Epigenetic Regulation of the Human Papillomavirus Life Cycle
Abstract
1. Introduction
2. The HPV Life Cycle
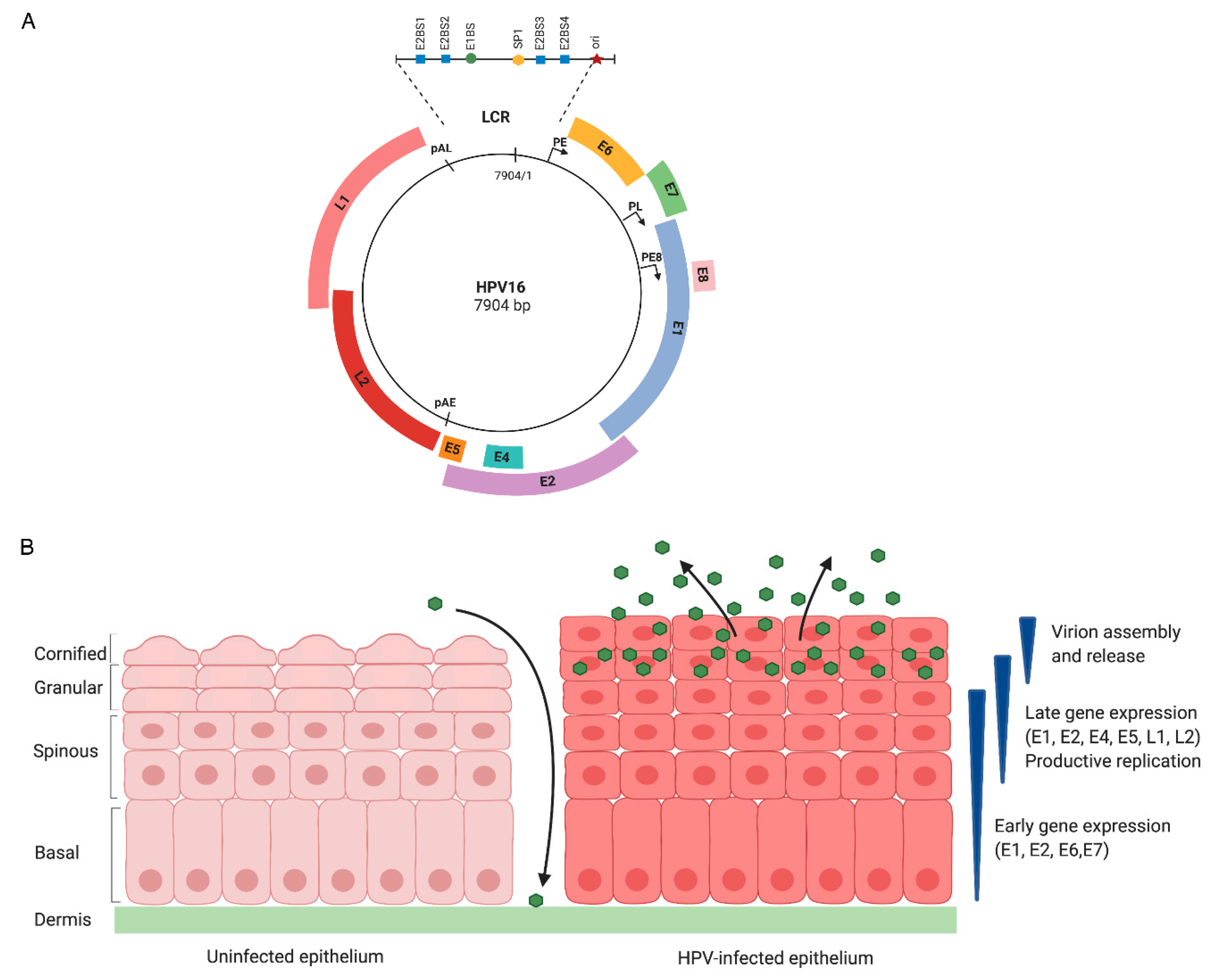
2.1. HPV Genome Structure
2.2. The HPV Life Cycle Consists of Three Stages of Replication
3. Epigenetic Regulation of HPV Gene Expression
3.1. Epigenetic Modifications of HPV Chromatin Regulate Viral Gene Expression Throughout the Viral Life Cycle
3.2. Histone Acetylation
3.3. DNA Methylation
4. HPV-mediated Epigenetic Modifications of Cellular Chromatin
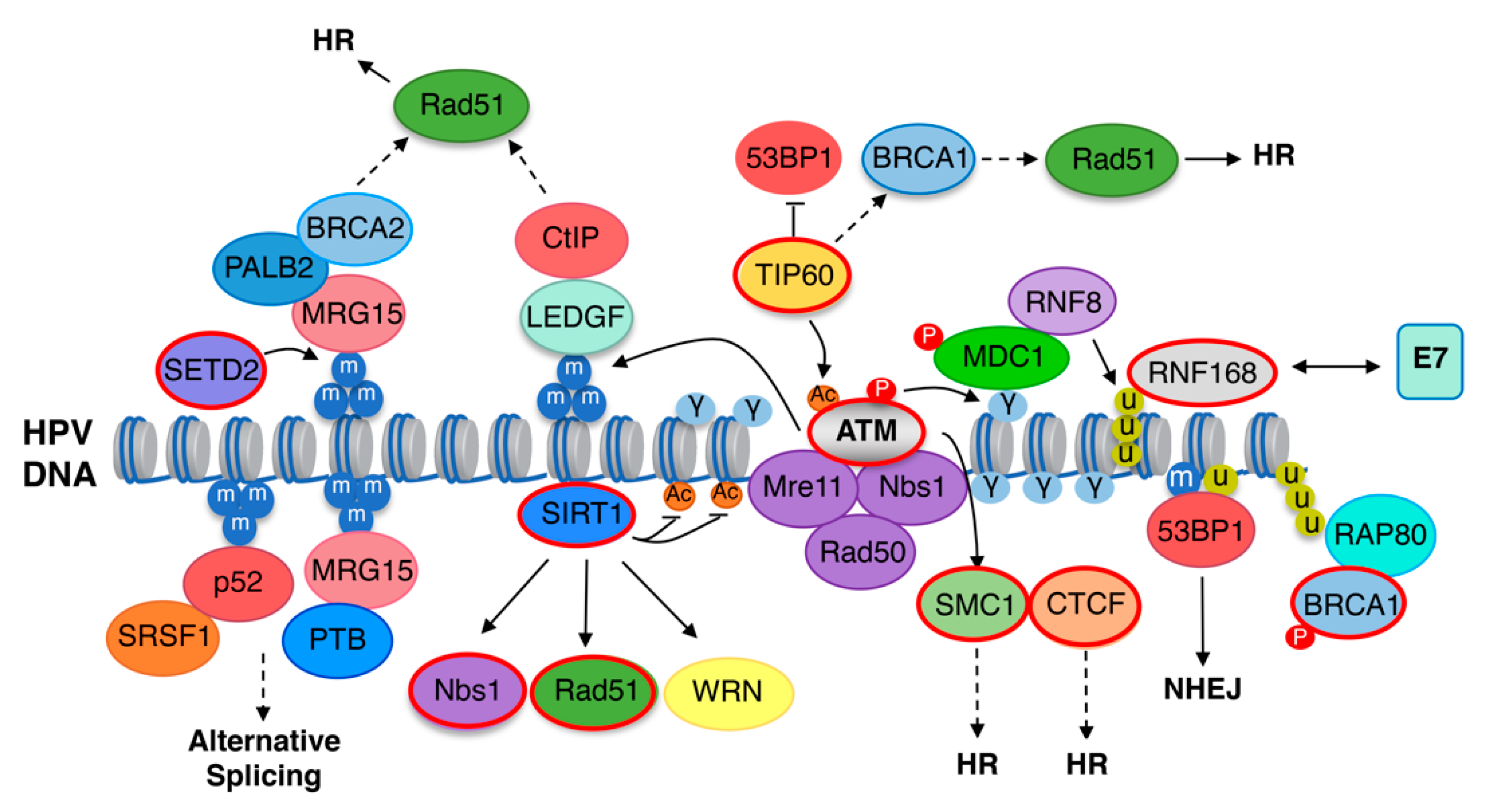
5. DNA Repair-Induced Epigenetic Modifications of HPV Chromatin
5.1. The DNA Damage Response
5.2. The DNA Damage Response Promotes Productive Replication through Viral Chromatin Modifications
5.3. TIP60 Acetyltransferase and SIRT1 Deacetylase Are Required for Productive Viral Replication
5.4. SMC1 and CTCF May Influence DNA Repair Factor Recruitment to Viral Chromatin
5.5. SETD2 and H3K36me3 Are Required for Productive Replication
5.6. Utilization of DDR Pathways during HPV Replication May Promote Viral Genome Integration and Genomic Instability
6. Summary and Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef] [PubMed]
- PaVE: Papilloma Virus Genome Database. Available online: https://pave.niaid.nih.gov/ (accessed on 6 January 2020).
- De Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; Zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Walboomers, J.M.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.F.; Peto, J.; Meijer, C.J.L.M.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Zur Hausen, H. Papillomavirus infections—A major cause of human cancers. Biochim. Biophys. Acta Rev. Cancer 1996, 1288, F55–F78. [Google Scholar] [CrossRef]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC), D. of C.P. and C. Cancers Associated with Human Papillomavirus, United States, 2011–2015; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018.
