Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Phylogenetic Analysis of MaWRKYs
2.3. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
2.4. Subcellular Localization Analysis
2.5. Dural Luciferase (LUC) Assay through Transient Expression
2.6. Chromatin Immunoprecipitation Quantitative Real-Time PCR (ChIP-qPCR)
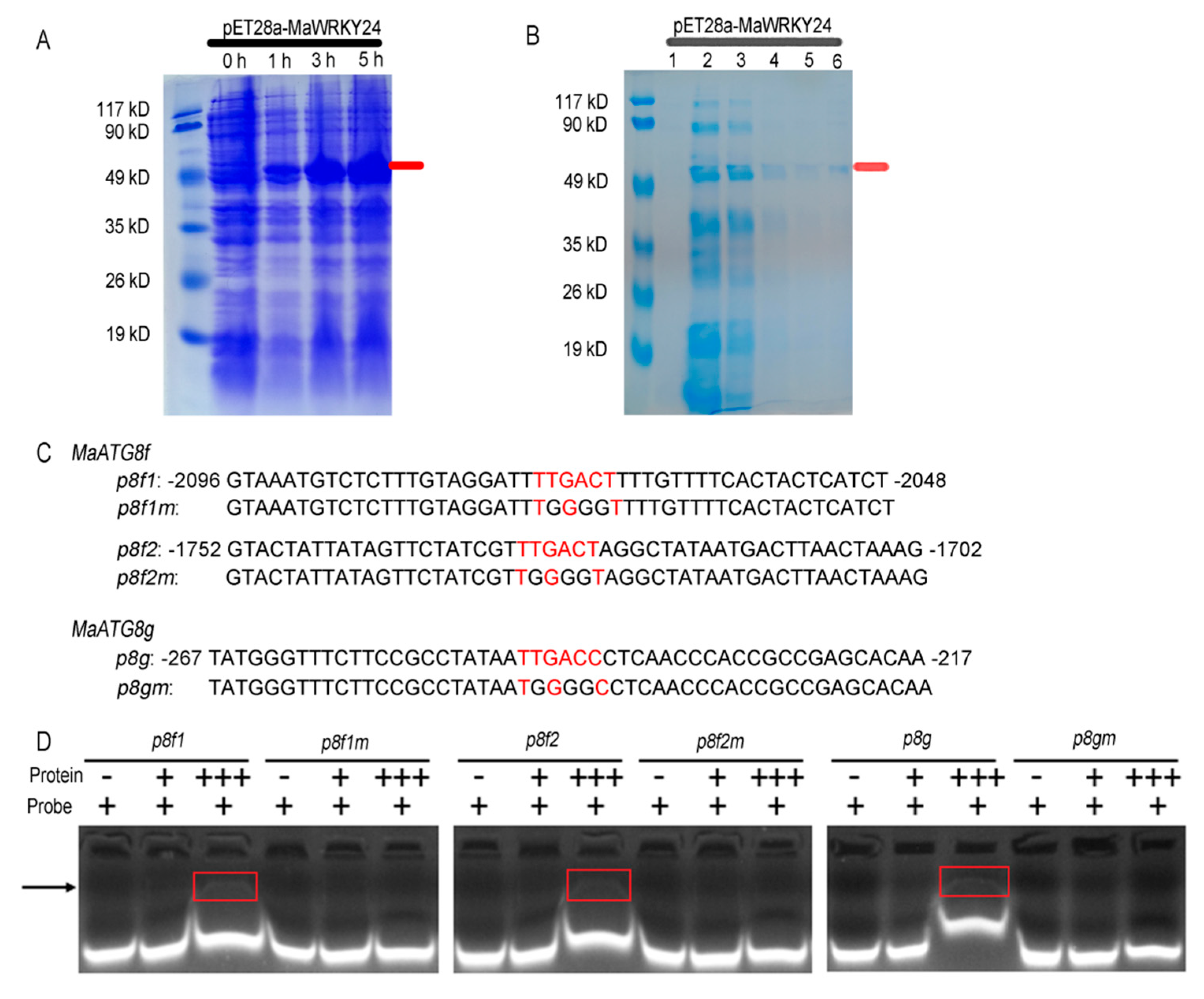
2.7. Electrophoretic Mobility Shift Assay (EMSA)
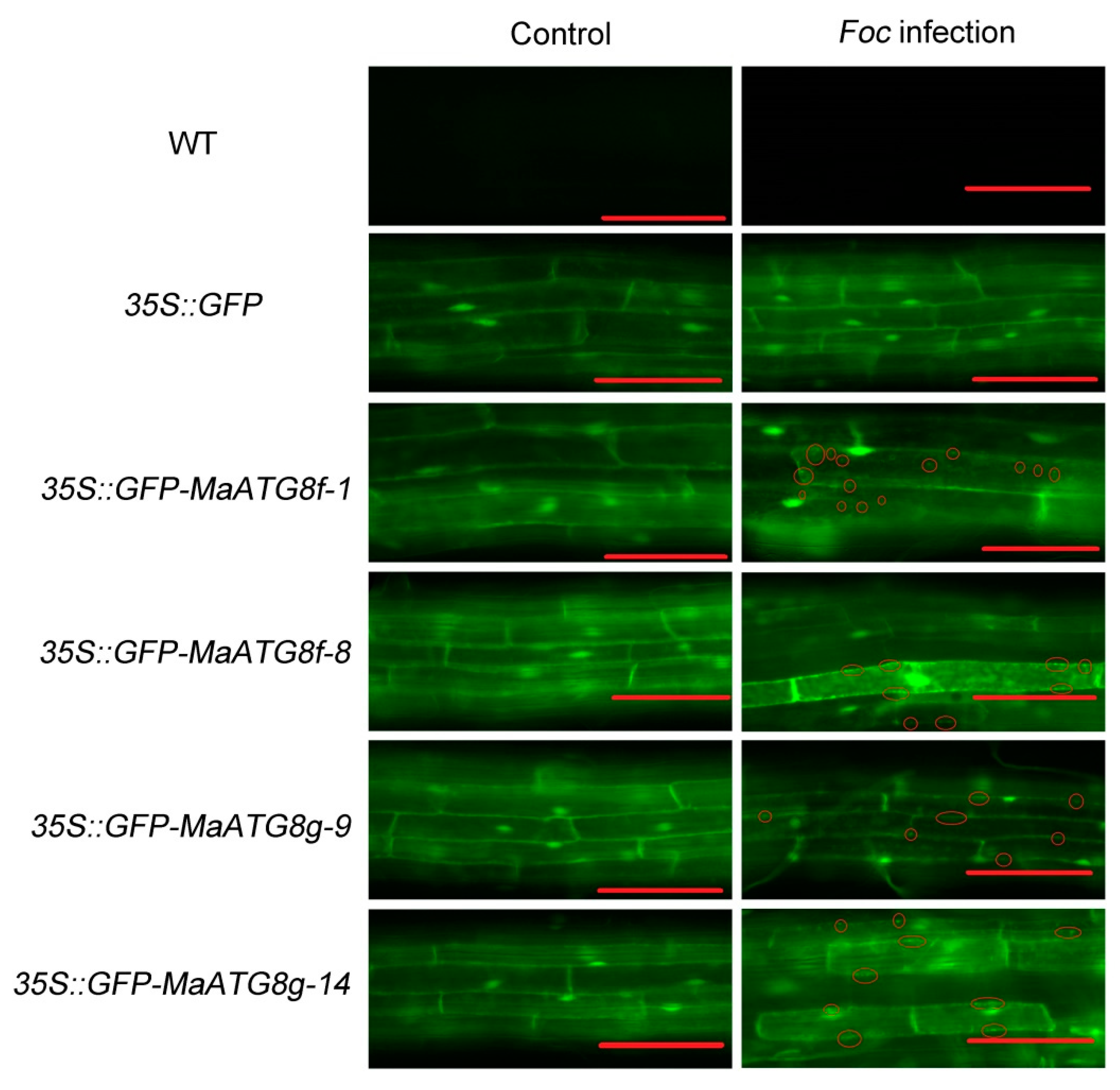
2.8. Generation of Transgenic Plants and Observation