Characterization of Pathogenic and Nonpathogenic Fusarium oxysporum Isolates Associated with Commercial Tomato Crops in the Andean Region of Colombia
Abstract
1. Introduction
2. Results
2.1. Fungal Isolates from the Central Colombian Andean Region
2.2. Identification of F. oxysporum Isolates
2.2.1. Morphology
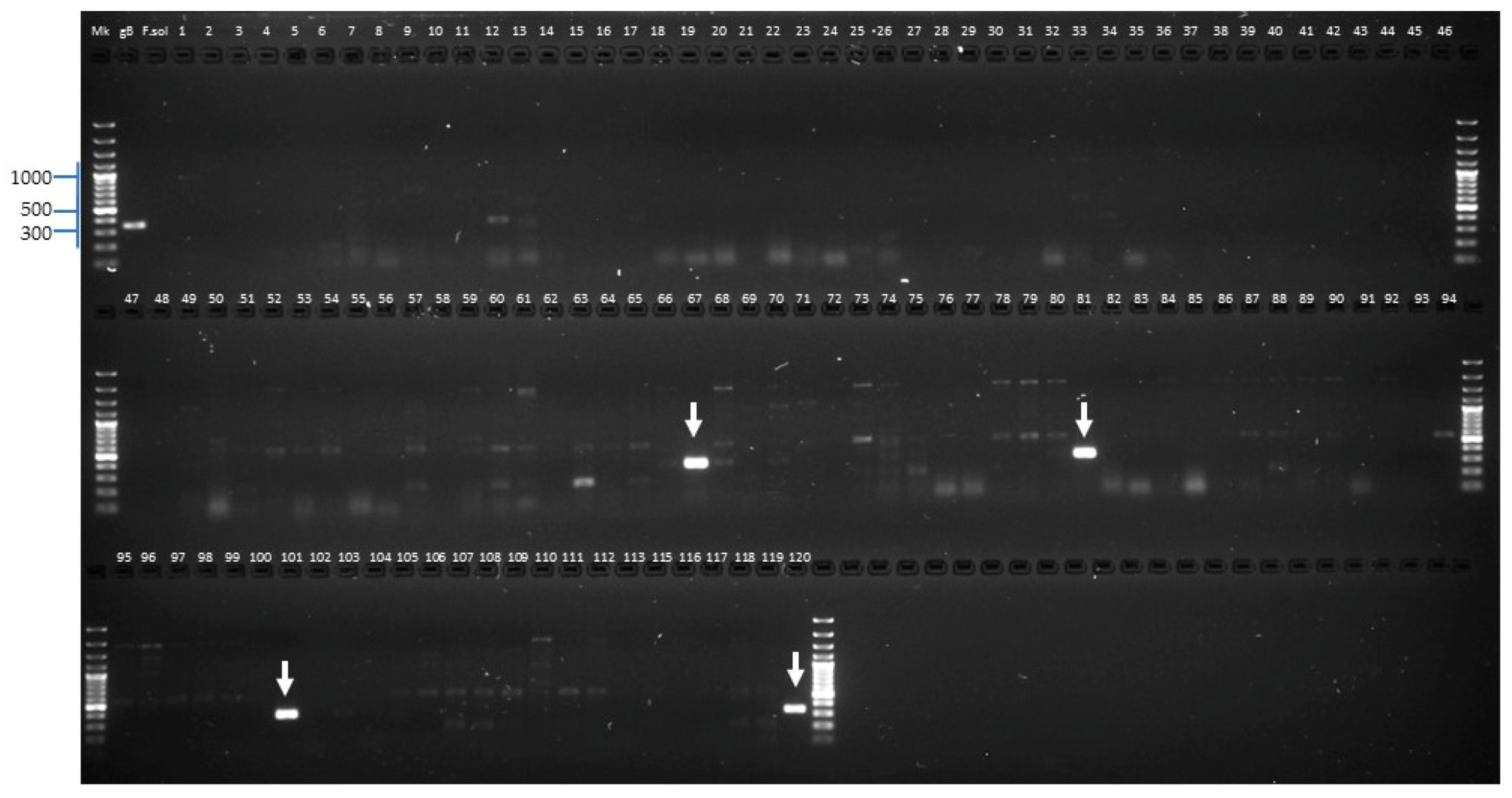
2.2.2. Molecular Identification of the Putative Fusarium Isolates
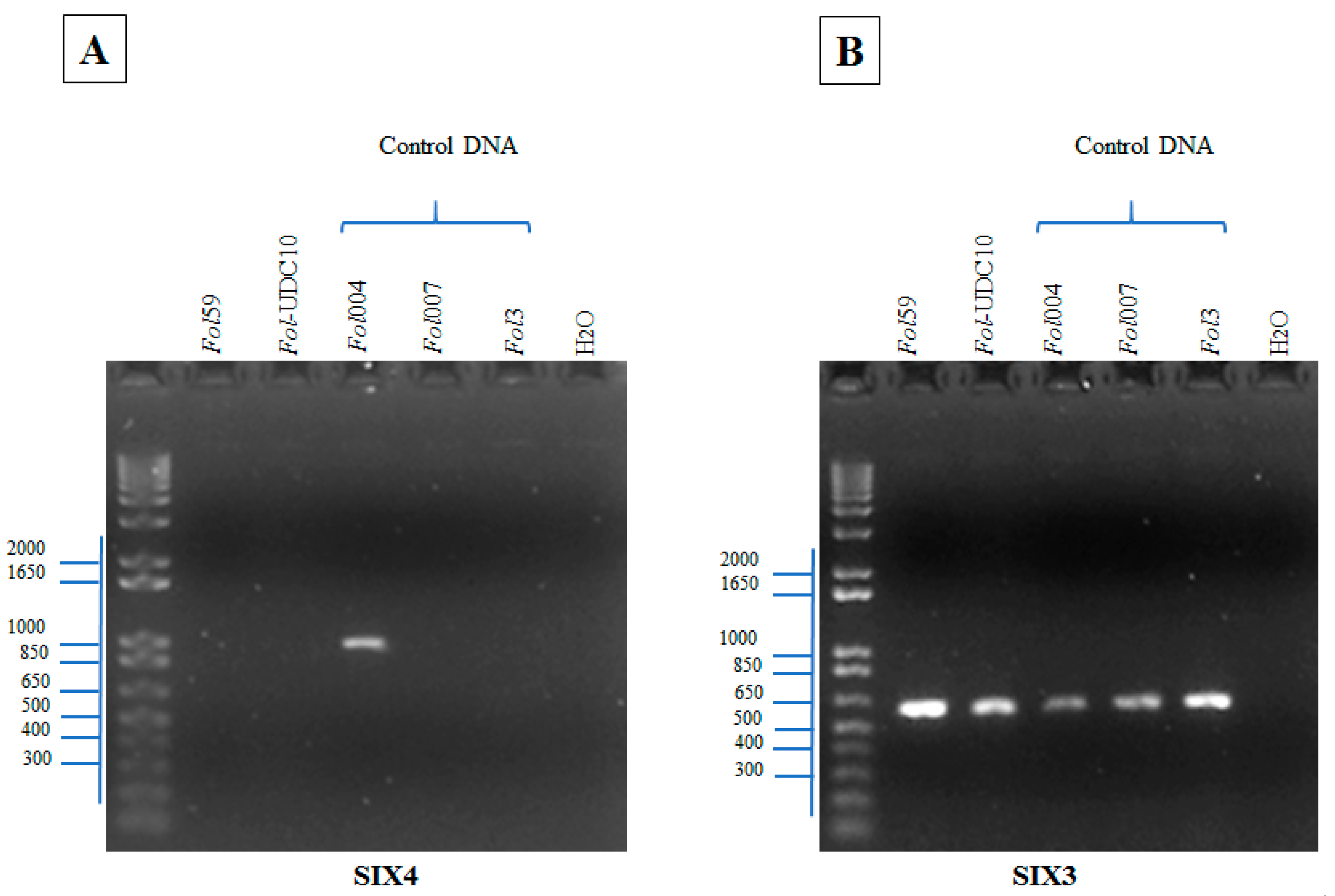
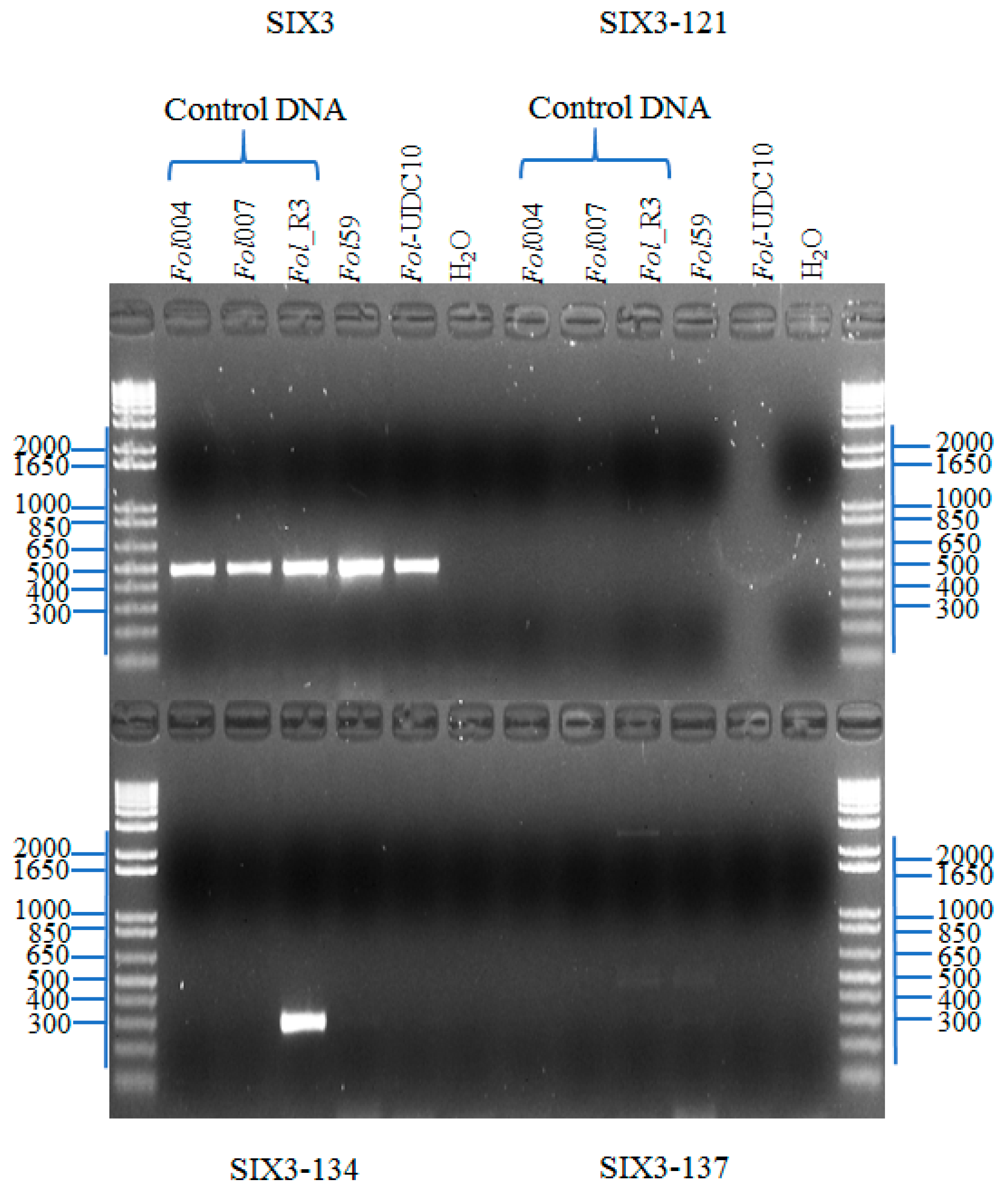
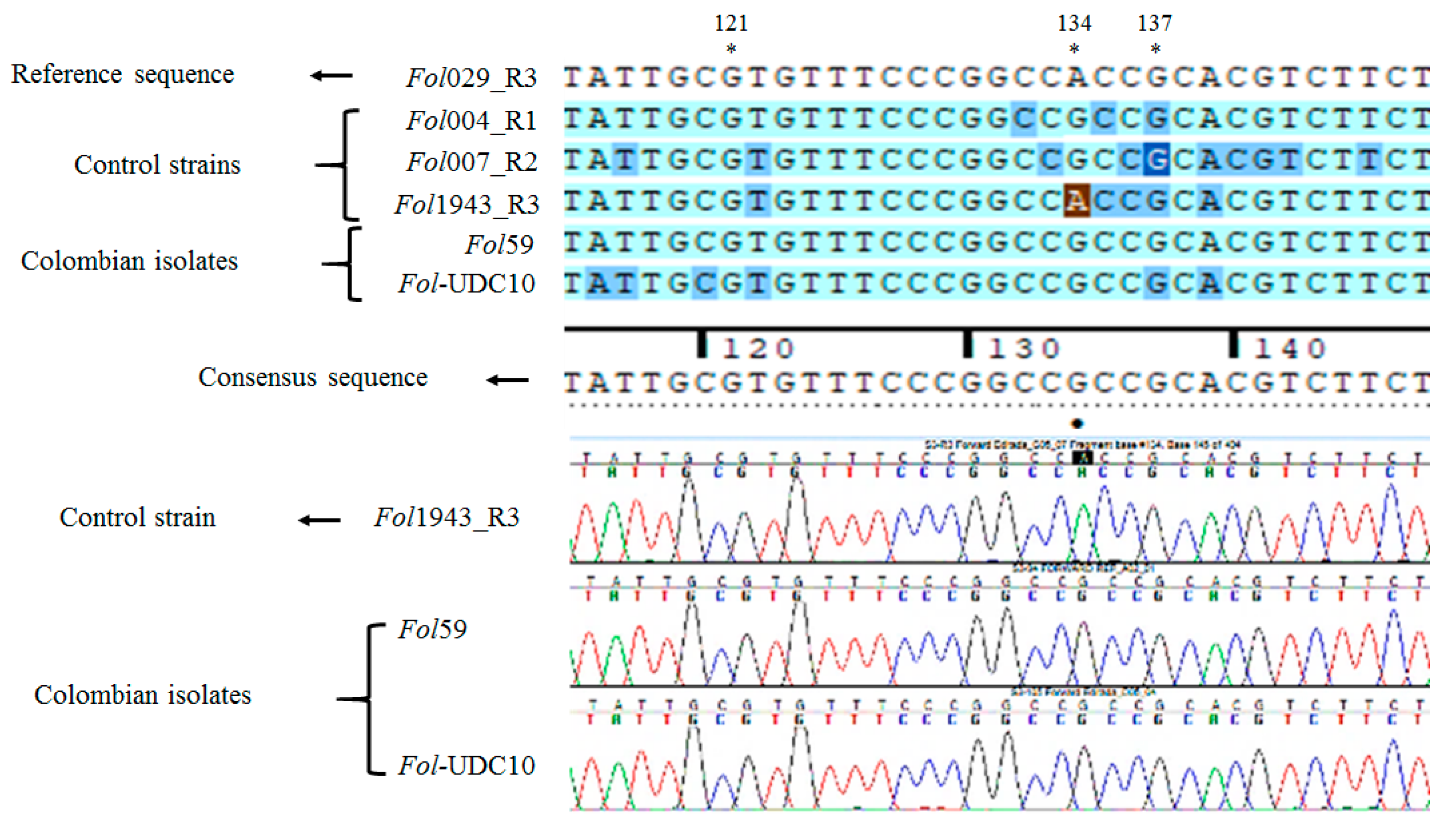
2.2.3. Identification of Fusarium oxysporum f. sp. lycopersici Candidate Isolates
2.2.4. Identification of Forl Candidates
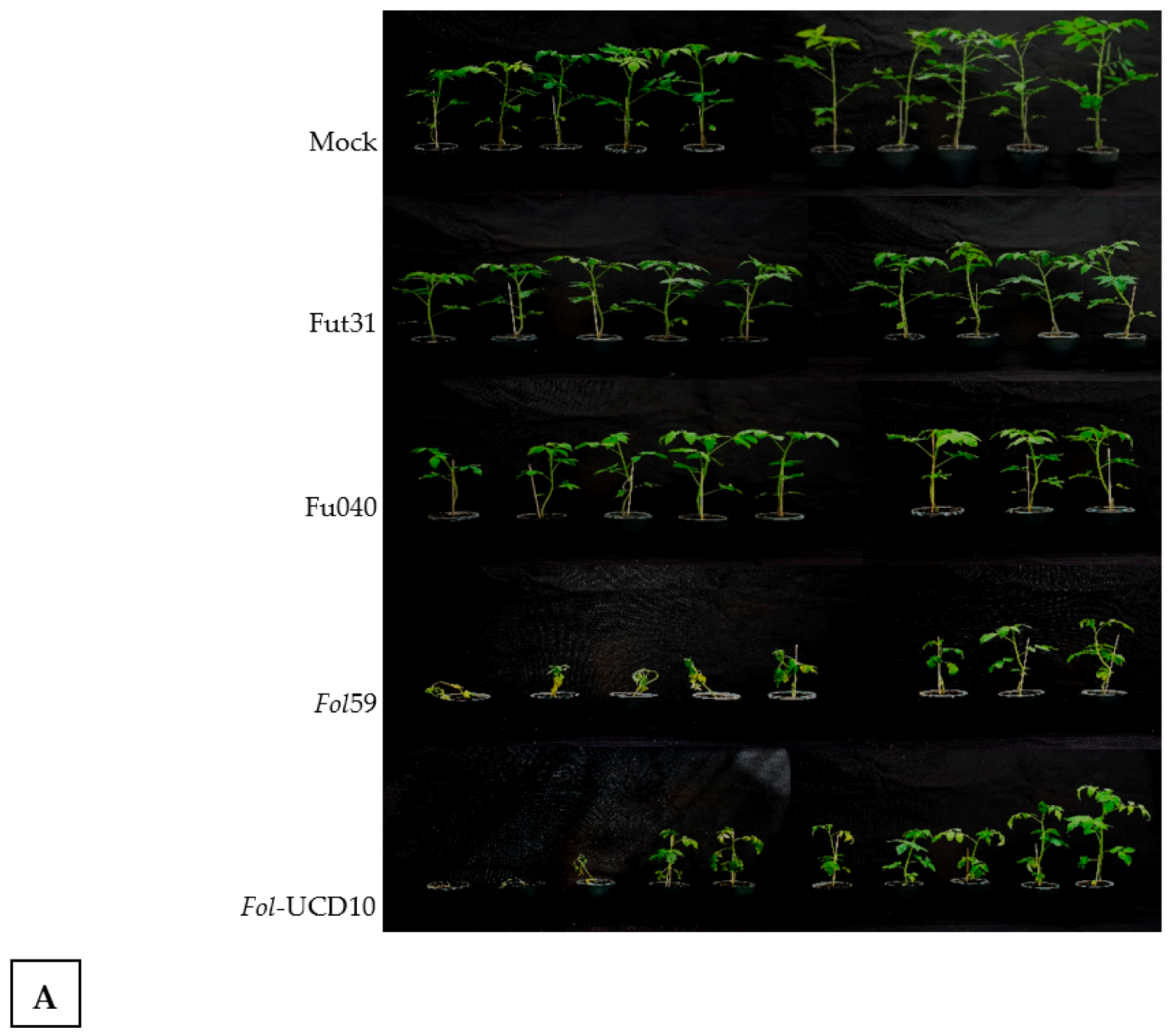
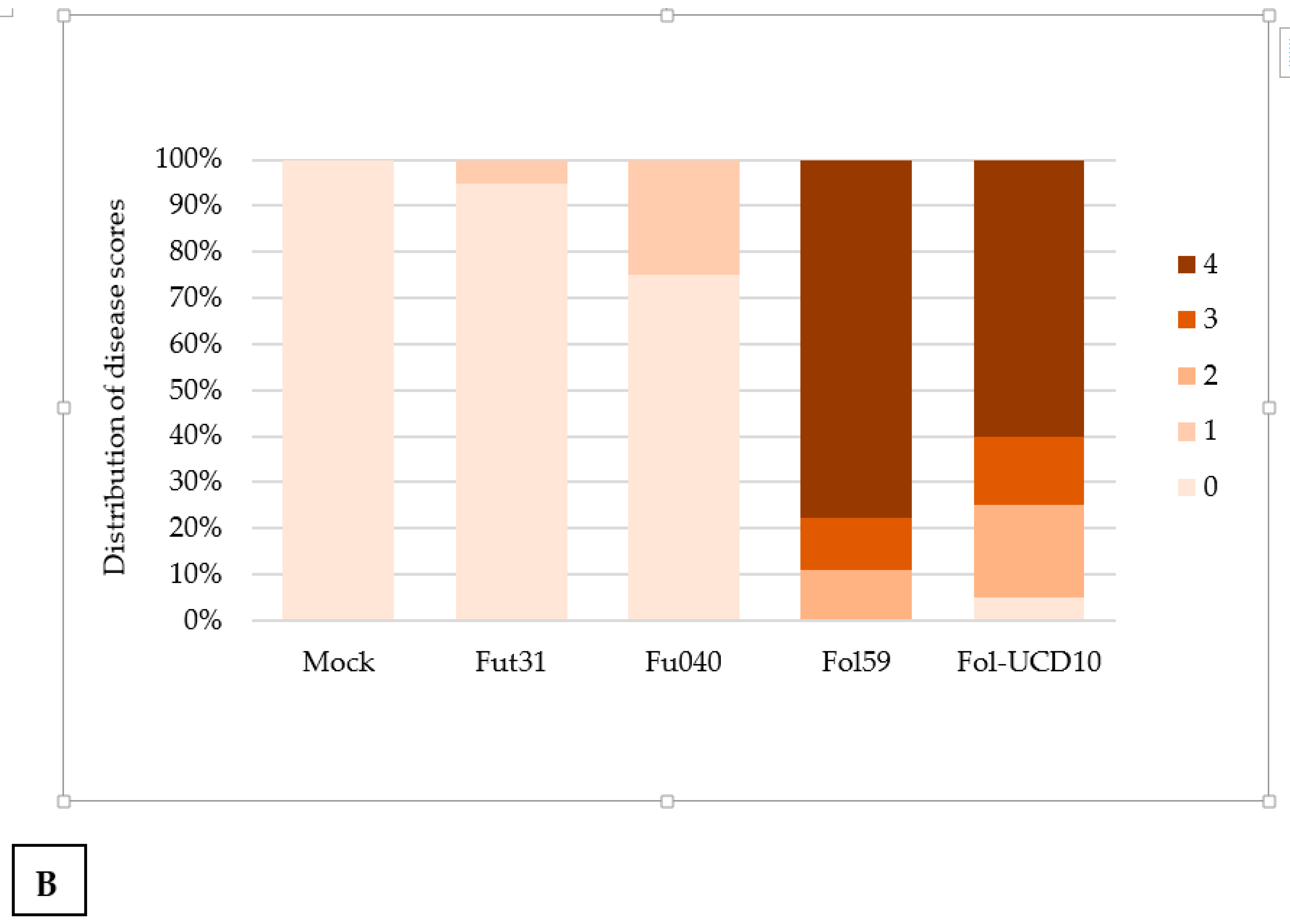
2.3. The Colombian Isolates Fol59 and Fol-UDC10 Were Highly Virulent on Susceptible Tomato Plants
2.4. Pathogenicity Assays with the Forl Candidate Isolates Showed Root Rot Symptoms in Tomato Plants
3. Discussion and Conclusions
4. Materials and Methods
4.1. Fusarium Isolates and Culture Conditions
4.2. Morphological Characterization
4.3. DNA Extraction and PCR Analysis
4.4. Sequencing Analysis of the EF1a Gene among the Fungal Isolates