- You, E.L.; Henry, M.; Zeitouni, A.G. Human papillomavirus–associated oropharyngeal cancer: Review of current evidence and management. Curr. Oncol. 2019, 26. [Google Scholar] [CrossRef]
- Garbuglia, A.R.; Lapa, D.; Sias, C.; Capobianchi, M.R.; Del Porto, P. The Use of Both Therapeutic and Prophylactic Vaccines in the Therapy of Papillomavirus Disease. Front. Immunol. 2020, 11, 188. [Google Scholar] [CrossRef]
- Moody, C.A. Mechanisms by which HPV induces a replication competent environment in differentiating keratinocytes. Viruses 2017, 9, 261. [Google Scholar] [CrossRef]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- Ozbun, M.A.; Meyers, C. Temporal Usage of Multiple Promoters during the Life Cycle of Human Papillomavirus Type 31b. J. Virol. 1998, 72, 2715–2722. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. Pathogenesis of Human Papillomaviruses in Differentiating Epithelia. Microbiol. Mol. Biol. Rev. 2004, 68, 362–372. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A. Mechanisms and strategies of papillomavirus replication. Biol. Chem. 2017, 398, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Anacker, D.C.; Moody, C.A. Modulation of the DNA damage response during the life cycle of human papillomaviruses. Virus Res. 2017, 231, 41–49. [Google Scholar] [CrossRef]
- Hummel, M.; Hudson, J.B.; Laimins, L.A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 1992, 66, 6070–6080. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, D.J.; Laimins, L.A. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 1999, 257, 239–246. [Google Scholar] [CrossRef]
- Bedell, M.A.; Hudson, J.B.; Golub, T.R.; Turyk, M.E.; Hosken, M.; Wilbanks, G.D.; Laimins, L.A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 1991, 65, 2254–2260. [Google Scholar] [CrossRef]
- Hummel, M.; Lim, H.B.; Laimins, L.A. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 1995, 69, 3381–3388. [Google Scholar] [CrossRef]
- Ozbun, M.A.; Meyers, C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 1997, 71, 5161–5172. [Google Scholar] [CrossRef]
- Longworth, M.S.; Wilson, R.; Laimins, L.A. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J. 2005, 24, 1821–1830. [Google Scholar] [CrossRef]
- Howie, H.L.; Katzenellenbogen, R.A.; Galloway, D.A. Papillomavirus E6 proteins. Virology 2009, 384, 324–334. [Google Scholar] [CrossRef]
- Münger, K.; Werness, B.A.; Dyson, N.; Phelps, W.C.; Harlow, E.; Howley, P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989, 8, 4099–4105. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998, 12, 2245–2262. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.; Howley, P.; Munger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [CrossRef] [PubMed]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Stünkel, W.; Bernard, H.-U. The Chromatin Structure of the Long Control Region of Human Papillomavirus Type 16 Represses Viral Oncoprotein Expression. J. Virol. 1999, 73, 1918–1930. [Google Scholar] [CrossRef] [PubMed]
- Favre, M.; Breitburd, F.; Croissant, O.; Orth, G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 1977, 21, 1205–1209. [Google Scholar] [CrossRef]
- You, J. Papillomavirus interaction with cellular chromatin. Biochim. Biophys. Acta Gene Regul. Mech. 2010, 1799, 192–199. [Google Scholar] [CrossRef]
- Soto, D.; Song, C.; McLaughlin-Drubin, M.E. Epigenetic alterations in human papillomavirus-associated cancers. Viruses 2017, 9, 248. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Wooldridge, T.R.; Laimins, L.A. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology 2008, 374, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Del Pena, L.M.; Laimins, L.A. Differentiation-Dependent Chromatin Rearrangement Coincides with Activation of Human Papillomavirus Type 31 Late Gene Expression. J. Virol. 2001, 75, 10005–10013. [Google Scholar] [CrossRef] [PubMed]
- Gunasekharan, V.; Hache, G.; Laimins, L. Differentiation-Dependent Changes in Levels of C/EBP Repressors and Activators Regulate Human Papillomavirus Type 31 Late Gene Expression. J. Virol. 2012, 86, 5393–5398. [Google Scholar] [CrossRef] [PubMed]
- Bauknecht, T.; Angel, P.; Royer, H.D.; zur Hausen, H. Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: Interaction with the transcriptional repressor YY1. EMBO J. 1992, 11, 4607–4617. [Google Scholar] [CrossRef]
- O’Connor, M.J.; Tan, S.H.; Tan, C.H.; Bernard, H.U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J. Virol. 1996, 70, 6529–6539. [Google Scholar] [CrossRef]
- Wilkinson, F.H.; Park, K.; Atchison, M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 2006, 103, 19296–19301. [Google Scholar] [CrossRef]
