Abstract
Grapevine Bois noir (BN) is caused by ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’) and is one of the most important phytoplasma diseases in the Euro-Mediterranean viticultural areas. The epidemiology of BN can include grapevine as a plant host and is usually transmitted via sap-sucking insects that inhabit herbaceous host plants. Tracking the spread of ‘Ca. P. solani’ strains is of great help for the identification of plant reservoirs and insect vectors involved in local BN outbreaks. The molecular epidemiology of ‘Ca. P. solani’ is primarily based on sequence analysis of the tuf housekeeping gene (which encodes elongation factor Tu). In this study, molecular typing of tuf, through restriction fragment length polymorphism and sequencing, was carried out on grapevine samples from Iranian vineyards. According to the molecular characterization, three molecular types—tuf b1, tuf b5 and tuf b6—were found, with tuf b1 being the most prominent. These data provide further knowledge of tuf gene diversity and question the ecological role of such “minor” tuf types in Iranian vineyards, which have been detected only in grapevines.
1. Introduction
Grapevine yellow diseases are widespread in many viticultural areas of the world, causing significant economic losses [1], and are very difficult to manage in the field [2]. The most widespread grapevine yellow disease is Bois noir (BN), which is caused by the phytoplasma ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’) [3]. This phytoplasma is transmitted from plant to plant mainly by polyphagous planthoppers (e.g., Hyalesthes obsoletus and Reptalus panzeri) [4,5]. Urtica dioica and Convolvulus arvensis are the main phytoplasma reservoir plants [6], while grapevine is a dead-end host.
The molecular epidemiology of ‘Ca. P. solani’ is based on the analysis of the house-keeping gene known as tuf, which encodes the elongation factor Tu. Molecular characterization of tuf by restriction fragment length polymorphism (RFLP) revealed two main types: tuf a, which infects stinging nettle (U. dioca), and tuf b (later called tuf b1), which is associated with field bindweed (C. arvensis) [7]. A third minor genotype called tuf c was detected in hedge bindweed (Calystegia sepium) in a restricted area of Germany as an alternative host plant [7,8].
The distribution of the tuf types in Europe has also been defined, with tuf a being the most common in Germany, Switzerland, Northern France, Northern Italy and Austria, while tuf b is spread across most of Europe and Asia. In Iranian vineyards, molecular typing of ‘Ca. P. solani’ strains associated with BN led to the identification of the dominant epidemiological cycle of tuf b and field bindweed [9].
Using sequence analysis, new tuf type variants associated with the nettle and called tuf b2 [9,10] or tuf ab [11,12], were recently detected in vineyards in Austria, Macedonia, Croatia and Montenegro. Additionally, tuf b3 variant was reported in vineyards in the Republic of Azerbaijan [13] and formerly detected in a diseased potato plant collected in Romania [14,15].
The aim of this study was a further characterization of ‘Ca. P. solani’ from BN-infected grapevines grown in Iranian vineyards, based on molecular analysis of the tuf gene.
2. Results
2.1. PCR-RFLP and Phylogenetic Analysis
According to the 16Sr molecular characterization using a nested PCR and the primer pair fStol/rStol, 85 samples were infected by ‘Ca. P. solani’. The positive samples were analyzed with the tuf gene primer pairs and further characterized by RFLP with HpaII. Seventy-five samples showed an RFLP profile typical of tuf b1, making it the predominant molecular type. Five isolates collected from the Azarbaijan region of Iran and Zanjan province of Iran showed different profiles, distinguishable from the tuf a (isolate 19–25) and tuf b (isolate P7) reference strains. To support these data, two samples that were representative of the different RFLP patterns were sequenced: BN-a8 and BN-DG23. The BLAST (Basic Local Alignment Search Tool) analysis of the two nucleotide sequences revealed a high sequence homology with ‘Ca. P. solani’. The comparison between the tuf sequences of the reference strains and the sequences of BN-a8 and BN-DG23 demonstrated two nucleotide substitutions: “A” in position 415 from tuf start codon for BN-a8, and “A” in position 444 for BN-DG23, plus a common change for position 727 from “A” to “G”, with respect to the classical tuf b1 (Table 1). On the basis of the molecular data, we propose that sequence BN-a8 is referred to as tuf b5, and sequence BN-DG23 as tuf b6. Phylogenetic analysis indicated significant divergence, due to mutations, among both these new strains and the previous strains (Figure 1).