4.5. Pathogenicity Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ma, L.-J.; Shea, T.; Young, S.; Zeng, Q.; Kistler, H.C. Genome sequence of Fusarium oxysporum f. sp. melonis strain NRRL 26406, a fungus Causing wilt disease on melon. Genome Announc. 2014, 2, e00730-14. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Gueidan, C.; Sink, S.; Johnston, P.R.; Crous, P.W.; Glenn, A.; Riley, R.; Zitomer, N.C.; Colyer, P.; Waalwijk, C.; et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009, 46, 936–948. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Di Pietro, A.; Madrid, M.P.; Caracuel, Z.; Delgado-Jarana, J.; Roncero, M.I.G. Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 2003, 4, 315–325. [Google Scholar] [CrossRef]
- Van Der Does, H.C.; Lievens, B.; Claes, L.; Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 2008, 10, 1475–1485. [Google Scholar] [CrossRef]
- FAOSTAT. Production—Crops—Area Harvested/Production Quantity—Tomatoes. Available online: www.fao.org/faostat/en (accessed on 1 September 2018).
- Menzies, J.G. Additions to the host range of Fusarium oxysporum f. sp. radicis-lycopersici. Plant Dis. 1990, 74, 569. [Google Scholar] [CrossRef]
- Lagopodi, A.L.; Ram, A.F.J.; Lamers, G.E.M.; Punt, P.J.; Van den Hondel, C.A.M.J.J.; Lugtenberg, B.J.J.; Bloemberg, G.V. Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant-Microbe Interact. 2002, 15, 172–179. [Google Scholar] [CrossRef]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Brayford, D. IMI descriptions of fungi and bacteria. Mycopathologia 1996, 133, 35–63. [Google Scholar] [CrossRef]
- Catanzariti, A.-M.; Lim, G.T.T.; Jones, D.A. The tomato I-3 gene: A novel gene for resistance to Fusarium wilt disease. New Phytol. 2015, 207, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Catanzariti, A.-M.; Do, H.T.T.; Bru, P.; de Sain, M.; Thatcher, L.F.; Rep, M.; Jones, D.A. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 2017, 89, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Cendales, Y.; Catanzariti, A.-M.; Baker, B.; Mcgrath, D.J.; Jones, D.A. Identification of I -7 expands the repertoire of genes for resistance to Fusarium wilt in tomato to three resistance gene classes. Mol. Plant Pathol. 2016, 17, 448–463. [Google Scholar] [CrossRef]
- Ori, N.; Eshed, Y.; Paran, I.; Presting, G.; Aviv, D.; Tanksley, S.; Zamir, D.; Fluhr, R. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 1997, 9, 521–532. [Google Scholar]