- Wilkinson, F.; Pratt, H.; Atchison, M.L. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 2010, 109, 478–486. [Google Scholar] [CrossRef]
- Srinivasan, L.; Atchison, M.L. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004, 18, 2596–2601. [Google Scholar] [CrossRef]
- Pentland, I.; Campos-León, K.; Cotic, M.; Davies, K.J.; Wood, C.D.; Groves, I.J.; Burley, M.; Coleman, N.; Stockton, J.D.; Noyvert, B.; et al. Disruption of CTCF-YY1-dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PLoS Biol. 2018, 16, e2005752. [Google Scholar] [CrossRef]
- Eberharter, A.; Becker, P.B. Histone acetylation: A switch between repressive and permissive chromatin. Second in review on chromatin dynamics. EMBO Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chang, W.; Jin, Y.; Feng, C.; Wu, S.; He, J.; Xu, T. The function of histone acetylation in cervical cancer development. Biosci. Rep. 2019, 29. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 1996, 384, 641–643. [Google Scholar] [CrossRef]
- Krüppel, U.; Müller-Schiffmann, A.; Baldus, S.E.; Smola-Hess, S.; Steger, G. E2 and the co-activator p300 can cooperate in activation of the human papillomavirus type 16 early promoter. Virology 2008, 377, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Bernat, A.; Avvakumov, N.; Mymryk, J.S.; Banks, L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 2003, 22, 7871–7881. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Hernández, A.; Cuevas-Bennett, C.; Garrido, E. Transcriptional regulation of human papillomavirus type 18 P105 promoter by the co-activator CBP. Intervirology 2008, 50, 418–425. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. The Binding of Histone Deacetylases and the Integrity of Zinc Finger-Like Motifs of the E7 Protein Are Essential for the Life Cycle of Human Papillomavirus Type 31. J. Virol. 2004, 78, 3533–3541. [Google Scholar] [CrossRef]
- Brehm, A.; Nielsen, S.J.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999, 18, 2449–2458. [Google Scholar] [CrossRef]
- Huang, B.H.; Laban, M.; Leung, C.H.W.; Lee, L.; Lee, C.K.; Salto-Tellez, M.; Raju, G.C.; Hooi, S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005, 12, 395–404. [Google Scholar] [CrossRef]
- Lace, M.J.; Anson, J.R.; Thomas, G.S.; Turek, L.P.; Haugen, T.H. The E8 E2 Gene Product of Human Papillomavirus Type 16 Represses Early Transcription and Replication but Is Dispensable for Viral Plasmid Persistence in Keratinocytes. J. Virol. 2008, 82, 10841–10853. [Google Scholar] [CrossRef]
- Sankovski, E.; Mannik, A.; Geimanen, J.; Ustav, E.; Ustav, M. Mapping of Betapapillomavirus Human Papillomavirus 5 Transcription and Characterization of Viral-Genome Replication Function. J. Virol. 2014, 88, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Stubenrauch, F.; Hummel, M.; Iftner, T.; Laimins, L.A. The E8^E2C Protein, a Negative Regulator of Viral Transcription and Replication, Is Required for Extrachromosomal Maintenance of Human Papillomavirus Type 31 in Keratinocytes. J. Virol. 2000, 74, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Kurg, R.; Uusen, P.; Võsa, L.; Ustav, M. Human papillomavirus E2 protein with single activation domain initiates HPV18 genome replication, but is not sufficient for long-term maintenance of virus genome. Virology 2010, 408, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Straub, E.; Dreer, M.; Fertey, J.; Iftner, T.; Stubenrauch, F. The Viral E8^E2C Repressor Limits Productive Replication of Human Papillomavirus 16. J. Virol. 2014, 88, 937–947. [Google Scholar] [CrossRef]
- Zobel, T.; Iftner, T.; Stubenrauch, F. The Papillomavirus E8 ∧ E2C Protein Represses DNA Replication from Extrachromosomal Origins. Mol. Cell. Biol. 2003, 23, 8352–8362. [Google Scholar] [CrossRef] [PubMed]
- Dreer, M.; Fertey, J.; van de Poel, S.; Straub, E.; Madlung, J.; Macek, B.; Iftner, T.; Stubenrauch, F. Interaction of NCOR/SMRT Repressor Complexes with Papillomavirus E8^E2C Proteins Inhibits Viral Replication. PLoS Pathog. 2016, 12, e1005556. [Google Scholar] [CrossRef]
- Ammermann, I.; Bruckner, M.; Matthes, F.; Iftner, T.; Stubenrauch, F. Inhibition of Transcription and DNA Replication by the Papillomavirus E8^E2C Protein Is Mediated by Interaction with Corepressor Molecules. J. Virol. 2008, 82, 5127–5136. [Google Scholar] [CrossRef]
- Powell, M.L.C.; Smith, J.A.; Sowa, M.E.; Harper, J.W.; Iftner, T.; Stubenrauch, F.; Howley, P.M. NCoR1 Mediates Papillomavirus E8^E2C Transcriptional Repression. J. Virol. 2010, 84, 4451–4460. [Google Scholar] [CrossRef]
- Smith, J.A.; White, E.A.; Sowa, M.E.; Powell, M.L.C.; Ottinger, M.; Harper, J.W.; Howley, P.M. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 3752–3757. [Google Scholar] [CrossRef]