Table 1.
Mutations identified for the different tuf types, with respect to tuf b1 (Mutation Surveyor (MS) analysis).
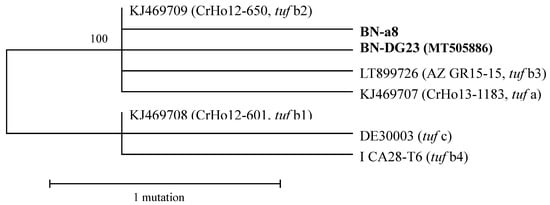
Figure 1.
Unrooted phylogenetic tree inferred from the ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’) strain nucleotide sequences of the tuf gene. Minimum evolution analysis was carried out using the maximum parsimony method. Percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches.
2.2. Mutation Detection
The sequence data were analyzed with the Mutation Surveyor software V5.1.2 (SoftGenetics, State College, PA, USA), using the 2D setting, as this increases the sensitivity and accuracy of the analysis. Figure S1 shows the Mutation Surveyor report that indicated the mutation positions and scores. Two novel mutations were detected in the samples, and some were also found in the reference strains. All of the mutations were substitution mutations. The most common mutations were detected for position 727. According to this position, there are two groups of tuf types: tuf a, tuf b2, tuf b3, tuf b5, and tuf b6 with a substitution from “A” to “G”, and tuf b1, tuf b4, and tuf c with a substitution from “G” to “A”.
3. Discussion
Prokaryotic and eukaryotic elongation factors are essential in the formation of peptide bonds during protein synthesis [16]. The bacterial elongation factor Tu is a GTP-binding protein that has a central role in protein synthesis, is highly conserved throughout bacteria and is homologous to its eukaryotic counterpart. The tuf gene is a single-copy gene in phytoplasmas that displays more variations than the 16S rRNA gene and is therefore a good candidate for phytoplasma classification [17]. This conserved gene encodes the elongation factor Tu that has a central function in translation processes [18].
Initially, characterization of ‘Ca. P. solani’ according to RFLP analysis of tuf identified three genetic types as tuf a, tuf b, and tuf c [7]. U. dioica is considered the dominant reservoir plant for tuf a, and C. sepium for tuf c [6]. However, for tuf b, while field bindweed is the dominant reservoir plant, several other herbaceous plants that can be found in and around infected vineyards have been reported as being hosts for ‘Ca. P. solani’, including Chenopodium album, Tussilago farfara and Malva sylvestris; therefore, these might all have key roles in the spread of BN [19,20,21,22,23]. The tuf b strain of ‘Ca. P. solani’ has also been identified in an epidemiological setting involving Vitex agnus-castus, with transmission to grapevine via associated populations of H. obsoletus [12,24]. Recent studies have also reported on a further independent epidemiological cycle for the tuf b strain of ‘Ca. P. solani’, detected in Crepis foetida and with transmission by H. obsoletus [25]. However, this study detected tuf b1 and two new genotypes that have not been reported previously that differ from tuf b1 in mutations. For BN-a8, the substitution in position 415 after the ATG start codon results in a more stable amino acid leading the synthesis of isoleucine instead of leucine. The substitution at position 444 for the BN-DG23 strain does not result in a change in amino acid. These new two tuf types were recorded in grapevine samples, and we can presumably hypothesize that they are present in wild plants and insects, which occasionally feed on grapevine. It is well known that diverse functions have been ascribed to the elongation factor Tu, many of which include important functions in the bacterial cytosol and for the cell surface [26,27,28,29]. Functions such as adhesion to host cells and molecules are fundamental to pathogenesis in many bacterial species as it facilitates colonization, invasion and host immune suppression [30]. Therefore, the different types of tuf gene generated by nucleotide modification might lead to a variation in amino acid level, hence playing a key role as fitness factors by extending or reducing the range of hosts.
These findings support the hypothesis that the tuf gene is involved in interactions of ‘Ca. P. solani’-related strains with their host plants and/or their insect vectors, which can drive the adaptation of diverse phytoplasma genetic lineages to varied vineyard ecosystems. In conclusion, in Iranian vineyards, the main tuf type is tuf b1, but we describe here the new tuf types of tuf b5 and tuf b6 that have emerged. Future research about their ecological and epidemiological roles in vineyard ecosystems should be conducted using innovative tools based on geostatistical analysis [31].