- Houterman, P.M.; Speijer, D.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C.; Rep, M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007, 8, 215–221. [Google Scholar] [CrossRef]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, 1–6. [Google Scholar] [CrossRef]
- Ma, L.; Houterman, P.M.; Gawehns, F.; Cao, L.; Sillo, F.; Richter, H.; Clavijo-Ortiz, M.J.; Schmidt, S.M.; Boeren, S.; Vervoort, J.; et al. The AVR2-SIX5 gene pair is required to activate I-2 -mediated immunity in tomato. New Phytol. 2015, 208, 507–518. [Google Scholar] [CrossRef]
- Carrillo-Fasio, J.A.; Rodríguez, T.D.J.M.; Estrada, I.M.; Ortega, J.E.C.; Zequera, I.M.; Barajas, A.J.S. Razas de Fusarium oxysporum f. sp lycopersici Snyder y Hansen, en tomate (Lycopersicon esculentum Mill.) en el Valle de Culiacán, Sinaloa, México. Rev. Mex. Fitopatol. 2003, 21, 123–127. [Google Scholar]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar]
- Buriticá, P. Las Enfermedades de las Plantas y su Ciencia en Colombia; Universidad Nacional de Colombia: Medellín, Colombia, 1999; Available online: http://bdigital.unal.edu.co/9340/1/17081124.%201999%20Parte1.pdf (accessed on 17 December 2019).