of the Autophagosome
2.9. Statistical Analysis
3. Results
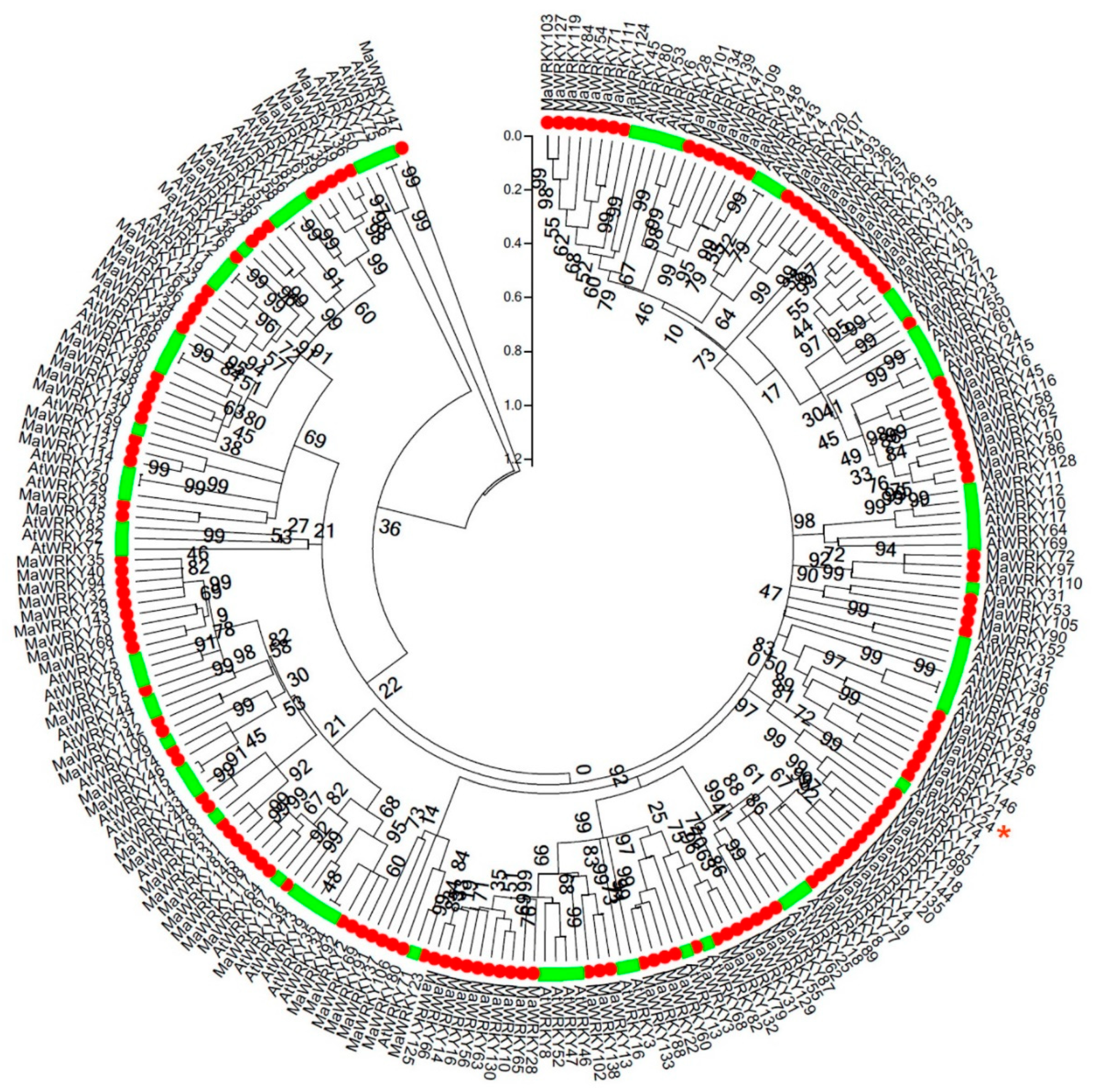
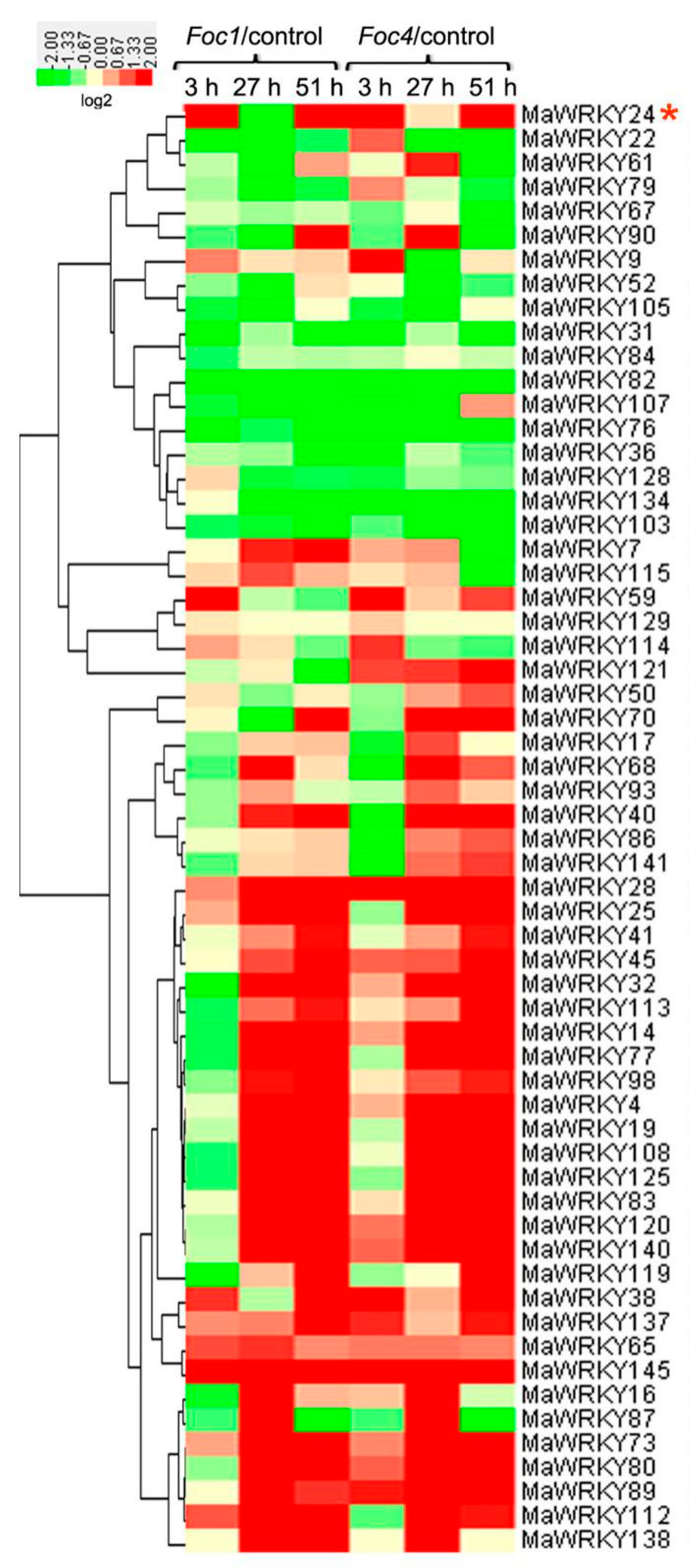
