Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense
Abstract
1. Introduction
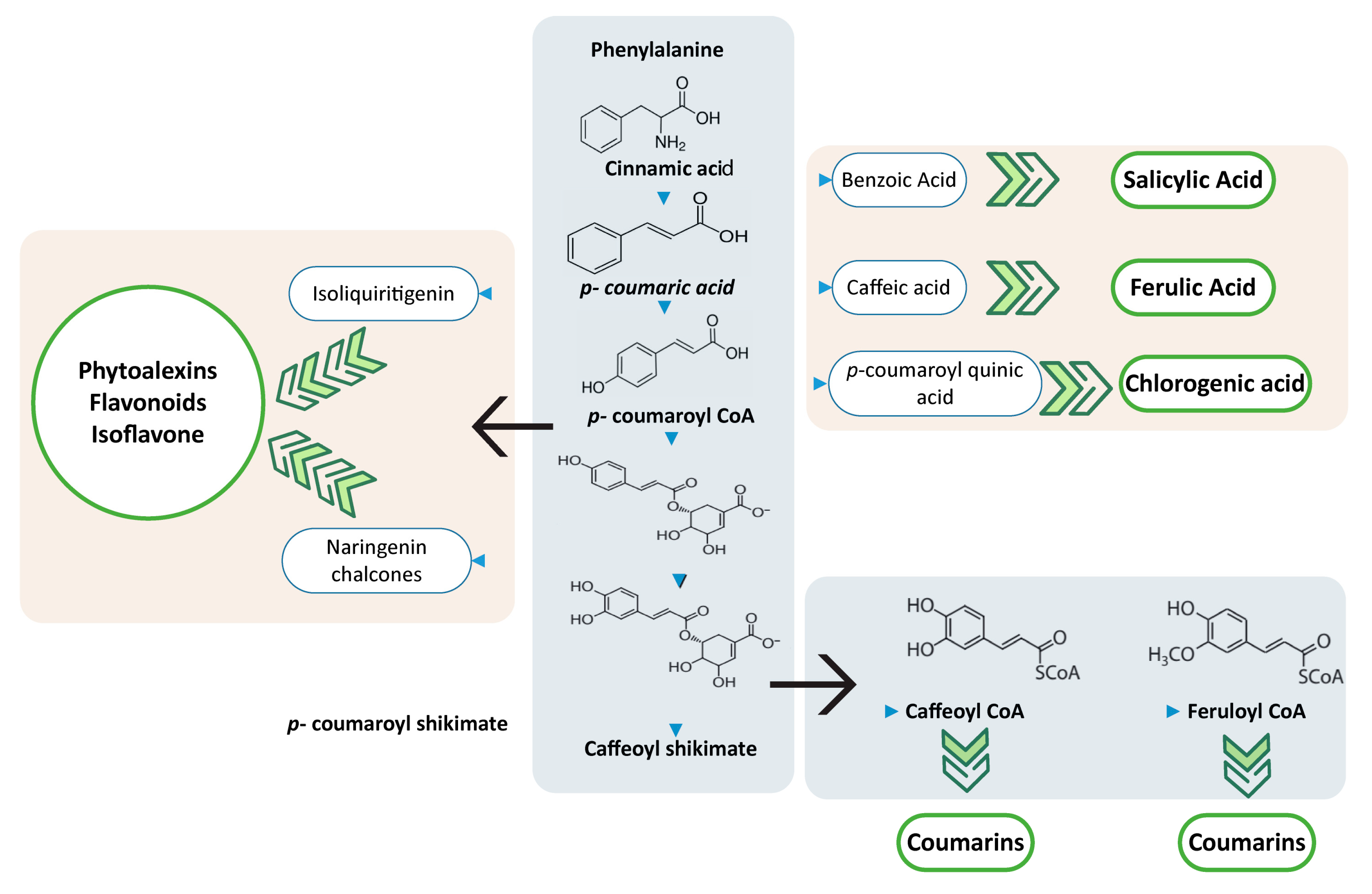
2. Phenylpropanoid Pathways—Biochemistry to Genetics
3. Phenylpropanoid and Lignin Pathway associated Genes in Plant Defense
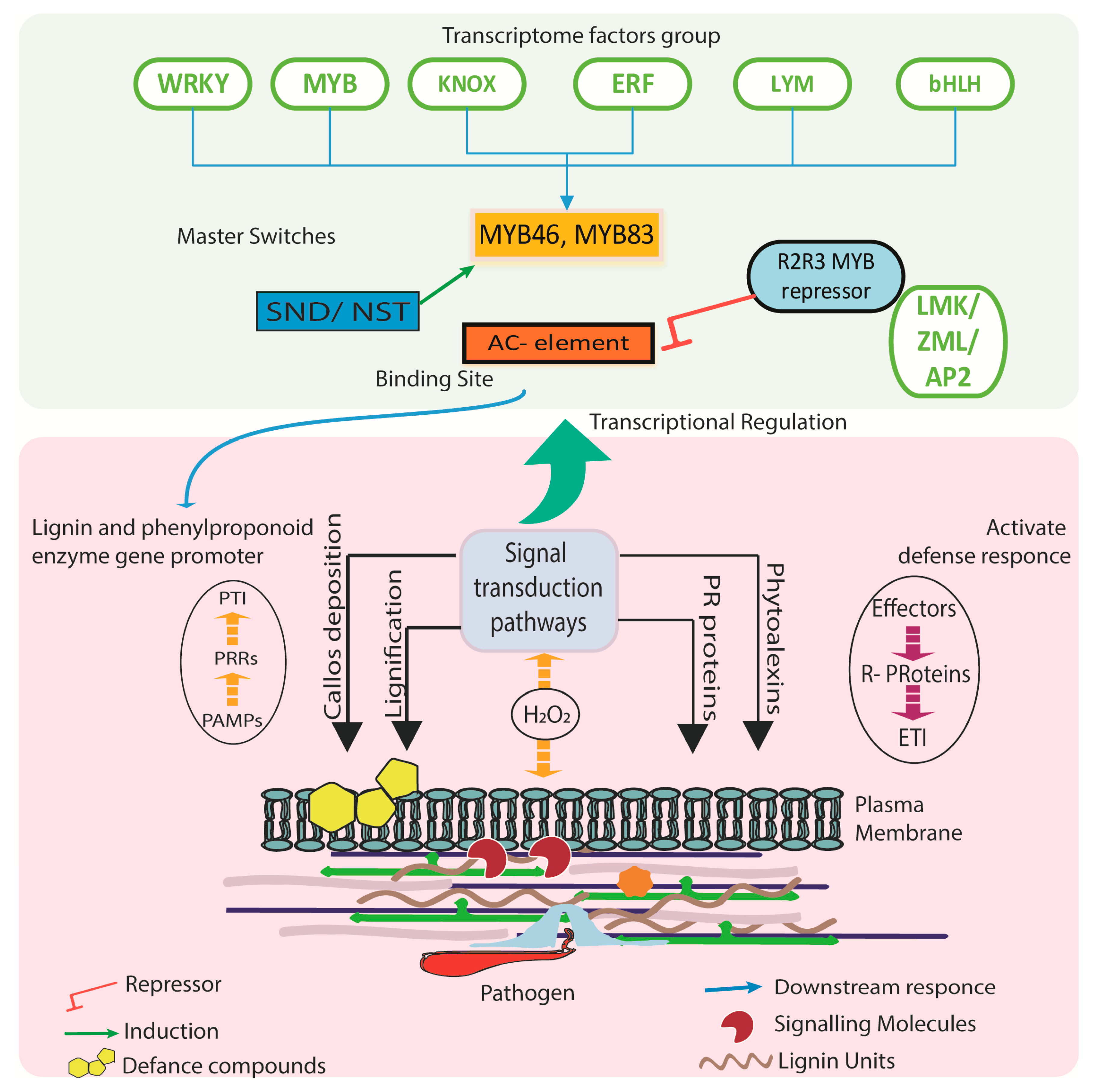
4. Transcriptional Regulation for Phenylpropanoid Pathways
5. Mechanism of Player Genes in Defense Response
5.1. Lignification
5.2. Coumarins, Flavonoid, Phytoalexins Naringenin (Metabolites)
5.2.1. Coumarins
5.2.2. Flavonoids
5.3. SA-Mediated Resistance
5.4. Signaling and Elicitor Based Pathway
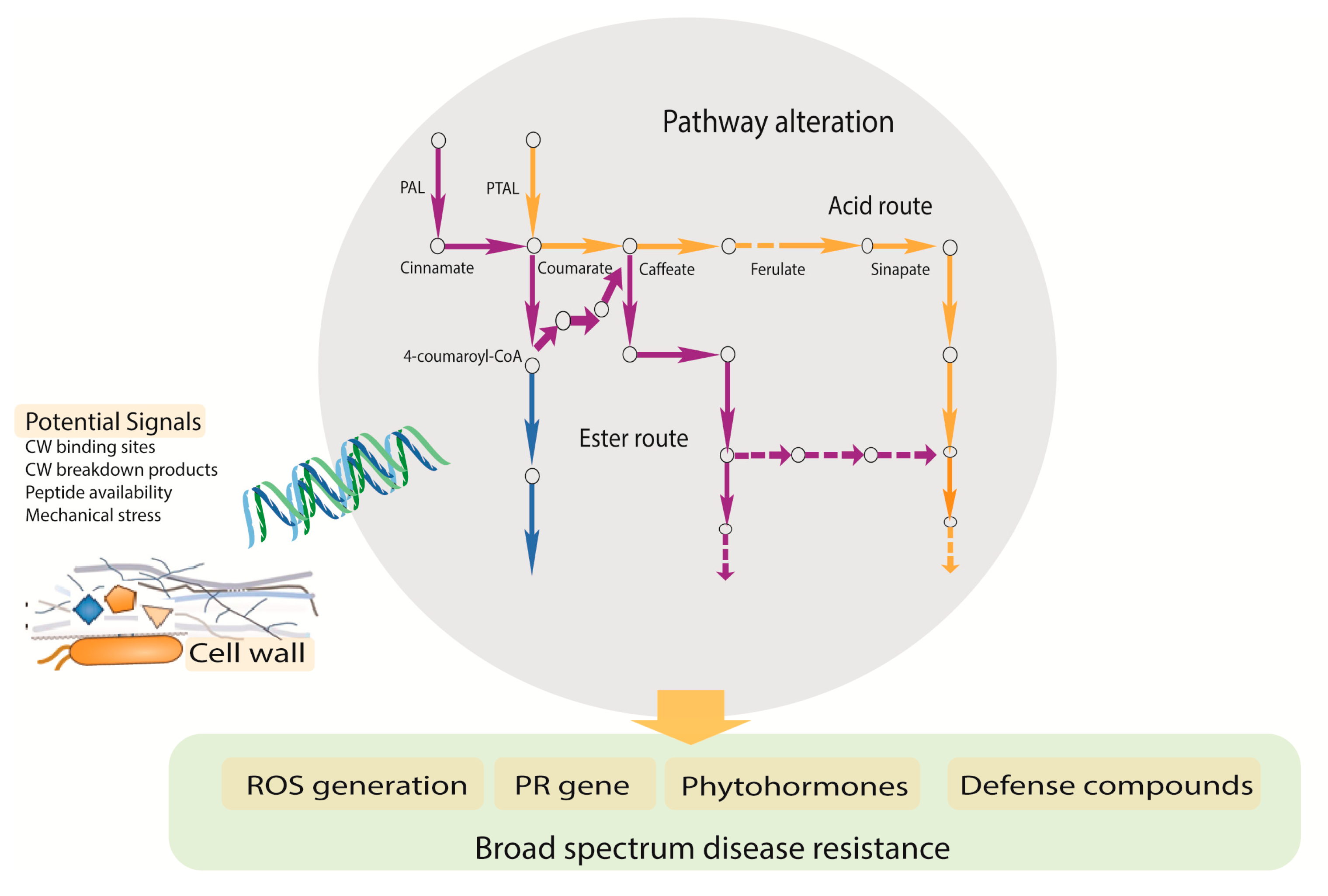
6. Virulence Pathogen Regulates Phenylpropanoid Pathway
7. PALs: Emerging Key Players in Broad Spectrum Disease Resistance
8. Research Questions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Lionetti, V.; Métraux, J.-P. Plant cell wall in pathogenesis, parasitism and symbiosis. Front. Plant Sci 2014, 5, 612. [Google Scholar] [CrossRef] [PubMed]
- Höfte, H.; Voxeur, A. Plant cell walls. Curr. Biol. 2017, 27, R865–R870. [Google Scholar] [CrossRef] [PubMed]
- Hoson, T.; Wakabayashi, K. Role of the plant cell wall in gravity resistance. Phytochemistry 2015, 112, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; Birch, P.R.J. Rotting softly and stealthily. Curr. Opin. Plant Biol. 2005, 8, 424–429. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Pogorelko, G.; Lionetti, V.; Bellincampi, D.; Zabotina, O. Cell wall integrity. Plant Signal. Behav. 2013, 8, e25435. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Ferrari, S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 2002, 5, 295–299. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef]
- Rui, Y.; Dinneny, J.R. A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef]
- Engelsdorf, T.; Hamann, T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann. Bot. 2014, 114, 1339–1347. [Google Scholar] [CrossRef]
- Wolf, S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017, 474, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Van der Does, D.; Zipfel, C.; Hamann, T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Phytopathol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Lion, C.; Biot, C.; Gierlinger, N.; Hawkins, S. Lignification and Advances in Lignin Imaging in Plant Cell Walls. Annu. Plant Rev. Online 2018, 1, 909–940. [Google Scholar] [CrossRef]
- Denancé, N.; Ranocha, P.; Oria, N.; Barlet, X.; Rivière, M.-P.; Yadeta, K.A.; Hoffmann, L.; Perreau, F.; Clément, G.; Maia-Grondard, A.; et al. Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 2013, 73, 225–239. [Google Scholar] [CrossRef]
- Jaeck, E.; Dumas, B.; Geoffroy, P.; Favet, N.; Inze, D.; Van Montagu, M.; Fritig, B.; Legrand, M. Regulation of enzymes involved in lignin biosynthesis: Induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol. Plant Microbe Int. 1992, 5, 294–300. [Google Scholar] [CrossRef]
- Chowdhury, J.; Henderson, M.; Schweizer, P.; Burton, R.A.; Fincher, G.B.; Little, A. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol. 2014, 204, 650–660. [Google Scholar] [CrossRef]
- Chowdhury, J.; Schober, M.S.; Shirley, N.J.; Singh, R.R.; Jacobs, A.K.; Douchkov, D.; Schweizer, P.; Fincher, G.B.; Burton, R.A.; Little, A. Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp. hordei. New Phytol. 2016, 212, 434–443. [Google Scholar] [CrossRef]
- Kwon, C.; Neu, C.; Pajonk, S.; Yun, H.S.; Lipka, U.; Humphry, M.; Bau, S.; Straus, M.; Kwaaitaal, M.; Rampelt, H.; et al. Co-option of a default secretory pathway for plant immune responses. Nature 2008, 451, 835–840. [Google Scholar] [CrossRef]
- Vorwerk, S.; Somerville, S.; Somerville, C. The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 2004, 9, 203–209. [Google Scholar] [CrossRef]
- Wuyts, N.; Swennen, R.; Waele, D.D. Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 2006, 8, 89. [Google Scholar] [CrossRef]
- Zhao, R.; Nakamura, T.; Fu, Y.; Lazar, Z.; Spector, D.L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 2011, 13, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-H.; Yang, Q.; Ma, R.-C. Erwinia carotovora ssp. carotovora Infection Induced “Defense Lignin” Accumulation and Lignin Biosynthetic Gene Expression in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). J. Integr. Plant Biol. 2007, 49, 993–1002. [Google Scholar] [CrossRef]
- Eynck, C.; Koopmann, B.; Karlovsky, P.; von Tiedemann, A. Internal Resistance in Winter Oilseed Rape Inhibits Systemic Spread of the Vascular Pathogen Verticillium longisporum. Phytopathology 2009, 99, 802–811. [Google Scholar] [CrossRef]
- Hano, C.; Martin, I.; Fliniaux, O.; Legrand, B.; Gutierrez, L.; Arroo, R.R.J.; Mesnard, F.; Lamblin, F.; Lainé, E. Pinoresinol–lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 2006, 224, 1291–1301. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic Interaction between Abscisic Acid and Jasmonate-Ethylene Signaling Pathways Modulates Defense Gene Expression and Disease Resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef]