4. Materials and Methods
4.1. Plant Samples and DNA Extraction
A total of 124 grapevine samples were collected during surveys carried out from mid-September to the beginning of October in 2016 and 2017 in local vineyards which constituted of white and red varieties. This intensified the phytosanitary monitoring for grapevine yellows carried out in 2015 in vineyards located in the Azarbaijan Gharbi, Azarbaijan Sharghi, Zanjan, Qazvin, Fars, Lorestan and Khorasan Razavi provinces of Iran, as previously reported by Jamshidi et al. [9]. Total nucleic acids were extracted from 1 g fresh petioles of symptomatic grapevines from old and young leaves using the cetyltrimethylammonium bromide method, as described by Daire et al. [32]. The concentrations and purities of the DNA samples were determined using a spectrophotometer (Biophotometer Plus, Eppendorf, Hamburg, Germany).
4.2. PCR Amplification, RFLP and Sequencing
The PCR was performed in a programmable Bio-Rad Cycler (Bio-Rad, Berkeley, CA, USA), using the universal P1/P7 phytoplasma primer pair, followed by a nested PCR with the rStol/fStol specific primer pair for stolbur phytoplasma [33]. DNA amplification was performed in 20 μL volumes and PCR products were separated on 1% agarose gel, stained with GelRed (Biotium Inc., Fremont, CA, USA) and visualized with a UV transilluminator. The tuf gene was amplified using Tuf1f/r, followed by the TufAYf/r primer in a nested PCR [6,7]. Nested PCR amplicons (940 bp) were subjected to restriction digestion using HpaII endonuclease (ThermoScientific, Darmstadt, Germany), following the manufacturer’s instructions. The restriction products were separated by electrophoresis in 2.5% agarose gel. Samples, amplified with tufAYf/r primer pairs and showing a different restriction pattern compared to tuf a and tuf b reference isolates (19–25 and P7) were selected and sent for sequence analysis at the Genewiz Genomics sequencing service (Genewiz UK, Takeley, UK: https://www.genewiz.com/).
4.3. Phylogenetic Analysis
The nucleotide sequences of BN-a8 and BN-DG23 were subjected to BLAST analysis to confirm their identities. Multiple alignments of the nucleotide and amino acid sequences were carried out using ClustalX [34]. The phylogenetic relationships were calculated using the maximum parsimony method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test, with 500 replicates, are shown next to the branches. The maximum parsimony tree was obtained using the subtree pruning and regrafting algorithm. A total of 901 positions were found in the final dataset. Evolutionary analyses were conducted using the MEGA (Molecular Evolutionary Genetics Analysis) 6 software (http://www.megasoftware.net/index.html).
4.4. Data Analysis
The sequence data obtained were loaded into Mutation Surveyor V5.0.1 (SoftGenetics, State College, PA, USA), a software specifically designed to detect low-frequency DNA variants from sequenced data. The mutation score was used by the software to recognize a mutation and to rank its confidence level, a measure of the probability of error, which was based on the ratios of the noise level, the overlapping factor and the dropping factor, as calculated by the software. Mutations were investigated in nucleotide reference strains and in the samples found in this study. The sequences of BN-a8 and BN-DG23 were analyzed and compared with reference sequences that were already available in the NCBI database (https://www.ncbi.nlm.nih.gov/), including: tuf a (CrHo13-1183), tuf b1 (CrHo12-601), tuf b2 (CrHo12-650) [8], tuf b3 (AZ_GR15-156 15) [12], tuf b4 (I CA28-T6) (Murolo and Romanazzi, unpublished) and tuf c (DE 30003) (Langer and Maixner, unpublished). The Mutation Surveyor software was set for 2D (bi-directional) small peaks; the mutation-calling parameters were set to the defaults, including for the overlapping factor and the dropping factor. The overlapping factor was calculated by the software from the two different bases in the reference and sample traces on either side of the mutation. The dropping factor was determined from the relative intensities of the four neighboring peaks (i.e., two peaks on each side) between the sample traces and the reference traces.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/6/508/s1, Figure S1: Sequence analysis results using Mutation Surveyor V5.1.2, Graphic Analysis display for single nucleotide changes in the a8 and DG23 samples, compared with the reference strains.
