, 581. https://doi.org/10.3390/pathogens9070581
Zhang B, Liu B, Zhou Y, Zhang X, Zou Q, Liu X. Contributions of Mass Spectrometry-Based Proteomics to Understanding Salmonella-Host Interactions. Pathogens. 2020; 9(7):581. https://doi.org/10.3390/pathogens9070581
Chicago/Turabian StyleZhang, Buyu, Bohao Liu, Yinglin Zhou, Xinxiang Zhang, Qinghua Zou, and Xiaoyun Liu. 2020. "Contributions of Mass Spectrometry-Based Proteomics to Understanding Salmonella-Host Interactions" Pathogens 9, no. 7: 581. https://doi.org/10.3390/pathogens9070581
APA StyleZhang, B., Liu, B., Zhou, Y., Zhang, X., Zou, Q., & Liu, X. (2020). Contributions of Mass Spectrometry-Based Proteomics to Understanding Salmonella-Host Interactions. Pathogens, 9(7), 581. https://doi.org/10.3390/pathogens9070581