- Sattler, S.E.; Funnell-Harris, D.L.; Pedersen, J.F. Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci. 2010, 178, 229–238. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 2009, 4, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.D.; Gries, T.; Sarath, G.; Palmer, N.A.; Baird, L.; Serapiglia, M.J.; Dien, B.S.; Boateng, A.A.; Ge, Z.; Funnell-Harris, D.L.; et al. Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in sorghum bicolor. Plant J. 2016, 85, 378–395. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Wang, G.-L. Breeding plant broad-spectrum resistance without yield penalties. Proc. Natl. Acad. Sci. USA 2018, 115, 2859–2861. [Google Scholar] [CrossRef]
- Ning, Y.; Liu, W.; Wang, G.-L. Balancing Immunity and Yield in Crop Plants. Trends Plant Sci. 2017, 22, 1069–1079. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Plant Physiol. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef]
- Hao, Z.; Mohnen, D. A review of xylan and lignin biosynthesis: Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 212–241. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl Shikimate Esterase (CSE) Is an Enzyme in the Lignin Biosynthetic Pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Buendgen, M.R.; Coors, J.G.; Grombacher, A.W.; Russell, W.A. European Corn Borer Resistance and Cell Wall Composition of Three Maize Populations. Crop Sci. 1990, 30, 505–510. [Google Scholar] [CrossRef]
- Bonello, P.; Blodgett, J.T. Pinus nigra–Sphaeropsis sapinea as a model pathosystem to investigate local and systemic effects of fungal infection of pines. Physiol. Mol. Plant Pathol. 2003, 63, 249–261. [Google Scholar] [CrossRef]
- Dicko, M.H.; Gruppen, H.; Barro, C.; Traore, A.S.; van Berkel, W.J.H.; Voragen, A.G.J. Impact of Phenolic Compounds and Related Enzymes in Sorghum Varieties for Resistance and Susceptibility to Biotic and Abiotic Stresses. J. Chem. Ecol. 2005, 31, 2671–2688. [Google Scholar] [CrossRef] [PubMed]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Ulanov, A.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 2007, 225, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; He, X.; Dixon, R.A. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 2008, 8, 132. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Liu, W.; Liu, G.; Selvaraj, G.; Wei, Y.; King, J. Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol. Biol. 2007, 64, 305–318. [Google Scholar] [CrossRef]
- Zhao, J.; Buchwaldt, L.; Rimmer, S.R.; Sha, A.; Mcgregor, L.; Bekkaoui, D.; Hegdues, D. Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol. Plant. Pathol. 2009, 10, 635–649. [Google Scholar] [CrossRef]
- Weng, J.-K.; Mo, H.; Chapple, C. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 2010, 64, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tobimatsu, Y.; Havkin-Frenkel, D.; Dixon, R.A.; Ralph, J. A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 2012, 109, 1772–1777. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.; Jung, W.; Shi, J.; Croes, R.A.; Fader, G.M.; McGonigle, B.; Odell, J.T. Production of the Isoflavones Genistein and Daidzein in Non-Legume Dicot and Monocot Tissues. Plant Physiol. 2000, 124, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.; Jez, J.M. Nature’s assembly line: Biosynthesis of simple phenylpropanoids and polyketides. Plant J. 2008, 54, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.-J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Dixon, L.J.; Castlebury, L.A.; Aime, M.C.; Glynn, N.C.; Comstock, J.C. Phylogenetic relationships of sugarcane rust fungi. Mycol. Prog. 2010, 9, 459–468. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Posé, S.; Pattathil, S.; Peralta, A.G.; Hahn, M.G.; Ayre, B.G.; Sunuwar, J.; Hernandez, J.; Patel, M.; Shah, J.; et al. Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol. 2018, 219, 1235–1251. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Altering the Cell Wall and Its Impact on Plant Disease: From Forage to Bioenergy. Annu. Rev. Phytopathol. 2014, 52, 69–91. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 2009, 60, 509–521. [Google Scholar] [CrossRef]
- Shadle, G.L.; Wesley, S.V.; Korth, K.L.; Chen, F.; Lamb, C.; Dixon, R.A. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of l-phenylalanine ammonia-lyase. Phytochemistry 2003, 64, 153–161. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Bukhman, Y.V.; Foster, C.E.; Thrower, N.; Bruno, L.C.; et al. Effects of phenylalanine ammonia lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015, 66, 4317–4335. [Google Scholar] [CrossRef] [PubMed]