Author Contributions
Conceptualization: E.J., S.M., M.S., G.R.; funding acquisition: M.S., G.R.; supervision: G.R.; formal analysis: E.J., S.M.; investigation: E.J., S.M., M.S.; data curation: E.J., S.M.; writing—original draft preparation: E.J.; writing—review and editing: S.M., M.S., G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Acknowledgments
The authors would like to thank Xavier Foissac for providing ‘Ca. Phytoplasma solani’ reference strains and for confirming the phylogenetic analysis.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Endeshaw, T.; Murolo, S.; Romanazzi, G.; Neri, D. Effects of Bois noir on carbon assimilation, transpiration, stomatal conductance of leaves and yield of grapevine (Vitis vinifera) cv. Chardonnay. Physiol. Plant 2012, 145, 286–295. [Google Scholar] [CrossRef]
- Romanazzi, G.; Murolo, S.; Feliziani, E. A new approach to manage phytoplasma diseases: Field treatments with resistance inducers to contain grapevine Bois noir. Phytopathology 2013, 103, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur- and Bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef] [PubMed]
- Maixner, M. Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis 1994, 33, 103–104. [Google Scholar]
- Cvrković, T.; Jović, J.; Mitrović, M.; Krstić, O.; Toševski, I. Experimental and molecular evidence of Reptalus panzeri as a natural vector of Bois noir. Plant Pathol. 2014, 63, 42–53. [Google Scholar] [CrossRef]
- Langer, M.; Maixner, M. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur-group based on RFLP-analysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar]
- Johannesen, J.; Foissac, X.; Kehrli, P.; Maixner, M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE 2012, 7, e51809. [Google Scholar] [CrossRef]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant ‘Candidatus Phytoplasma solani’ tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef]
- Jamshidi, E.; Murolo, S.; Ravari, S.B.; Salehi, M.; Romanazzi, G. Molecular typing of ‘Candidatus Phytoplasma solani’ in Iranian vineyards. Plant Dis. 2019, 103, 2412–2416. [Google Scholar] [CrossRef]
- Plavec, J.; Križanac, I.; Budinšćak, Ž.; Škorić, D.; Musić, M.Š. A case study of FD and BN phytoplasma variability in Croatia: Multigene sequence analysis approach. Eur. J. Plant Pathol. 2015, 142, 591–601. [Google Scholar] [CrossRef]
- Atanasova, B.; Jakovljević, M.; Spasov, D.; Jović, J.; Mitrović, M.; Toševski, I.; Cvrković, T. The molecular epidemiology of Bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur. J. Plant Pathol. 2015, 142, 759–770. [Google Scholar] [CrossRef]
- Kosovac, A.; Radonjić, S.; Hrnčić, S.; Krstić, O.; Toševski, I.; Jović, J. Molecular tracing of the transmission routes of Bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitex agnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant Pathol. 2016, 65, 285–298. [Google Scholar] [CrossRef]
- Balakishiyeva, G.; Bayramova, J.; Mammadov, A.; Salar, P.; Danet, J.-L.; Ember, I.; Verdin, E.; Foissac, X.; Huseynova, I. Important genetic diversity of ‘Candidatus Phytoplasma solani’ related strains associated with Bois noir grapevine yellows and planthoppers in Azerbaijan. Eur. J. Plant Pathol. 2018, 151, 937–946. [Google Scholar] [CrossRef]
- Ember, I.; Bodor, P.; Zsófi, Z.; Pálfi, Z.; Ladányi, M.; Pásti, G.; Bisztray, G.D. Bois noir affects the yield and wine quality of Vitis vinifera L. cv. ‘Chardonnay’. Eur. J. Plant Pathol. 2018, 152, 185–197. [Google Scholar] [CrossRef]
- Foissac, X.; Carle, P.; Fabre, A.; Salar, P.; Danet, J.L. Candidatus Phytoplasma solani’ genome project and genetic diversity in Mediterranean basin. In Proceedings of the 3rd European Bois noir Workshop, Barcelona, Spain, 22–23 March 2013; pp. 11–13. [Google Scholar]