- Smith, J.A.; Haberstroh, F.S.; White, E.A.; Livingston, D.M.; DeCaprio, J.A.; Howley, P.M. SMCX and components of the TIP60 complex contribute to E2 regulation of the HPV E6/E7 promoter. Virology 2014, 468, 311–321. [Google Scholar] [CrossRef][Green Version]
- Jha, S.; Vande Pol, S.; Banerjee, N.S.; Dutta, A.B.; Chow, L.T.; Dutta, A. Destabilization of TIP60 by Human Papillomavirus E6 Results in Attenuation of TIP60-Dependent Transcriptional Regulation and Apoptotic Pathway. Mol. Cell 2010, 38, 700–711. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Jang, M.K. Current understanding of the role of the Brd4 protein in the papillomavirus lifecycle. Viruses 2013, 5, 1374–1394. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation, methyltransferases, and cancer. Oncogene 2001, 20, 3139–3155. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Garner-Hamrick, P.A.; Fisher, C.; Lee, D.; Lambert, P.F. Methylation Patterns of Papillomavirus DNA, Its Influence on E2 Function, and Implications in Viral Infection. J. Virol. 2003, 77, 12450–12459. [Google Scholar] [CrossRef]
- Thain, A.; Jenkins, O.; Clarke, A.R.; Gaston, K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996, 70, 7233–7235. [Google Scholar] [CrossRef]
- Leung, T.W.; Liu, S.S.; Leung, R.C.Y.; Chu, M.M.Y.; Cheung, A.N.Y.; Ngan, H.Y.S. HPV 16 E2 binding sites 1 and 2 become more methylated than E2 binding site 4 during cervical carcinogenesis. J. Med. Virol. 2015, 87, 1022–1033. [Google Scholar] [CrossRef]
- Vinokurova, S.; von Knebel Doeberitz, M. Differential Methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE 2011, 6, e24451. [Google Scholar] [CrossRef]
- Hegde, R.S. The papillomavirus E2 proteins: Structure, function, and biology. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 343–360. [Google Scholar] [CrossRef]
- Stubenrauch, F.; Lim, H.B.; Laimins, L.A. Differential Requirements for Conserved E2 Binding Sites in the Life Cycle of Oncogenic Human Papillomavirus Type 31. J. Virol. 1998, 72, 1071–1077. [Google Scholar] [CrossRef]
- Lu, Q.; Ma, D.; Zhao, S. DNA methylation changes in cervical cancers. Methods Mol. Biol. 2012, 863, 155–176. [Google Scholar] [CrossRef]
- Clarke, M.A.; Wentzensen, N.; Mirabello, L.; Ghosh, A.; Wacholder, S.; Harari, A.; Lorincz, A.; Schiffman, M.; Burk, R.D. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2125–2137. [Google Scholar] [CrossRef]
- Bowden, S.J.; Kalliala, I.; Veroniki, A.A.; Arbyn, M.; Mitra, A.; Lathouras, K.; Mirabello, L.; Chadeau-Hyam, M.; Paraskevaidis, E.; Flanagan, J.M.; et al. The use of human papillomavirus DNA methylation in cervical intraepithelial neoplasia: A systematic review and meta-analysis. EBioMedicine 2019, 50, 246–259. [Google Scholar] [CrossRef]
- Jin-tao, W.; Ling, D.; Shi-Wen, J.; Junxia, H.; Wei-min, Z.; Qin, Z.; Zuo-kai, Y.; Li, Z. Folate Deficiency and Aberrant Expression of DNA Methyltransferase 1 were Associated with Cervical Cancerization. Curr. Pharm. Des. 2014, 20, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.M.; Wei, W.; Collins, S.I.; Pereira, M.; Diyaf, A.; Constandinou-Williams, C.; Young, L.S.; Roberts, S.; Woodman, C.B. Oncogenic human papillomavirus imposes an instructive pattern of dna methylation changes which parallel the natural history of cervical hpv infection in young women. Carcinogenesis 2012, 33, 1286–1293. [Google Scholar] [CrossRef]
- Sartor, M.A.; Dolinoy, D.C.; Jones, T.R.; Colacino, J.A.; Prince, M.E.P.; Carey, T.E.; Rozek, L.S. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 2011, 6, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Burgers, W.A.; Blanchon, L.; Pradhan, S.; De Launoit, Y.; Kouzarides, T.; Fuks, F. Viral oncoproteins target the DNA methyltransferases. Oncogene 2007, 26, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Yeung, A.; Lam, C.; Tsang, W.P.; Tsang, T.Y.; Co, N.N.; Yau, P.L.; Kwok, T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010, 24. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Crum, C.P.; Münger, K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. USA 2011, 108, 2130–2135. [Google Scholar] [CrossRef]
- Hyland, P.L.; McDade, S.S.; McCloskey, R.; Dickson, G.J.; Arthur, K.; McCance, D.J.; Patel, D. Evidence for Alteration of EZH2, BMI1, and KDM6A and Epigenetic Reprogramming in Human Papillomavirus Type 16 E6/E7-Expressing Keratinocytes. J. Virol. 2011, 85, 10999–11006. [Google Scholar] [CrossRef]