- Shine, M.B.; Yang, J.-W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.-Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Si, J.; Cui, X.; Peng, H.; Chen, X.; Xing, H.; Dou, D. The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant Growth Regul. 2019, 88, 139–149. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, M.; Hou, C.; Lu, T.; Liu, L.; Wei, H.; Cheng, Y.; Wei, Z. Functional characterization of populus psnshn2 in coordinated regulation of secondary wall components in tobacco. Sci. Rep. 2017, 7, 42. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Escamilla-Trevino, L.; Jackson, L.A.; Dixon, R.A. Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20814–20819. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Jikumaru, Y.; Kamiya, Y.; Tang, Y.; Dixon, R.A. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 2011, 190, 627–639. [Google Scholar] [CrossRef]
- Eynck, C.; Seguin-Swartz, G.; Clarke, W.E.; Parkin, I.A.P. Monolignol biosynthesis is associated with resistance to Sclerotinia sclerotiorum in Camelina sativa. Mol. Plant Pathol. 2012, 13, 887–899. [Google Scholar] [CrossRef]
- Yang, Q.; He, Y.; Kabahuma, M.; Chaya, T.; Kelly, A.; Borrego, E.; Bian, Y.; El Kasmi, F.; Yang, L.; Teixeira, P.; et al. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 2017, 49, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, X.; Wang, K.; Lu, C.; Luo, M.; Shan, T.; Zhang, Z. A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 2018, 8, 6543. [Google Scholar] [CrossRef] [PubMed]
- Quentin, M.; Allasia, V.; Pegard, A.; Allais, F.; Ducrot, P.-H.; Favery, B.; Levis, C.; Martinet, S.; Masur, C.; Ponchet, M.; et al. Imbalanced Lignin Biosynthesis Promotes the Sexual Reproduction of Homothallic Oomycete Pathogens. PLOS Pathog. 2009, 5, e1000264. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef]
- Maury, S.; Delaunay, A.; Mesnard, F.; Crônier, D.; Chabbert, B.; Geoffroy, P.; Legrand, M. O-methyltransferase(s)-suppressed plants produce lower amounts of phenolic vir inducers and are less susceptible to Agrobacterium tumefaciens infection. Planta 2010, 232, 975–986. [Google Scholar] [CrossRef]
- Funnell-Harris, D.L.; Pedersen, J.F.; Sattler, S.E. Alteration in Lignin Biosynthesis Restricts Growth of Fusarium spp. in Brown Midrib Sorghum. Phytopathology 2010, 100, 671–681. [Google Scholar] [CrossRef]
- Wróbel-Kwiatkowska, M.; Starzycki, M.; Zebrowski, J.; Oszmiański, J.; Szopa, J. Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J. Biotechnol. 2007, 128, 919–934. [Google Scholar] [CrossRef]
- Sibout, R.; Eudes, A.; Mouille, G. Cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 2005, 17. [Google Scholar] [CrossRef]
- Huang, J.; Bhinu, V.S.; Li, X.; Dallal Bashi, Z.; Zhou, R.; Hannoufa, A. Pleiotropic changes in Arabidopsis f5h and sct mutants revealed by large-scale gene expression and metabolite analysis. Planta 2009, 230, 1057–1069. [Google Scholar] [CrossRef]
- König, S.; Feussner, K.; Kaever, A.; Landesfeind, M.; Thurow, C.; Karlovsky, P.; Gatz, C.; Polle, A.; Feussner, I. Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum. New Phytol. 2014, 202, 823–837. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.M.M.; Langlois-Meurinne, M.; Henry, Y.; Saindrenan, P. Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: Functional and evolutionary implications. Plant Mol. Biol. 2005, 58, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; Zhao, Q.; Gallego-Giraldo, L.; Dai, X.; Zhao, P.X.; Dixon, R.A. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010, 11, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Dixon, R.A. Current Models for Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2019, 56, 82–87. [Google Scholar] [CrossRef]
- Lacombe, E.; Van Doorsselaere, J.; Boerjan, W.; Boudet, A.M.; Grima-Pettenati, J. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein–DNA complex formation. Plant J. 2000, 23, 663–676. [Google Scholar] [CrossRef]
- Legay, S.; Lacombe, E.; Goicoechea, M.; Brière, C.; Séguin, A.; Mackay, J.; Grima-Pettenati, J. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 2007, 173, 542–549. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef]
- Zhao, Q.; Gallego-Giraldo, L.; Wang, H.; Zeng, Y.; Ding, S.-Y.; Chen, F.; Dixon, R.A. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 2010, 63, 100–114. [Google Scholar] [CrossRef]
- McCarthy, R.L.; Zhong, R.; Ye, Z.-H. MYB83 Is a Direct Target of SND1 and Acts Redundantly with MYB46 in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef] [PubMed]
- Patzlaff, A.; McInnis, S.; Courtenay, A.; Surman, C.; Newman, L.J.; Smith, C.; Bevan, M.W.; Mansfield, S.; Whetten, R.W.; Sederoff, R.R.; et al. Characterisation of a pine MYB that regulates lignification. Plant J. 2003, 36, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef] [PubMed]