- Garcia-Bastidas, F.; Quintero-Vargas, C.; Ayala-Vasquez, M.; Seidl, M.; Schermer, T.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Sørensen, A.; et al. First report of Fusarium wilt tropical race 4 in Cavendish bananas caused by Fusarium odoratissimum in Colombia. Plant Dis. 2019, 1–5. [Google Scholar] [CrossRef]
- Arbeláez, G.; de Granada, E.G.; de Amézquita, M.O.; Calderón, O.L. Respuesta de algunas variedades de clavel estándar a cuatro razas fisiológicas de Fusarium oxysporum f. sp. Dianthi. Agron. Colomb. 1996, 13, 117–127. [Google Scholar]
- Forero-Reyes, C.M.; Alvarado-Fernández, A.M.; Ceballos-Rojas, A.M.; González-Carmona, L.C.; Linares-Linares, M.Y.; Castañeda-Salazar, R.; Pulido-Villamarín, A.; Góngora-Medina, M.E.; Cortés-Vecino, J.A.; Rodríguez-Bocanegra, M.X. Evaluación de la capacidad patogénica de Fusarium spp. en modelos vegetal y murino. Rev. Argent. Microbiol. 2018, 50, 90–96. [Google Scholar] [CrossRef] [PubMed]
- AGRONET, Red de Información y Comunicación del Sector Agropecuario Colombiano. Available online: http://www.agronet.gov.co/estadistica/Paginas/default.aspx (accessed on 14 May 2018).
- Marín-Serna, S.; González-Guzmán, J.; Castaño-Zapata, J.; Ceballos-Aguirre, N. Respuesta de quince introducciones de tomate tipo cereza (Solanum spp.) a la marchitez vascular (Fusarium oxysporum f. sp. lycopersici Snyder & Hansen). Agron. Colomb. 2015, 22, 48–59. [Google Scholar]
- Jaramillo, J.; Rodriguez, V.P.; Gil, L.F.; García, M.C.; Hio, J.C.; Quevedo, D.; Guzmán, M.; Sánchez, G.D.; Aguilar, P.; Pinzón, L.; et al. Tecnologías del Cultivo del Tomate Bajo Condiciones Protegidas, 1st ed.; Corpoica: Bogotá, Colombia, 2012; Volume 1, ISBN 978-958-740-120-2. [Google Scholar]
- Leslie, J.; Summerel, B. The Fusarium Laboratory Manual, 1st ed.; Blackwell: Ames, IA, USA, 2006; ISBN 978-0-8138-1919-8. [Google Scholar]
- Cobo-Díaz, J.F.; Baroncelli, R.; Le Floch, G.; Picot, A. A novel metabarcoding approach to investigate Fusarium species composition in soil and plant samples. FEMS Microbiol. Ecol. 2019, 95, 1–13. [Google Scholar] [CrossRef]
- Imazaki, I.; Kadota, I. Molecular phylogeny and diversity of Fusarium endophytes isolated from tomato stems. FEMS Microbiol. Ecol. 2015, 91, 1–16. [Google Scholar] [CrossRef]
- Hirano, Y.; Arie, T. PCR-based differentiation of Fusarium oxysporum ff. sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 2006, 72, 273–283. [Google Scholar] [CrossRef]
- Manikandan, R.; Harish, S.; Karthikeyan, G.; Raguchander, T. Comparative proteomic analysis of different isolates of Fusarium oxysporum f.sp. lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front. Microbiol. 2018, 9, 420. [Google Scholar] [CrossRef]
- van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.J.; Kistler, H.C.; Ma, L.-J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef]
- Ma, L.-J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.C.; Takken, F.L.W.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Carvajalino, J.; Alzate, D.; Ramirez, A.; Santa-Sepulveda, J.; Fajardo-Rojas, A.; Soto-Suárez, M. Evaluating late blight severity in potato crops using unmanned aerial vehicles and machine learning algorithms. Remote Sens. 2018, 10, 1513. [Google Scholar] [CrossRef]
- Vásquez Ramírez, L.M.; Castaño Zapata, J. Manejo integrado de la marchitez vascular del tomate [Fusarium oxysporum f. sp. lycopersici (sacc.) W.C. Snyder & H.N. Hansen]: Una revisión. Rev. U.D.C.A Act. Div. Cient. 2017, 20, 363–374. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species: An Illustrated Manual for Identification; Pennsylvania State University Press: University Park, PA, USA, 1983; ISBN 978-0-271-00349-8. [Google Scholar]
- Moreno, C.A.; Castillo, F.; González, A.; Bernal, D.; Jaimes, Y.; Chaparro, M.; González, C.; Rodriguez, F.; Restrepo, S.; Cotes, A.M. Biological and molecular characterization of the response of tomato plants treated with Trichoderma koningiopsis. Physiol. Mol. Plant Pathol. 2009, 74, 111–120. [Google Scholar] [CrossRef]
- Nirmaladevi, D.; Venkataramana, M.; Srivastava, R.K.; Uppalapati, S.R.; Gupta, V.K.; Yli-Mattila, T.; Clement Tsui, K.M.; Srinivas, C.; Niranjana, S.R.; Chandra, N.S. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Sci. Rep. 2016, 6, 21367. [Google Scholar] [CrossRef]
- Jelinski, N.A.; Broz, K.; Jonkers, W.; Ma, L.-J.; Kistler, H.C. Effector gene suites in some soil isolates of Fusarium oxysporum are not sufficient predictors of vascular wilt in tomato. Phytopathology 2017, 107, 842–851. [Google Scholar] [CrossRef]
- Debbi, A.; Boureghda, H.; Monte, E.; Hermosa, R. Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in Algeria and a biocontrol strategy with indigenous Trichoderma spp. Front. Microbiol. 2018, 9, 282. [Google Scholar] [CrossRef]
- Alabouvette, C.; Olivain, C.; Migheli, Q.; Steinberg, C. Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 2009, 184, 529–544. [Google Scholar] [CrossRef]
- Aimé, S.; Alabouvette, C.; Steinberg, C.; Olivain, C. The Endophytic strain Fusarium oxysporum Fo47: A good candidate for priming the defense responses in tomato roots. Mol. Plant-Microbe Interact. 2013, 26, 918–926. [Google Scholar]
- Griffith, G.W.; Shaw, D.S. Polymorphisms in Phytophthora infestans: Four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Appl. Environ. Microbiol. 1998, 64, 4007–4014. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Geneious Biologics. Available online: Geneious.com (accessed on 8 May 2018).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- MAFFT (Multiple Alignment Using Fast Fourier Transform). Available online: https://www.ebi.ac.uk/Tools/msa/mafft/ (accessed on 9 February 2018).
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- BEAST. Bayesian Evolutionary Analysis Sampling Trees. Available online: https://beast.community/ (accessed on 9 August 2018).
- Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 October 2018).