3.1. Evolutionary Analysis and Expression Profiles of MaWRKYs
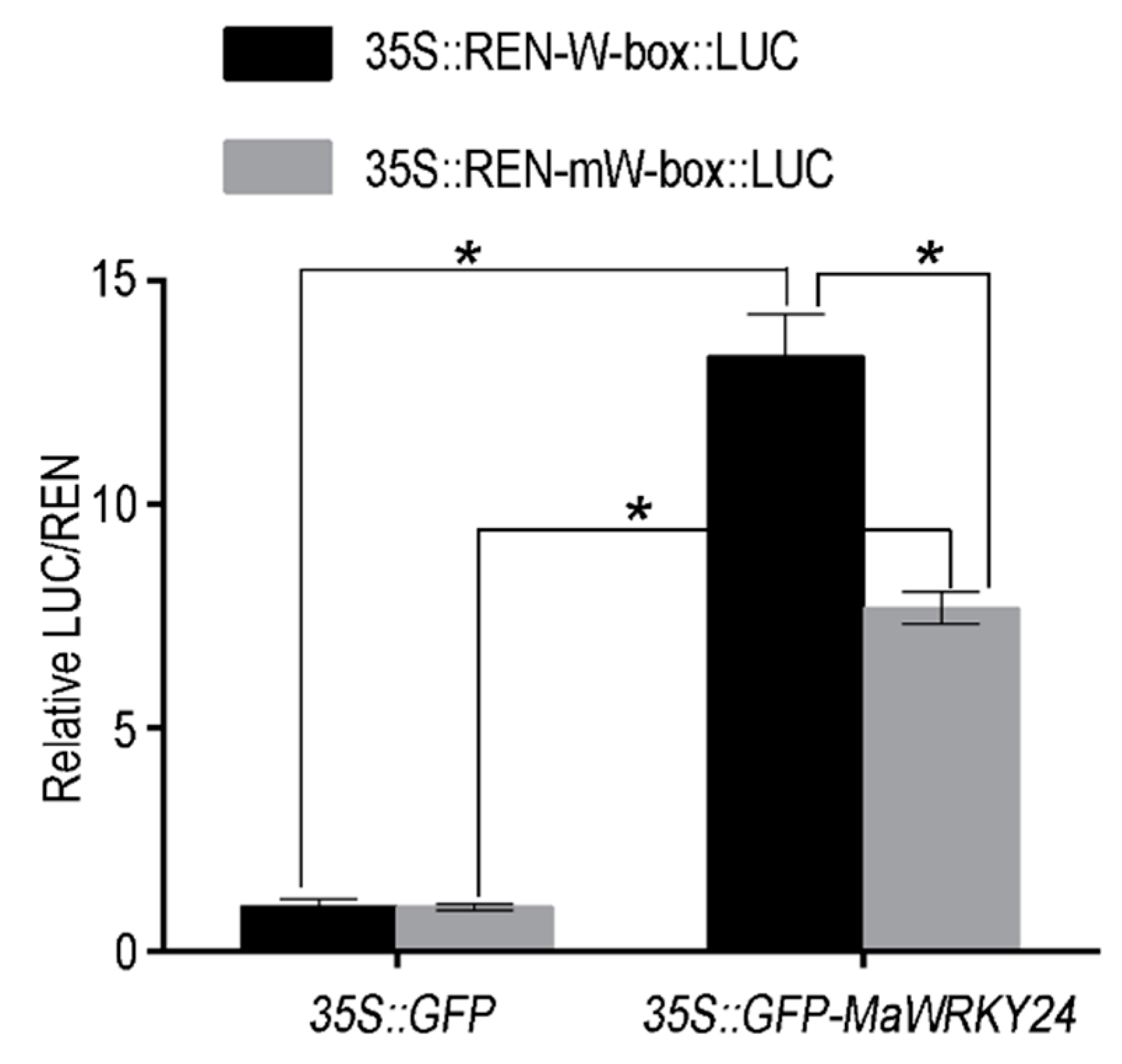
3.2. Subcellular Localization and Transcriptional Activated Activity of MaWRKY24