- Mele, G.; Ori, N.; Sato, Y.; Hake, S. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 2003, 17, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Sonbol, F.-M.; Fornalé, S.; Capellades, M.; Encina, A.; Touriño, S.; Torres, J.-L.; Rovira, P.; Ruel, K.; Puigdomènech, P.; Rigau, J.; et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 2009, 70, 283. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 Transcription Factors from Antirrhinum Regulate Phenylpropanoid and Lignin Biosynthesis in Transgenic Tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Liang, Y.-K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB Transcription Factor Controlling Stomatal Aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; Zhao, Y.; Gong, X.; Guo, L.; Zhu, G.; Wang, X.; Gong, Z.; Schumaker, K.S.; Guo, Y. SAD2, an Importin β-Like Protein, Is Required for UV-B Response in arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 2007, 19, 3805–3818. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, Z.; Wang, C.; Zhang, X.; Tang, Y.; Zhu, X.; Shao, J.; Wu, Y. Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant Cell 2015, 84, 395–403. [Google Scholar] [CrossRef]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant Cell 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Chezem, W.R.; Memon, A.; Li, F.-S.; Weng, J.-K.; Clay, N.K. SG2-Type R2R3-MYB Transcription Factor MYB15 Controls Defense-Induced Lignification and Basal Immunity in Arabidopsis. Plant Cell 2017, 29, 1907–1926. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; He, X.; Poovaiah, C.R.; Wuddineh, W.A.; Ma, J.; Mann, D.G.J.; Wang, H.; Jackson, L.; Tang, Y.; Neal Stewart, C., Jr.; et al. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012, 193, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Shi, X.; Chai, C.; Encina, A.; Irar, S.; Capellades, M.; Fuguet, E.; Torres, J.-L.; Rovira, P.; Puigdomènech, P.; et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Bermúdez, I.-C.; Salazar-Henao, J.E.; Fornalé, S.; López-Vidriero, I.; Franco-Zorrilla, J.-M.; Grotewold, E.; Gray, J.; Solano, R.; Schmidt, W.; Pagés, M.; et al. A MYB/ZML Complex Regulates Wound-Induced Lignin Genes in Maize. Plant Cell 2015, 27, 3245–3259. [Google Scholar] [CrossRef]
- Fornalé, S.; Sonbol, F.-M.; Maes, T.; Capellades, M.; Puigdomènech, P.; Rigau, J.; Caparrós-Ruiz, D. Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 2006, 62, 809–823. [Google Scholar] [CrossRef]
- Bedon, F.; Bomal, C.; Caron, S.; Levasseur, C.; Boyle, B.; Mansfield, S.D.; Schmidt, A.; Gershenzon, J.; Grima-Pettenati, J.; Séguin, A.; et al. Subgroup 4 R2R3-MYBs in conifer trees: Gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 2010, 61, 3847–3864. [Google Scholar] [CrossRef]
- Bomal, C.; Duval, I.; Giguère, I.; Fortin, É.; Caron, S.; Stewart, D.; Boyle, B.; Séguin, A.; MacKay, J.J. Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. J. Exp. Bot. 2013, 65, 495–508. [Google Scholar] [CrossRef]
- Li, M.; Yasuda, M.; Yamaya-Ito, H.; Maeda, M.; Sasaki, N.; Nagata, M.; Suzuki, A.; Okazaki, S.; Sekimoto, H.; Yamada, T.; et al. Involvement of programmed cell death in suppression of the number of root nodules formed in soybean induced by Bradyrhizobium infection. Soil Sci. Plant Nutr. 2017, 63, 561–577. [Google Scholar] [CrossRef][Green Version]
- Liu, J.; Ding, P.; Sun, T.; Nitta, Y.; Dong, O.; Huang, X.; Yang, W.; Li, X.; Botella, J.R.; Zhang, Y. Heterotrimeric G Proteins Serve as a Converging Point in Plant Defense Signaling Activated by Multiple Receptor-Like Kinases. Plant Physiol. 2013, 161, 2146–2158. [Google Scholar] [CrossRef]
- Omer, S.; Kumar, S.; Khan, B.M. Over-expression of a subgroup 4 R2R3 type MYB transcription factor gene from Leucaena leucocephala reduces lignin content in transgenic tobacco. Plant Cell Rep. 2013, 32, 161–171. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Wang, C.; Zhu, H.-H. TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie 2011, 93, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, X.-r.; Zeng, J.-k.; Ge, H.; Song, M.; Xu, C.-j.; Li, X.; Ferguson, I.B.; Chen, K.-s. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, X.; Li, C.; Lu, W.; Yang, L.; Jiang, Y.; Luo, K. Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS ONE 2013, 8, e76369. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, X.; Ran, L.; Li, C.; Fan, D.; Luo, K. PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 2017, 7, 41209. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, X.; Yang, F.; Fan, D.; Jiang, Y.; Luo, K. PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 2016, 6, 18643. [Google Scholar] [CrossRef] [PubMed]
- Yogendra, K.N.; Kumar, A.; Sarkar, K.; Li, Y.; Pushpa, D.; Mosa, K.A.; Duggavathi, R.; Kushalappa, A.C. Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 2015, 66, 7377–7389. [Google Scholar] [CrossRef]