- Mes, J.J.; Weststeijn, E.A.; Herlaar, F.; Lambalk, J.J.M.; Wijbrandi, J.; Haring, M.A.; Cornelissen, B.J.C. Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 1999, 89, 156–160. [Google Scholar] [CrossRef]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef]
- Simbaqueba, J.; Catanzariti, A.-M.; González, C.; Jones, D.A. Evidence for horizontal gene transfer and separation of effector recognition from effector function revealed by analysis of effector genes shared between cape gooseberry- and tomato-infecting formae speciales of Fusarium oxysporum. Mol. Plant Pathol. 2018, 19, 2302–2318. [Google Scholar] [CrossRef]
- Çakır, B.; Gül, A.; Yolageldi, L.; Özaktan, H. Response to Fusarium oxysporum f.sp. radicis-lycopersici in tomato roots involves regulation of SA- and ET-responsive gene expressions. Eur. J. Plant Pathol. 2014, 139, 379–391. [Google Scholar]
- Akhter, A.; Hage-Ahmed, K.; Soja, G.; Steinkellner, S. Compost and biochar alter mycorrhization, tomato root exudation, and development of Fusarium oxysporum f. sp. lycopersici. Front. Plant Sci. 2015, 6, 529. [Google Scholar] [CrossRef]
- Rongai, D.; Pulcini, P.; Pesce, B.; Milano, F. Antifungal activity of pomegranate peel extract against Fusarium wilt of tomato. Eur. J. Plant Pathol. 2017, 147, 229–238. [Google Scholar] [CrossRef]
- Chiang, K.S.; Liu, H.I.; Tsai, J.W.; Tsai, J.R.; Bock, C.H. A discussion on disease severity index values. Part II: Using the disease severity index for null hypothesis testing. Ann. Appl. Biol. 2017, 171, 490–505. [Google Scholar] [CrossRef]
- Rep, M.; Meijer, M.; Houterman, P.M.; van der Does, H.C.; Cornelissen, B.J.C. Fusarium oxysporum evades I-3 -mediated resistance without altering the matching avirulence gene. Mol. Plant-Microbe Interact. 2005, 18, 15–23. [Google Scholar] [CrossRef] [PubMed]
- VassarStats: Website for Statistical Computation. Available online: http://vassarstats.net/ (accessed on 21 October 2018).
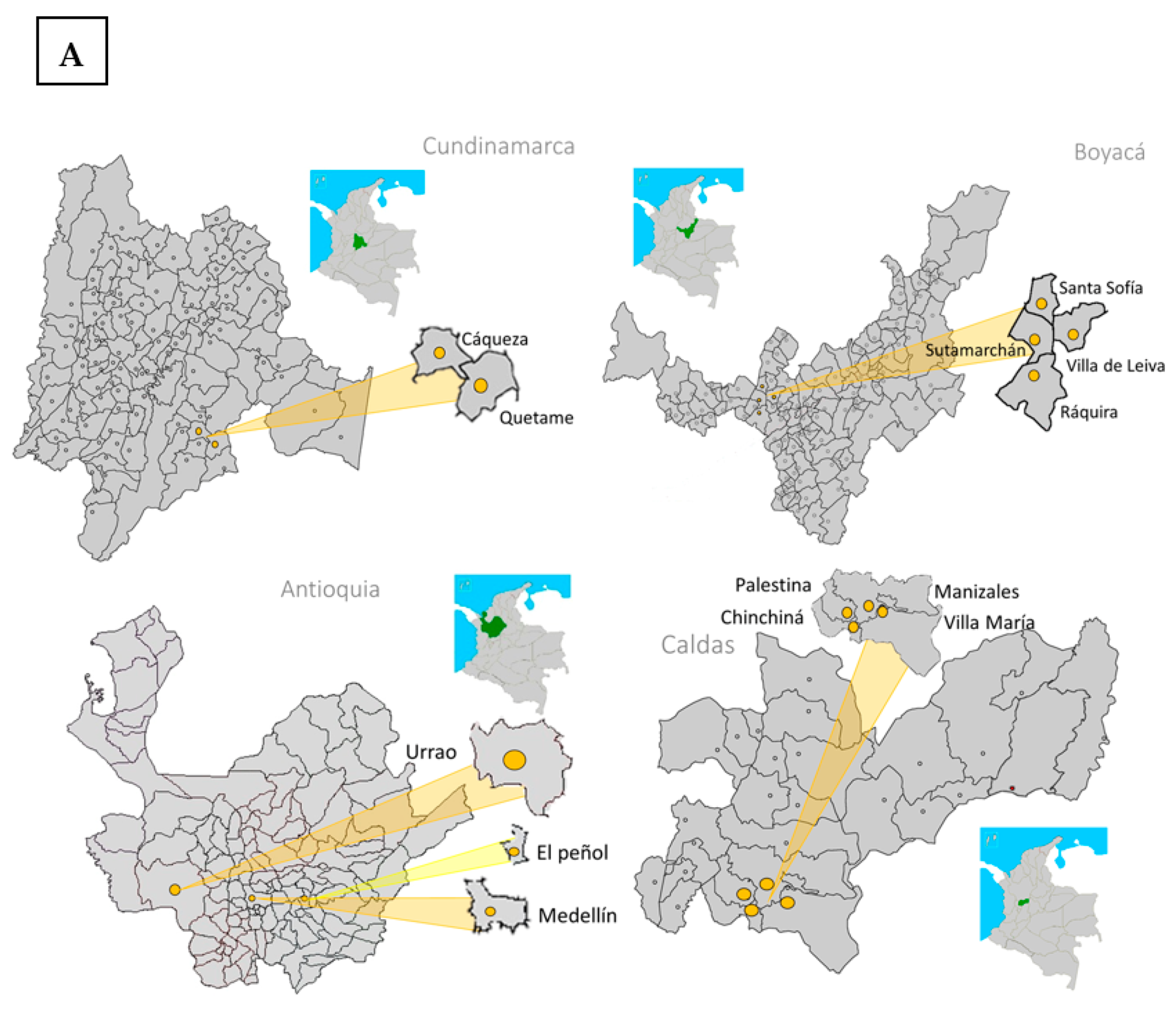



| Department | Field Location | Coordinates | Tomato Cultivar | Fol Resistance | Isolates and Tissue of Sample Collection | ||
|---|---|---|---|---|---|---|---|
| Town | Latitude | Longitude | Isolates from Stem | Isolates from Roots | |||
| Cundinamarca | Cáqueza | N.A. | N.A. | Libertador a | R1, R2 | 36_Cundinamarca | |
| Cundinamarca | Cáqueza | N.A. | N.A. | Nicolas b | R1, R2 | 37_Cundinamarca | |
| Cundinamarca | Cáqueza | 4°39′84.46″ | 73°94′90.32″ | Nicolas b | R1, R2 | 1_Cundinamarca | 38_Cundinamarca |
| Cundinamarca | Cáqueza | N.A. | N.A. | Libertador a | R1, R2 | 2_Cundinamarca | 39_Cundinamarca |
| Cundinamarca | Cáqueza | N.A. | N.A. | Libertador a | R1, R2 | 3_Cundinamarca | 40_Cundinamarca |
| Cundinamarca | Quetame | N.A. | N.A. | Nicolas b | R1, R2 | 4_Cundinamarca | 41_Cundinamarca |
| Cundinamarca | Quetame | N.A. | N.A. | Nicolas b | R1, R2 | 5_Cundinamarca | 42_Cundinamarca |
| Cundinamarca | Quetame | N.A. | N.A. | Nicolas b | R1, R2 | 6_Cundinamarca | 43_Cundinamarca |
| Cundinamarca | Quetame | N.A. | N.A. | Nicolas b | R1, R2 | 7_Cundinamarca | 44_Cundinamarca |
| Cundinamarca | Quetame | N.A. | N.A. | Nicolas b | R1, R2 | 8_Cundinamarca | 45_Cundinamarca |
| Boyacá | Villa de Leyva | N.A. | N.A. | Aslam a | R1, R2, R3 | 46_Boyacá | |
| Boyacá | Villa de Leyva | N.A. | N.A. | Aslam a | R1, R2, R4 | 9_Boyacá | 58_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Aslam a | R1, R2, R3 | 10_Boyacá | 47_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Libertador a | R1, R2 | 11_Boyacá | 48_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Roble a | R1, R2, R3 | 12_Boyacá | 49_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Roble a | R1, R2, R3 | 13_Boyacá | 50_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Roblea a | R1, R2, R3 | 14_Boyacá | 71_Boyacá |
| Boyacá | Villa de Leyva | N.A. | N.A. | Libertador a | R1, R2 | 117_Boyacá | |
| Boyacá | Sutamarchán | N.A. | N.A. | Libertador a | R1, R2 | 15_Boyacá | 51_Boyacá |
| Cundinamarca | Mosquera | 4°41′44″ | 74°12′12″ | Santa Clara a | N.D. | 16_Cundinamarca (Fu40) e | |