- Gameiro, S.F.; Kolendowski, B.; Zhang, A.; Barrett, J.W.; Nichols, A.C.; Torchia, J.; Mymryk, J.S. Human papillomavirus dysregulates the cellular apparatus controlling the methylation status of H3K27 in different human cancers to consistently alter gene expression regardless of tissue of origin. Oncotarget 2017, 8, 72564–72576. [Google Scholar] [CrossRef]
- Iancu, I.V.; Botezatu, A.; Plesa, A.; Huica, I.; Socolov, D.; Anton, G. Histone lysine demethylases as epigenetic modifiers in HPV-induced cervical neoplasia. Roman Biotechnol. Lett. 2015, 20, 10236–10244. [Google Scholar]
- McLaughlin-Drubin, M.E.; Huh, K.-W.; Munger, K. Human Papillomavirus Type 16 E7 Oncoprotein Associates with E2F6. J. Virol. 2008, 82, 8695–8705. [Google Scholar] [CrossRef] [PubMed]
- Soto, D.R.; Barton, C.; Munger, K.; McLaughlin-Drubin, M.E. KDM6A addiction of cervical carcinoma cell lines is triggered by E7 and mediated by p21CIP1suppression of replication stress. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Laribee, R.N.; Klemsz, M.J.; Roman, A. Human papillomavirus type 16 E7 protein increases acetylation of histone H3 in human foreskin keratinocytes. Virology 2004, 329, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, N.S.; Moore, D.; Parker, C.J.; Broker, T.R.; Chow, L.T. Targeting DNA damage response as a strategy to treat HPV infections. Int. J. Mol. Sci. 2019, 20, 5455. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009, 5, 1000605. [Google Scholar] [CrossRef]
- Hong, S.; Cheng, S.; Iovane, A.; Laimins, L.A. STAT-5 Regulates transcription of the topoisomerase IIβ-Binding protein 1 (TopBP1) gene to activate the ATR pathway and promote human papillomavirus Replication. MBio 2015, 6. [Google Scholar] [CrossRef]
- Anacker, D.C.; Aloor, H.L.; Shepard, C.N.; Lenzi, G.M.; Johnson, B.A.; Kim, B.; Moody, C.A. HPV31 utilizes the ATR-Chk1 pathway to maintain elevated RRM2 levels and a replication-competent environment in differentiating Keratinocytes. Virology 2016, 499, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.G.; Helmus, M.J.; Koeller, K.; Bashkin, J.K.; Fisher, C. Human Papillomavirus Episome Stability Is Reduced by Aphidicolin and Controlled by DNA Damage Response Pathways. J. Virol. 2013, 87, 3979–3989. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, N.S.; Wang, H.K.; Broker, T.R.; Chow, L.T. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J. Biol. Chem. 2011, 286, 15473–15482. [Google Scholar] [CrossRef] [PubMed]
- Fradet-Turcotte, A.; Bergeron-Labrecque, F.; Moody, C.A.; Lehoux, M.; Laimins, L.A.; Archambault, J. Nuclear Accumulation of the Papillomavirus E1 Helicase Blocks S-Phase Progression and Triggers an ATM-Dependent DNA Damage Response. J. Virol. 2011, 85, 8996–9012. [Google Scholar] [CrossRef]
- Sakakibara, N.; Mitra, R.; McBride, A.A. The Papillomavirus E1 Helicase Activates a Cellular DNA Damage Response in Viral Replication Foci. J. Virol. 2011, 85, 8981–8995. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Chen, S.; Fernandes, N.; Price, B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 2005, 102, 13182–13187. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM Phosphorylates Histone H2AX in Response to DNA Double-strand Breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair (Amst) 2015, 36, 8–12. [Google Scholar] [CrossRef]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; Van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef]
- Mattiroli, F.; Vissers, J.H.A.; Van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Faustrup, H.; Melander, F.; Bartek, J.; Lukas, C.; Lukas, J. RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins. Cell 2007, 131, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.Y.; Grant, R.; Manke, I.; Minn, K.; Yu, X.; Yaffe, M.B.; Chen, J. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell 2007, 131, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Doil, C.; Mailand, N.; Bekker-Jensen, S.; Menard, P.; Larsen, D.H.; Pepperkok, R.; Ellenberg, J.; Panier, S.; Durocher, D.; Bartek, J.; et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell 2009, 136, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Smeenk, G.; Mailand, N. Writers, readers, and erasers of histone ubiquitylation in DNA double-strand break repair. Front. Genet. 2016, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Luijsterburg, M.S.; Typas, D.; Caron, M.C.; Wiegant, W.W.; Van Den Heuvel, D.; Boonen, R.A.; Couturier, A.M.; Mullenders, L.H.; Masson, J.Y.; Van Attikum, H. A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation. Elife 2017, 6. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Ismail, A.; Klement, K.; Goodarzi, A.A.; Conrad, S.; Freire, R.; Shibata, A.; Lobrich, M.; Jeggo, P.A. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013, 41, 9719–9731. [Google Scholar] [CrossRef]