- Guillaumie, S.; Mzid, R.; Méchin, V.; Léon, C.; Hichri, I.; Destrac-Irvine, A.; Trossat-Magnin, C.; Delrot, S.; Lauvergeat, V. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 2009, 72, 215. [Google Scholar] [CrossRef]
- Robischon, M.; Du, J.; Miura, E.; Groover, A. The Populus Class III HD ZIP, popREVOLUTA, Influences Cambium Initiation and Patterning of Woody Stems. Plant Physiol. 2011, 155, 1214–1225. [Google Scholar] [CrossRef]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP Transcription Factor POPCORONA Affects Cell Differentiation during Secondary Growth of Woody Stems. PLoS ONE 2011, 6, e17458. [Google Scholar] [CrossRef]
- Sonoda, T.; Koita, H.; Nakamoto-Ohta, S.; Kondo, K.; Suezaki, T.; Kato, T.; Kato, T.; Nagai, K.; Iida, N.; Sato, S.; et al. Increasing fiber length and growth in transgenic tobacco plants overexpressing a gene encoding the Eucalyptus camaldulensis HD-Zip class II transcription factor. Plant Biotechnol. 2009, 26, 115–120. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Lin, Y.-C.; Sun, Y.-H.; Song, J.; Chen, H.; Zhang, X.-H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-mediated alternative splicing of wood-associated nac transcription factor1b regulates cell wall thickening during fiber development in populus species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Hou, Y.; Zhao, X.; Lu, W.; Li, Y.; Yang, F.; Tang, S.; Luo, K. Identification and characterization of a wood-associated NAC domain transcription factor PtoVNS11 from Populus tomentosa Carr. Trees 2015, 29, 1091–1101. [Google Scholar] [CrossRef]
- Nuhse, T. Cell wall integrity signaling and innate immunity in plants. Front. Plant Sci. 2012, 3, 280. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, L.A. Lignification and lignin topochemistry—an ultrastructural view. Phytochemistry 2001, 57, 859–873. [Google Scholar] [CrossRef]
- Bechinger, C.; Giebel, K.-F.; Schnell, M.; Leiderer, P.; Deising, H.B.; Bastmeyer, M. Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 1999, 285, 1896–1899. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Rong, W.; Yang, J.; Li, Z.; Wu, L.; Zhang, G.; Ma, Z. Histochemical analyses reveal that stronger intrinsic defenses in gossypium barbadense than in g. hirsutum are associated with resistance to verticillium dahliae. Front. Plant Sci. 2017, 30, 984–996. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Voigt, C.A. Cellulose/callose glucan networks: The key to powdery mildew resistance in plants? New Phytol. 2016, 212, 303–305. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.u.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Santiago, R.; Barros-Rios, J.; Malvar, R.A. Impact of cell wall composition on maize resistance to pests and diseases. Int. J. Mol. Sci. 2013, 14, 6960–6980. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Kar, I.; Mukherjee, A.K.; Acharya, P. Elicitor-induced defense responses in Solanum lycopersicum against Ralstonia solanacearum. Sci. World J. 2013, 2013, 561056. [Google Scholar] [CrossRef] [PubMed]
- Tronchet, M.; Balague, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-F.; He, Y.; Strauch, R.; Olukolu, B.A.; Nielsen, D.; Li, X.; Balint-Kurti, P.J. Maize homologs of hydroxycinnamoyltransferase, a key enzyme in lignin biosynthesis, bind the nucleotide binding leucine-rich repeat rp1 proteins to modulate the defense response. Plant Physiol. 2015, 169, 2230–2243. [Google Scholar] [CrossRef]
- Liu, H.; Guo, Z.; Gu, F.; Ke, S.; Sun, D.; Dong, S.; Liu, W.; Huang, M.; Xiao, W.; Yang, G.; et al. 4-Coumarate-CoA Ligase-Like Gene OsAAE3 Negatively Mediates the Rice Blast Resistance, Floret Development and Lignin Biosynthesis. Front. Plant Sci. 2017, 7, 2041. [Google Scholar] [CrossRef]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Stringlis, I.A.; de Jonge, R.; Pieterse, C.M.J. The age of coumarins in plant–microbe interactions. Plant Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef]
- Kai, K.; Shimizu, B.-i.; Mizutani, M.; Watanabe, K.; Sakata, K. Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry 2006, 67, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.; Baltz, R.; Saindrenan, P. Over-expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol. Biol. 2004, 54, 137–146. [Google Scholar] [CrossRef]
- Shimizu, B.; Miyagawa, H.; Ueno, T.; Sakata, K.; Watanabe, K.; Ogawa, K. Morning Glory Systemically Accumulates Scopoletin and Scopolin after Interaction with Fusarium oxysporum. Z. Für Naturforschung C 2014, 60, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Baltz, R.; Schmitt, C.; Beffa, R.; Fritig, B.; Saindrenan, P. Downregulation of a pathogen-responsive tobacco udp-glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 2002, 14, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Tanguy, J.; Martin, C. Phenolic compounds and the hypersensitivity reaction in nicotiana tabacum infected with tobacco mosaic virus. Phytochemistry 1972, 11, 19–28. [Google Scholar] [CrossRef]