| Boyacá | Santa Sofía | 5°43′2.36″ | 73°36′7.38″ | Conquistador a | R1, R2 | 17_Boyacá | 52_Boyacá |
| Boyacá | Santa Sofía | 5°43′15.36″ | 73°35′28.9″ | Monterone a | R1, R2 | 18_Boyacá | 53_Boyacá |
| Boyacá | Santa Sofía | 5°43′10.57″ | 73°35′41.05″ | Nicolás b | R2 | 19_Boyacá | 54_Boyacá |
| Boyacá | Santa Sofía | 5°41′42.72″ | 73°35′19.73″ | Libertador a | R1, R2 | 20_Boyacá | 55_Boyacá |
| Boyacá | Santa Sofía | 5°41′42.72″ | 73°35′19.73″ | Aslam a | R1, R2 | 21_Boyacá | 56_Boyacá |
| Antioquia | El Peñol | 6°15′25.39″ | 75°16′28.18″ | Aslam a | R1, R2, R3 | 22_Antioquia | 57_Antioquia |
| Antioquia | El Peñol | 6°15′25.39″ | 75°16′28.18″ | Aslam a | R1, R2, R3 | 23_Antioquia | 59_Antioquia |
| Antioquia | El Peñol | N.A. | N.A. | Aslam a | R1, R2, R3 | 24_Antioquia | 72_Antioquia |
| Antioquia | El Peñol | 6°15′25.39″ | 75°16′28.18″ | Gem 604 a | R1, R2, R3 | 25_Antioquia (Fut31) | 60_Antioquia |
| Antioquia | El Peñol | 6°16′04.98″ | 75°14′54.27″ | Aslam a | R1, R2 | 26_Antioquia | 61_Antioquia |
| Antioquia | El Peñol | 6°16′04.98″ | 75°14′54.27″ | Aslam a | R1, R2, R3 | 27_Antioquia | 62_Antioquia |
| Antioquia | Urrao | 6°15′8.41″ | 75°14′48″ | Venanzio a | R1, R2, R3 | 28_Antioquia | 63_Antioquia |
| Antioquia | Urrao | 6°15′48.78″ | 76°07′39.20″ | Venanzio a | N.D. | 29_Antioquia | 64_Antioquia |
| Antioquia | Urrao | 6°15′48.78″ | 76°07′39.20″ | Torrano a | N.D. | 30_Antioquia | 65_Antioquia |
| Antioquia | Urrao | 6°16′41.74″ | 76°05′46.18″ | Torrano a | R1, R2 | 31_Antioquia | 66_Antioquia |
| Antioquia | Urrao | 6°16′41.74″ | 76°05′46.18″ | Torrano a | R1, R2 | 32_Antioquia | 67_Antioquia (Fur38) g |
| Antioquia | Urrao | 6°20′38.56″ | 76°05′30.03″ | Torrano a | R1, R2 | 33_Antioquia | 68_Antioquia |
| Antioquia | Urrao | 6°20′38.56″ | 76°05′30.03″ | Torrano a | R1, R2 | 34_Antioquia | 69_Antioquia |
| Antioquia | Urrao | 6°20′38.56″ | 76°05′30.03″ | Torrano a | R1, R2 | 35_Antioquia | 70_Antioquia |
| Antioquia | Urrao | 6°20′29.3″ | 76°07′62.1″ | Torrano a | R1, R2 | 118_Antioquia | |
| Antioquia | Urrao | 6°20′11.5″ | 76°08′30.50″ | Roble a | R1, R2, R3 | 119_Antioquia | |
| Antioquia | Medellín | 6°18′33.3″ | 75°67′54.54″ | Roble a | R1, R2, R3 | 120_Antioquia (Fut64) g | |
| Cundinamarca | Fómeque | 4°28.094′ | 73°31.868′ | Roble a | R1, R2, R3 | 73_Cundinamarca | 88_Cundinamarca |
| Cundinamarca | Fómeque | 4°28.094′ | 73°31.868′ | Roble a | R1, R2, R3 | 74_Cundinamarca | 89_Cundinamarca |
| Cundinamarca | Fómeque | 4°28.053′ | 33°51.980′ | Libertador a | R1, R2 | 90_Cundinamarca | |
| Cundinamarca | Fómeque | N.A. | N.A. | Nicolás b | R1, R2 | 75_Cundinamarca | 91_Cundinamarca |
| Cundinamarca | Fómeque | N.A. | N.A. | Nicolás b | R1, R2 | 76_Cundinamarca | 92_Cundinamarca |
| Antioquia | Rionegro | 6°9.735′ | 75°24.307′ | Tropical c | R1, R2 | 77_Antioquia | 93_Antioquia |
| Antioquia | Rionegro | 6°9.735′ | 75°24.307′ | Tropical c | R1, R2 | 78_Antioquia | 95_Antioquia |
| Antioquia | Rionegro | 6°9.735′ | 75°24.307′ | Tropical c | R1, R2 | 79_Antioquia | 96_Antioquia |
| Antioquia | Rionegro | 6°9.735′ | 75°24.307′ | Tropical c | R1, R2 | 80_Antioquia | 97_Antioquia |
| Antioquia | Rionegro | 6°9.735′ | 75°24.307′ | Tropical c | R1, R2 | 81_Antioquia (Fut52) g | 98_Antioquia |
| Antioquia | San Vicente | 6°16.222′ | 75°22.361′ | N.A. | N.D. | 82_Antioquia | 99_Antioquia |
| Antioquia | San Vicente | 6°16.222′ | 75°22.361′ | N.A. | N.D. | 83_Antioquia | 100_Antioquia |
| Antioquia | San Vicente | 6°16.222′ | 75°22.361′ | N.A. | N.D. | 84_Antioquia | 101_Antioquia (Fur55) g |
| Antioquia | San Vicente | 6° 16.222′ | 75°22.361′ | N.A. | N.D. | 85_Antioquia | 102_Antioquia |
| Antioquia | Guarne | N.A. | N.A. | N.A. | N.D. | 86_Antioquia | 103_Antioquia |
| Antioquia | Guarne | N.A. | N.A. | N.A. | N.D. | 87_Antioquia | 104_Antioquia |
| Caldas | Chinchiná | 4° 56′33″ | 75°37′10″ | Calima a | R1, R2 | 116_Antioquia | 94_Caldas (Fol59) f |
| Caldas | Manizales | 5°3′14″ | 75°29′30″ | N.A. | N.D. | 105_Caldas (Fol-UDC10) f | |
| Caldas | Manizales | N.A. | N.A. | N.A. | N.D. | 106_Caldas | |
| Caldas | Chinchiná | 4°59′7″ | 75°39′58″ | Armada Carguero d | R1, R2 | 107_Caldas | |