- Gautam, D.; Moody, C.A. Impact of the DNA Damage Response on Human Papillomavirus Chromatin. PLoS Pathog. 2016, 12, 6–12. [Google Scholar] [CrossRef]
- Gillespie, K.A.; Mehta, K.P.; Laimins, L.A.; Moody, C.A. Human Papillomaviruses Recruit Cellular DNA Repair and Homologous Recombination Factors to Viral Replication Centers. J. Virol. 2012, 86, 9520–9526. [Google Scholar] [CrossRef]
- Langsfeld, E.S.; Bodily, J.M.; Laimins, L.A. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog. 2015, 11, e1005181. [Google Scholar] [CrossRef]
- Chappell, W.H.; Gautam, D.; Ok, S.T.; Johnson, B.A.; Anacker, D.C.; Moody, C.A. Homologous Recombination Repair Factors Rad51 and BRCA1 Are Necessary for Productive Replication of Human Papillomavirus 31. J. Virol. 2016, 90, 2639–2652. [Google Scholar] [CrossRef]
- Anacker, D.C.; Gautam, D.; Gillespie, K.A.; Chappell, W.H.; Moody, C.A. Productive Replication of Human Papillomavirus 31 Requires DNA Repair Factor Nbs1. J. Virol. 2014, 88, 8528–8544. [Google Scholar] [CrossRef] [PubMed]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Laimins, L. Human papillomaviruses preferentially recruit DNA repair factors to viral genomes for rapid repair and amplification. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Dutta, A.; Laimins, L.A. The Acetyltransferase Tip60 Is a Critical Regulator of the Differentiation-Dependent Amplification of Human Papillomaviruses. J. Virol. 2015, 89, 4668–4675. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Cho, N.W.; Cui, G.; Manion, E.M.; Shanbhag, N.M.; Botuyan, M.V.; Mer, G.; Greenberg, R.A. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013, 20, 317–325. [Google Scholar] [CrossRef]
- Uhl, M.; Csernok, A.; Aydin, S.; Kreienberg, R.; Wiesmüller, L.; Gatz, S.A. Role of SIRT1 in homologous recombination. DNA Repair (Amst) 2010, 9, 383–393. [Google Scholar] [CrossRef]
- Oberdoerffer, P.; Michan, S.; McVay, M.; Mostoslavsky, R.; Vann, J.; Park, S.K.; Hartlerode, A.; Stegmuller, J.; Hafner, A.; Loerch, P.; et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell 2008, 135, 907–918. [Google Scholar] [CrossRef]
- Allison, S.J.; Jiang, M.; Milner, J. Oncogenic viral protein HPV E7 up-regulates the SIRT1 longevity protein in human cervical cancer cells. Aging (Albany. NY) 2009, 1, 316–327. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, X.; Sengupta, N.; Lane, W.S.; Seto, E. SIRT1 Regulates the Function of the Nijmegen Breakage Syndrome Protein. Mol. Cell 2007, 27, 149–162. [Google Scholar] [CrossRef]
- Li, K.; Casta, A.; Wang, R.; Lozada, E.; Fan, W.; Kane, S.; Ge, Q.; Gu, W.; Orren, D.; Luo, J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J. Biol. Chem. 2008, 283, 7590–7598. [Google Scholar] [CrossRef]
- Das, D.; Bristol, M.L.; Smith, N.W.; James, C.D.; Wang, X.; Pichierri, P.; Morgana, I.M. Werner helicase control of human papillomavirus 16 E1-E2 DNA replication is regulated by SIRT1 deacetylation. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Xu, B.; Kastan, M.B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002, 16, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, P.T.; Wang, Y.; Zhao, S.; Patel, N.; Lee, E.Y.H.P.; Qin, J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002, 16, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Gunasekharan, V.; Satsuka, A.; Laimins, L.A. Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators. PLoS Pathog. 2015, 11, e1004763. [Google Scholar] [CrossRef] [PubMed]
- Braccioli, L.; De Wit, E. CTCF: A Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 2019, 63, 157–165. [Google Scholar] [PubMed]
- Wendt, K.S.; Yoshida, K.; Itoh, T.; Bando, M.; Koch, B.; Schirghuber, E.; Tsutsumi, S.; Nagae, G.; Ishihara, K.; Mishiro, T.; et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008, 451, 796–801. [Google Scholar] [CrossRef]
- Hilmi, K.; Jangal, M.; Marques, M.; Zhao, T.; Saad, A.; Zhang, C.; Luo, V.M.; Syme, A.; Rejon, C.; Yu, Z.; et al. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci. Adv. 2017, 3. [Google Scholar] [CrossRef]
- Lang, F.; Li, X.; Zheng, W.; Li, Z.; Lu, D.; Chen, G.; Gong, D.; Yang, L.; Fu, J.; Shi, P.; et al. CTCF prevents genomic instability by promoting homologous recombination-directed DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2017, 114, 10912–10917. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Kang, M.A.; Baik, C.J.; Lee, Y.; Hang, N.T.; Kim, B.G.; Han, J.S.; Jeong, J.H.; Park, D.; Myung, K.; et al. CTCF cooperates with CtIP to drive homologous recombination repair of double-strand breaks. Nucleic Acids Res. 2019, 47, 9160–9179. [Google Scholar] [CrossRef]