- Dutsadee, C.; Nunta, C. Induction of peroxidase, scopoletin, phenolic compounds and resistance in Hevea brasiliensis by elicitin and a novel protein elicitor purified from phytophthora palmivora. Physiol. Mol. Plant Pathol. 2008, 72, 179–187. [Google Scholar] [CrossRef]
- Barilli, E.; Rubiales, D.; Amalfitano, C.; Evidente, A.; Prats, E. BTH and BABA induce resistance in pea against rust (Uromyces pisi) involving differential phytoalexin accumulation. Planta 2015, 242, 1095–1106. [Google Scholar] [CrossRef]
- Tortosa, M.; Cartea, M.E.; Rodríguez, V.M.; Velasco, P. Unraveling the metabolic response of Brassica oleracea exposed to Xanthomonas campestris pv. campestris. J. Sci. of Food Agric. 2018, 98, 3675–3683. [Google Scholar] [CrossRef]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- El Oirdi, M.; Trapani, A.; Bouarab, K. The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environ. Microbiol. 2010, 12, 239–253. [Google Scholar] [CrossRef]
- Sun, H.; Wang, L.; Zhang, B.; Ma, J.; Hettenhausen, C.; Cao, G.; Sun, G.; Wu, J.; Wu, J. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 2014, 65, 4305–4315. [Google Scholar] [CrossRef]
- Santhanam, R.; Menezes, R.C.; Grabe, V.; Li, D.; Baldwin, I.T.; Groten, K. A suite of complementary biocontrol traits allows a native consortium of root-associated bacteria to protect their host plant from a fungal sudden-wilt disease. Mol. Ecol. 2019, 28, 1154–1169. [Google Scholar] [CrossRef]
- Chaouch, S.; Queval, G.; Noctor, G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012, 69, 613–627. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Zhang, H.; Pieterse, C.M.J.; Bolton, M.D.; de Jonge, R. Microbial small molecules—weapons of plant subversion. Nat. Prod. Rep. 2018, 35, 410–433. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Environ. Chem. Lett. 2005, 7, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Iinuma, M. Reduction of membrane fluidity by antibacterial sophoraflavanone G isolated from sophora exigua. Phytomedicine 2000, 7, 161–165. [Google Scholar] [CrossRef]
- Tamba, Y.; Ohba, S.; Kubota, M.; Yoshioka, H.; Yoshioka, H.; Yamazaki, M. Single GUV method reveals interaction of tea catechin (−)-epigallocatechin gallate with lipid membranes. Biophys. J. 2007, 92, 3178–3194. [Google Scholar] [CrossRef]
- Katsumata, S.; Hamana, K.; Horie, K.; Toshima, H.; Hasegawa, M. Identification of sternbin and naringenin as detoxified metabolites from the rice flavanone phytoalexin sakuranetin by pyricularia oryzae. Chem. Biodivers. 2017, 14, e1600240. [Google Scholar] [CrossRef]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef]
- Lu, H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 2009, 4, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z.; et al. Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Zhang, F.; Dong, L.; Wu, J.; Cheng, Q.; Qi, D.; Yan, X.; Jiang, L.; Fan, S.; et al. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 7242. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Deng, X.; Jia, S.; Huang, J.; Miao, X.; Huang, Y. Role of Salicylic Acid in Tomato Defense against Cotton Bollworm, Helicoverpa armigera Hubner. Z. Für Naturforschung C 2004, 59, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Slusarenko, A.J. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of arabidopsis to peronospora parasitica. Plant Cell 1996, 8, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yu, J.; Bai, J.; Zhu, Z.; Wang, X. Induced defense responses in rice plants against small brown planthopper infestation. Crop J. 2014, 2, 55–62. [Google Scholar] [CrossRef]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef]
- Hibino, T.; Takabe, K.; Kawazu, T. Increase of Cinnamaldehyde Groups in Lignin of Transgenic Tobacco Plants Carrying an Antisense Gene for Cinnamyl Alcohol Dehydrogenase. Biosci Biotech Biochem 1995, 59, 929. [Google Scholar] [CrossRef][Green Version]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Anil, K.; Das, S.N.; Podile, A.R. Induced defense in plants: A short overview. Proc. Natl. Acad. Sci. USA India Sect. B Biol. Sci. 2014, 84, 669–679. [Google Scholar] [CrossRef]
- Abdul Malik, N.A.; Kumar, I.; Nadarajah, S. Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef] [PubMed]
- Boureau, T.; Siamer, S.; Perino, C.; Gaubert, S.; Patrit, O.; Degrave, A.; Fagard, M.; Chevreau, E.; Barny, M.-A. The HrpN effector of erwinia amylovora, Which is involved in Type III translocation, contributes directly or indirectly to callose elicitation on apple leaves. Mol. Plant Microbe Inter. 2011, 24, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Boureau, T.; ElMaarouf-Bouteau, H.; Garnier, A.; Brisset, M.-N.; Perino, C.; Pucheu, I.; Barny, M.-A. DspA/E, a type iii effector essential for erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol. Plant Microbe Inter. 2006, 19, 16–24. [Google Scholar] [CrossRef]