- Browning, K.S. The plant translational apparatus. Plant Mol. Biol. 1996, 32, 107–144. [Google Scholar] [CrossRef]
- Schneider, B.; Gibb, K.S.; Seemuller, E. Sequence and RFLP analysis of the gene coding for the elongation factor TU of several phytoplasma strains for differentiation and classification of phytoplasmas. Microbiology 1997, 143, 3381–3389. [Google Scholar] [CrossRef]
- Filer, D.; Furano, A.V. Portions of the gene encoding elongation factor Tu are highly conserved in prokaryotes. J. Biol. Chem. 1980, 255, 728–734. [Google Scholar]
- Credi, R.; Terlizzi, F.; Milanesi, L.; Bondavalli, R.; Cavallini, G.; Montermini, A.; Dradi, D. Wild host plants of stolbur phytoplasma and its vector, Hyalesthes obsoletus, at sites of grapevine Bois noir occurrence in Emilia-Romagna, Italy. In Proceedings of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine, Stellenbosch, South Africa, 3–7 April 2006. [Google Scholar]
- Romanazzi, G.; D’Ascenzo, D.; Murolo, S. Tussilago farfara: A new natural host of stolbur phytoplasma. Plant Pathol. 2009, 58, 392. [Google Scholar] [CrossRef]
- Marchi, G.; Cinelli, T.; Rizzo, D.; Stefani, L.; Goti, E.; Della Bartola, M.; Materazzi, A. Occurrence of different phytoplasma infections in wild herbaceous dicots growing in vineyards affected by Bois noir in Tuscany (Italy). Phytopathol. Mediterr. 2015, 54, 504–515. [Google Scholar]
- Mori, N.; Quaglino, F.; Tessari, F.; Pozzebon, A.; Bulgari, D.; Casati, P.; Bianco, P.A. Investigation on ‘Bois noir’ epidemiology in northeastern Italian vineyards through a multidisciplinary approach. Ann. Appl. Biol. 2015, 166, 75–89. [Google Scholar] [CrossRef]
- Oliveri, C.; Pacifico, D.; D’Urso, V.; La Rosa, R.; Marzachì, C.; Tessitori, M. Bois noir phytoplasma variability in a Mediterranean vineyard system: New plant host and putative vectors. Australas. Plant Pathol. 2015, 44, 235–244. [Google Scholar] [CrossRef]
- Sharon, R.; Soroker, V.; Wesley, S.; Zahavi, T.; Harari, A.; Weintraub, P. Vitex agnus-castus is a preferred host plant for Hyalesthes obsoletus. J. Chem. Ecol. 2005, 31, 1051–1063. [Google Scholar] [CrossRef]
- Kosovac, A.; Jakovljević, M.; Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I.; Jović, J. Role of plant-specialized Hyalesthes obsoletus associated with Convolvulus arvensis and Crepis foetida in the transmission of ‘Candidatus Phytoplasma solani’-inflicted Bois noir disease of grapevine in Serbia. Eur. J. Plant Pathol. 2019, 153, 183–195. [Google Scholar] [CrossRef]
- Sanchez, B.; Bressollier, P.; Urdaci, M.C. Exported proteins in probiotic bacteria: Adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol. Med. Microbiol. 2008, 54, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kainulainen, V.; Korhonen, T.K. Dancing to another tune-adhesive moonlighting proteins in bacteria. Biology 2014, 3, 178–204. [Google Scholar] [CrossRef] [PubMed]
- Ebner, K.; Singewald, N. The role of substance P in stress and anxiety responses. Amino Acids 2006, 31, 251–272. [Google Scholar] [CrossRef]
- Stones, D.H.; Krachler, A.M. Against the tide: The role of bacterial adhesion in host colonization. Biochem. Soc. Trans. 2016, 44, 1571–1580. [Google Scholar] [CrossRef]
- Harvey Kate, L.; Jarocki Veronica, M.; Charles Ian, G.; Djordjevic Steven, P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Murolo, S.; Garbarino, M.; Mancini, V.; Romanazzi, G. Spatial pattern of Bois noir: Case study of a delicate balance between disease progression and recovery. Sci. Rep. 2020. [Google Scholar] [CrossRef]
- Daire, X.; Clair, D.; Reinert, W. Boudon-Padieu, E. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. Eur. J. Plant Pathol. 1997, 103, 507–514. [Google Scholar] [CrossRef]
- Maixner, M.; Ahrens, U.; Seemüller, E. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 1995, 101, 241–250. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).