- Paris, C.; Pentland, I.; Groves, I.; Roberts, D.C.; Powis, S.J.; Coleman, N.; Roberts, S.; Parish, J.L. CCCTC-Binding Factor Recruitment to the Early Region of the Human Papillomavirus 18 Genome Regulates Viral Oncogene Expression. J. Virol. 2015, 89, 4770–4785. [Google Scholar] [CrossRef]
- Gautam, D.; Johnson, B.; Mac, M.; Moody Id, C.A. SETD2-dependent H3K36me3 plays a critical role in epigenetic regulation of the HPV31 life cycle. PLoS Pathog. 2018, 14, e1007367. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Duns, G.; Westers, H.; Sijmons, R.; van den Berg, A.; Kok, K. SETD2: An epigenetic modifier with tumor suppressor functionality. Oncotarget 2016, 7, 50719–50734. [Google Scholar] [CrossRef]
- Hacker, K.E.; Fahey, C.C.; Shinsky, S.A.; Chiang, Y.C.J.; DiFiore, J.V.; Jha, D.K.; Vo, A.H.; Shavit, J.A.; Davis, I.J.; Strahl, B.D.; et al. Structure/function analysis of recurrent mutations in SETD2 protein reveals a critical and conserved role for a SET domain residue in maintaining protein stability and histone H3 Lys-36 trimethylation. J. Biol. Chem. 2016, 291, 21283–21295. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, S.L.; Strahl, B.D. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 2017, 74, 3317–3334. [Google Scholar] [CrossRef] [PubMed]
- Pradeepa, M.M.; Sutherland, H.G.; Ule, J.; Grimes, G.R.; Bickmore, W.A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Bleuyard, J.-Y.; Fournier, M.; Nakato, R.; Couturier, A.M.; Katou, Y.; Ralf, C.; Hester, S.S.; Dominguez, D.; Rhodes, D.; Humphrey, T.C.; et al. MRG15-mediated tethering of PALB2 to unperturbed chromatin protects active genes from genotoxic stress. Proc. Natl. Acad. Sci. USA 2017, 114, 7671–7676. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef]
- Pfister, S.X.; Ahrabi, S.; Zalmas, L.P.; Sarkar, S.; Aymard, F.; Bachrati, C.Z.; Helleday, T.; Legube, G.; LaThangue, N.B.; Porter, A.C.G.; et al. SETD2-Dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell Rep. 2014, 7, 2006–2018. [Google Scholar] [CrossRef]
- Graham, S.V.; Faizo, A.A.A. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2017, 231, 83–95. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Duensing, S.; Münger, K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002, 62, 7075–7082. [Google Scholar] [PubMed]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Warburton, A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef]
- Thorland, E.C.; Myers, S.L.; Gostout, B.S.; Smith, D.I. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 2003, 22, 1225–1237. [Google Scholar] [CrossRef]
- Sarni, D.; Kerem, B. The complex nature of fragile site plasticity and its importance in cancer. Curr. Opin. Cell Biol. 2016, 40, 131–136. [Google Scholar] [CrossRef]
- Dooley, K.E.; Warburton, A.; McBride, A.A. Tandemly integrated HPV16 can form a brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. MBio 2016, 7. [Google Scholar] [CrossRef]
- Groves, I.J.; Knight, E.L.A.; Ang, Q.Y.; Scarpini, C.G.; Coleman, N. HPV16 oncogene expression levels during early cervical carcinogenesis are determined by the balance of epigenetic chromatin modifications at the integrated virus genome. Oncogene 2016, 35, 4773–4786. [Google Scholar] [CrossRef]
- McBride, A.A. Playing with fire: Consequences of human papillomavirus DNA replication adjacent to genetically unstable regions of host chromatin. Curr. Opin. Virol. 2017, 26, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Russo, D.; Merolla, F.; Varricchio, S.; Salzano, G.; Zarrilli, G.; Mascolo, M.; Strazzullo, V.; Di Crescenzo, R.M.; Celetti, A.; Ilardi, G. Epigenetics of oral and oropharyngeal cancers. Biomed. Rep. 2018, 9, 275–283. [Google Scholar] [CrossRef]
- Morgan, I.M.; Dinardo, L.J.; Windle, B. Integration of human papillomavirus genomes in head and neck cancer: Is it time to consider a paradigm shift? Viruses 2017, 9, 208. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mac, M.; Moody, C.A. Epigenetic Regulation of the Human Papillomavirus Life Cycle. Pathogens 2020, 9, 483. https://doi.org/10.3390/pathogens9060483
Mac M, Moody CA. Epigenetic Regulation of the Human Papillomavirus Life Cycle. Pathogens. 2020; 9(6):483. https://doi.org/10.3390/pathogens9060483
Chicago/Turabian StyleMac, Michelle, and Cary A. Moody. 2020. "Epigenetic Regulation of the Human Papillomavirus Life Cycle" Pathogens 9, no. 6: 483. https://doi.org/10.3390/pathogens9060483
APA StyleMac, M., & Moody, C. A. (2020). Epigenetic Regulation of the Human Papillomavirus Life Cycle. Pathogens, 9(6), 483. https://doi.org/10.3390/pathogens9060483