- Facchini, P.J.; Hagel, J.; Zulak, K.G. Hydroxycinnamic acid amide metabolism: Physiology and biochemistry. Can. J. Bot. 2002, 80, 577–589. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Thilmony, R.; Underwood, W.; He, S.Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen pseudomonas syringae pv. tomato DC3000 and the human pathogen escherichia coli O157:H7. Plant J. 2006, 46, 34–53. [Google Scholar] [CrossRef]
- Djamei, A.; Kahmann, R. Ustilago maydis: Dissecting the molecular interface between pathogen and plant. PLOS Pathog. 2012, 8, e1002955. [Google Scholar] [CrossRef]
- Jacobs, J.M.; Milling, A.; Mitra, R.M.; Hogan, C.S.; Ailloud, F.; Prior, P.; Allen, C. Ralstonia solanacearum requires pops, an ancient avre-family effector, for virulence and to overcome salicylic acid-mediated defenses during tomato pathogenesis. mBio 2013, 4, e00875-13. [Google Scholar] [CrossRef]
- Jelenska, J.; Yao, N.; Vinatzer, B.A.; Wright, C.M.; Brodsky, J.L.; Greenberg, J.T. AJ domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. CB 2007, 17, 499–508. [Google Scholar] [CrossRef]
- Zhou, H.; Lin, J.; Johnson, A.; Morgan, R.L.; Zhong, W.; Ma, W. Pseudomonas syringae type III effector hopz1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 2011, 9, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Venisse, J.-S.; Malnoy, M.; Faize, M.; Paulin, J.-P.; Brisset, M.-N. Modulation of Defense Responses of Malus spp. During Compatible and Incompatible Interactions with Erwinia amylovora. Mol. Plant Microbe Inter. 2002, 15, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Degrave, A.; Siamer, S.; Boureau, T.; Barny, M.-A. The AvrE superfamily: Ancestral type III effectors involved in suppression of pathogen-associated molecular pattern-triggered immunity. Mol. Plant Pathol. 2015, 16, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Asselin, J.A.E.; Lin, J.; Perez-Quintero, A.L.; Gentzel, I.; Majerczak, D.; Opiyo, S.O.; Zhao, W.; Paek, S.-M.; Kim, M.G.; Coplin, D.L.; et al. Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii. Plant Physiol. 2015, 167, 1117–1135. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- An, C.; Mou, Z. Salicylic Acid and its Function in Plant ImmunityF. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef]
- Fu, D.; Chen, L.; Yu, G.; Liu, Y.; Lou, Q.; Mei, H.; Xiong, L.; Li, M.; Xu, X.; Luo, L. QTL mapping of sheath blight resistance in a deep-water rice cultivar. Euphytica 2011, 180, 209–218. [Google Scholar] [CrossRef]
- Zhou, Y.-L.; Xie, X.-W.; Xu, M.-R.; Zang, J.-P.; Zhu, L.-H.; Xu, J.-L.; Li, Z.-K. Genetic Overlap in the Quantitative Resistance of Rice at the Seedling and Adult Stages to Xanthomonas oryzae pv. oryzae. J. Plant Biol. 2012, 55, 102–113. [Google Scholar] [CrossRef]
- Fukuoka, S.; Okuno, K. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor. Appl. Genet. 2001, 103, 185–190. [Google Scholar] [CrossRef]
- Tonnessen, B.W.; Manosalva, P.; Lang, J.M.; Baraoidan, M.; Bordeos, A.; Mauleon, R.; Oard, J.; Hulbert, S.; Leung, H.; Leach, J.E. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015, 87, 273–286. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Xiao, J.; Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: Application in study of rice-bacterium interaction. Plant Methods 2012, 8, 2. [Google Scholar] [CrossRef]
- Shen, X.; Yuan, B.; Liu, H.; Li, X.; Xu, C.; Wang, S. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 2010, 64, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic wrky genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Chuma, I.; Isobe, C.; Hotta, Y.; Ibaragi, K.; Futamata, N.; Kusaba, M.; Yoshida, K.; Terauchi, R.; Fujita, Y.; Nakayashiki, H.; et al. Multiple translocation of the avr-pita effector gene among chromosomes of the rice blast fungus magnaporthe oryzae and related species. PLOS Pathog. 2011, 7, e1002147. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Shen, X.; Li, X.; Xu, C.; Wang, S. Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 2007, 226, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Ke, Y.; Deng, H.; Wang, S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017, 90, 738–748. [Google Scholar] [CrossRef]
- Brown, J.K.M. Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 2002, 5, 339–344. [Google Scholar] [CrossRef]
- Xiao, S.; Calis, O.; Patrick, E.; Zhang, G.; Charoenwattana, P.; Muskett, P. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005, 42, 95–110. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Crop | Pathogen Tested | Expression | Immune Response | References |
|---|---|---|---|---|---|
| PAL | Linum uslerotiorum | Multiple pathogens | [26] | ||
| Triticum aestivum | Powdery mildew | Suppression | S | [60] | |
| Nicotiana tabacum | Cercosporanicotianae | Overexpression | R | [61] | |