| Caldas | Chinchiná | 4°59′7″ | 75°39′58″ | Armada Carguero d | R1, R2 | 108_Caldas | |
| Caldas | Palestina | 4°56′29″ | 75°39′25″ | Roble a | R1, R2, R3 | 109_Caldas | |
| Caldas | Manizales | 5°5′55″ | 75°37′13″ | Venanzio a | N.D. | 110_Caldas | |
| Caldas | Villamaría | 4°57′297″ | 75°31.371′ | Roble a | R1, R2, R3 | 111_Caldas | |
| Caldas | Chinchiná | 05°00.447′ | 75°35.831′ | Calima a | R1, R2 | 112_Caldas | |
| Caldas | Villamaría | 4° 57′397′ | 75° 31.377′ | Roble a | R1, R2, R3 | 113_Caldas | |
| Caldas | Chinchiná | 04°56′ 34″ | 75°37′7″ | Calima a | R1, R2 | 114_Caldas | |
| Caldas | Palestina | 05°1′ 6″ | 75°36′40″ | Ciénaga a | R1, R2 | 115_Caldas | |
| Primer | Sequence | Region Amplified | Predicted Size |
|---|---|---|---|
| Uni F | ATCATCTTGTGCCAACTTCAG | Pg1a | 670 bp a |
| Uni R | GTTTGTGATCTTTGAGTTGCCA | ||
| EF1-F | ATGGGTAAGGAGGACAAGACTCA | EF1Ab | 776 bp b |
| EF1-R | TGGAGATACCAGCCTCGAAC | ||
| P12-F2B | GTATCCCTCCGGATTTTGAGC | SIX1c | 992 bp |
| P12-R1 | AATAGAGCCTGCAAAGCATG | ||
| SIX3-F1 | CCAGCCAGAAGGCCAGTTT | SIX3c | 608 bp c |
| SIX3-R2 | GGCAATTAACCACTCTGCC | ||
| G121A-F2 e | ACGGGGTAACCCATATTGCA | 429 bp d | |
| G134A-F2 e | TTGCGTGTTTCCCGGCCA | 414 bp d | |
| G137C-F1 e | GCGTGTTTCCCGGCCGCCC | 412 bp d | |
| SIX4-F1 | TCAGGCTTCACTTAGCATAC | SIX4c | 967 bp |
| SIX4-R1 | GCCGACCGAAAAACCCTAA | ||
| Forl_155.3F | GGTGAGGTTGCCACATTTCT | GCA_000260155.3 f | 337 bp |
| Forl_155.3R | TCTTTGTTCATTCCCCAAGC |
| GenBank Accession | Fusarium Species | Isolate/Code | Reference |
|---|---|---|---|
| KY365602.1 | avenaceum | UBOCC-A-109 096 | [30] |
| MG550947.1 | verticillioides | UBOCC-A-109 122 | |
| MH496628.1 | proliferatum | SL0018 | [31] |
| MK604519.1 | fujikuroi | SL0089 | |
| KY283870.1 | graminearum | 17a | GenBank |
| KX034250.1 | falciforme | RRK20 | [33] |
| MG857253.1 | solani | SL0002 | [31] |
| MG707130.1 | solani | SL0016 | |
| MH716809.1 | commune | SL0021 | |
| JF740825.1 | foetens | FOSC-21 | |
| AB917000.1 | Fusarium sp. | SL0584 | GenBank |
| AB817202.1 | Fusarium sp. | FOSC-1065 | |
| Fusarium oxysporum Species Complex | |||
| MALH01000103.1 | f. sp. lycopersici race 1 | Fol004 | [34] |
| MALV01000081.1 | f. sp. lycopersici race 2 | Fol4287 | |
| RBXW01000019.1 | f. sp. lycopersici race 3 | D11 | GenBank |
| FOCG_09093 | f. sp. radicis-lycopersici | 26381 | Ensembl fungi |
| FOZG_08502 | oxysporum | Fo047 | |
| JX258849.1 | oxysporum | Bra2 | GenBank |
| MK172058.1 | oxysporum | FR3 | |
| MK226334.1 | oxysporum | SL0001 | [31] |
| MK414772.1 | oxysporum | SL0035 | |
| MK799835.1 | oxysporum | SL0019 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmona, S.L.; Burbano-David, D.; Gómez, M.R.; Lopez, W.; Ceballos, N.; Castaño-Zapata, J.; Simbaqueba, J.; Soto-Suárez, M. Characterization of Pathogenic and Nonpathogenic Fusarium oxysporum Isolates Associated with Commercial Tomato Crops in the Andean Region of Colombia. Pathogens 2020, 9, 70. https://doi.org/10.3390/pathogens9010070
Carmona SL, Burbano-David D, Gómez MR, Lopez W, Ceballos N, Castaño-Zapata J, Simbaqueba J, Soto-Suárez M. Characterization of Pathogenic and Nonpathogenic Fusarium oxysporum Isolates Associated with Commercial Tomato Crops in the Andean Region of Colombia. Pathogens. 2020; 9(1):70. https://doi.org/10.3390/pathogens9010070
Chicago/Turabian StyleCarmona, Sandra L., Diana Burbano-David, Magda R. Gómez, Walter Lopez, Nelson Ceballos, Jairo Castaño-Zapata, Jaime Simbaqueba, and Mauricio Soto-Suárez. 2020. "Characterization of Pathogenic and Nonpathogenic Fusarium oxysporum Isolates Associated with Commercial Tomato Crops in the Andean Region of Colombia" Pathogens 9, no. 1: 70. https://doi.org/10.3390/pathogens9010070
APA StyleCarmona, S. L., Burbano-David, D., Gómez, M. R., Lopez, W., Ceballos, N., Castaño-Zapata, J., Simbaqueba, J., & Soto-Suárez, M. (2020). Characterization of Pathogenic and Nonpathogenic Fusarium oxysporum Isolates Associated with Commercial Tomato Crops in the Andean Region of Colombia. Pathogens, 9(1), 70. https://doi.org/10.3390/pathogens9010070