| Arabidopsis thaliana | Pseudomonas syringae | S | [62,63] | ||
| Brachypodium | Magnaporthe Fusarium cuimorum | Knockdown | S | [63] | |
| Glycine max | Pseudomonas syringae | Silencing | S | [64] | |
| Capsicum annum | Xanthomonas | Suppression | S | [65] | |
| C4H | Glycine max | Phytophthora sojae | Overexpression | R | [66] |
| 4CL | Oryza sativa | Rice blast | S | [67] | |
| HCT | Arabidopsis thaliana | Colletotrichum trifolli | [68] | ||
| Medicago sativa | Multiple pathogens | [69] | |||
| CCR | Camelina sativa | Sclerotinia sclerotiorum | [70] | ||
| CCoAOMT | Triticum aestivum | Powdery mildew | [60] | ||
| Zea mays | Multiple pathogens | R | [71] | ||
| COMT | Triticum aestivum | Rhizoctonia cerealis | Silences | S | [72] |
| Triticum aestivum | Powdery mildew | [60] | |||
| Arabidopsis thaliana | X. campestris, P. syringae, Hyaloperonospora | R | [73] | ||
| Alternaria brassicicola B. cinerea, Blumeria graminis, | S | [74] | |||
| Nicotiana tabacum | Agrobacterium tumefaciens | Antisense | R | [75] | |
| Sorghum bicolor | F. thapsinum, F. proliferatum, | R | [76] | ||
| CAD | Triticum aestivum | Powdery mildew | S | [49] | |
| Linum usitatissimum | F. oxysporum | RNAi | [77] | ||
| Arabidopsis | Pseudomonas syringae | S | [78] | ||
| Sorghum bicolor | Alternaria alternate F. verticillioides, F. proliferatum, Fusarium thapsinum, | R | [76] | ||
| F5H | Arabidopsis thaliana | Verticillium longisporum Sclerotinia sclerotiorum | S S | [79,80] |
| Class | TFs | Gene Pathway/Enzyme Gene | Crop | Reference |
|---|---|---|---|---|
| MYB | AtMYB4 | Sinapate esters | Arabidopsis thaliana | [93,98,99] |
| AtMYB32 | Reduction of lignin, CoMT | Arabidopsis thaliana | [100] | |
| AtMYB15 | Lignification | Arabidopsis thaliana | [101] | |
| AmMYB330 | Lignin and increased G/S ratio 4Cl, CAD | Antirrhinum majus | [96] | |
| PvMYB4 | Reduced lignin, PAL CCoAOMT | Panicum virgatum L. | [102] | |
| ZmMYB8, ZmMYB11, ZmMYB31, ZmMYB42 | Reduced lignin, COMT, PAL and 4CL | Zea mays/Arabidopsis thaliana | [103,104,105] | |
| PtMYB14 | 4CL | Pinus taeda | [106,107] | |
| CsMYB4a | Reduced lignin content And phenylalanine PAL, CCoAMT, 4CL, COMT | Camellia sinensis/Nicotiana tabacum | [108] | |
| SmMYB39 | Reduced 4-coumaric acid, PAL. 4CL | Salvia miltiorrhiza | [109] | |
| LlMYB1 | Reduced lignin PAL, 4CL | Leucaena leucocephala | [110] | |
| TaMYB4 | Reduced lignin CCD, CCR | Triticum aestivum L. | [111] | |
| AtMYB052/AtMYB054/ AtMYB069 | Cell wall thickening | Arabidopsis thaliana | [112] | |
| EjMYB1 | Activate lignin biosynthetic genes | Eriobotrya japonica | [113] | |
| PtoMYB216 | Lignin biosynthetic pathway | Populus tomentosa | [114] | |
| PtoMYB156 | Repress phenylpropanoid biosynthesis | Populus tomentosa | [115] | |
| WRKY | PtrWRKY19 | Negatively regulate pith SCW | Populus trichocarpa | [116] |
| StWRKY8 | Phenylpropanoid metabolites | Solanum tuberosum | [117] | |
| VvWRKY2 | Affect S/G ratio. | Vitis vinifera L | [118] | |
| HD-Zip | popREVOLUTA (PRE) | Secondary vascular tissues | Populus trichocarpa | [119] |
| POPCORONA (PCN) | SCW lignification | Populus tremula × alba | [120] | |
| EcHB1 | Lignin and hemicellulose content | Eucalyptus camaldulensis | [121] | |
| SWN | PtrSND1-A2 (PtrWND1B) | Cell wall thickening | Populus trichocarpa | [122,123] |
| PtoVNS11 | Regulate lignin deposition | Populus tomentosa | [124] | |
| ERF | PsnSHN2 | Negatively regulate lignin biosynthesis | Populus simonii × nigra | [67] |
| Plant | Plant Tissue | Disease | Coumarins | Reference |
|---|---|---|---|---|
| Hevea brasiliensi | Leaves | Phytophthora palmivora | Scopoletin | [146] |
| Pisum sativum | Leaves | Uromyces pisi | Scopoletin | [147] |
| Brassica oleracea | Leaves | Xanthomonas campestris | Coumarin | [148] |
| Solanum lycopersicum | Leaves | Tomato yellow leaf curl virus | Scopoletin | [149] |
| Nicotiana tabacum | Leaves, roots | Botrytis cinereal | Scopoletin, Esculin, Fraxetin | [150,151,152] |
| Arabidopsis thaliana | Leaves, roots | Paeni bacilluspolymyxa Pseudomonas fluorescens Pythium sylvaticum | Scopoletin, Esculin, Esculetin | [153] [100] [154] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens 2020, 9, 312. https://doi.org/10.3390/pathogens9040312
Yadav V, Wang Z, Wei C, Amo A, Ahmed B, Yang X, Zhang X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens. 2020; 9(4):312. https://doi.org/10.3390/pathogens9040312
Chicago/Turabian StyleYadav, Vivek, Zhongyuan Wang, Chunhua Wei, Aduragbemi Amo, Bilal Ahmed, Xiaozhen Yang, and Xian Zhang. 2020. "Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense" Pathogens 9, no. 4: 312. https://doi.org/10.3390/pathogens9040312
APA StyleYadav, V., Wang, Z., Wei, C., Amo, A., Ahmed, B., Yang, X., & Zhang, X. (2020). Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens, 9(4), 312. https://doi.org/10.3390/pathogens9040312