3.3. Overexpression of MaWRKY24 Regulated Expression Level of MaATG8s
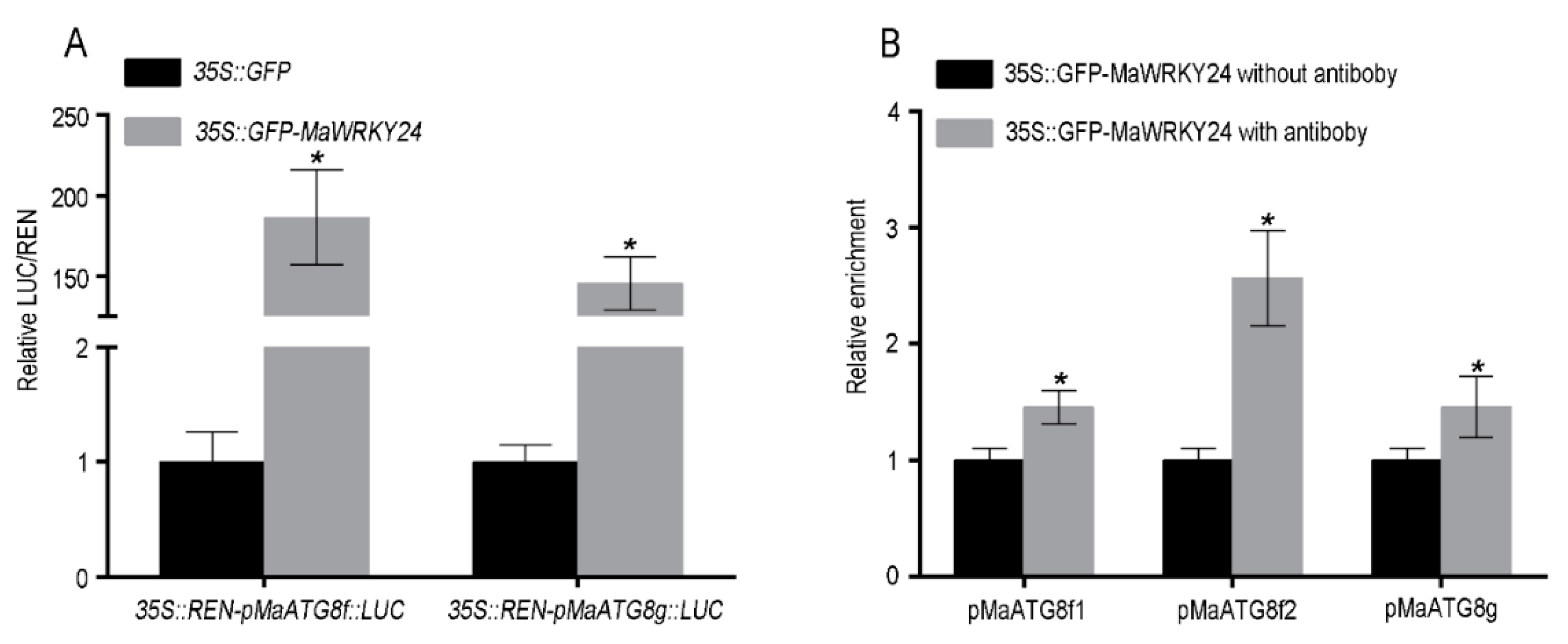
3.4. MaWRKY24 were Transcriptional Activators of MaATG8f/g
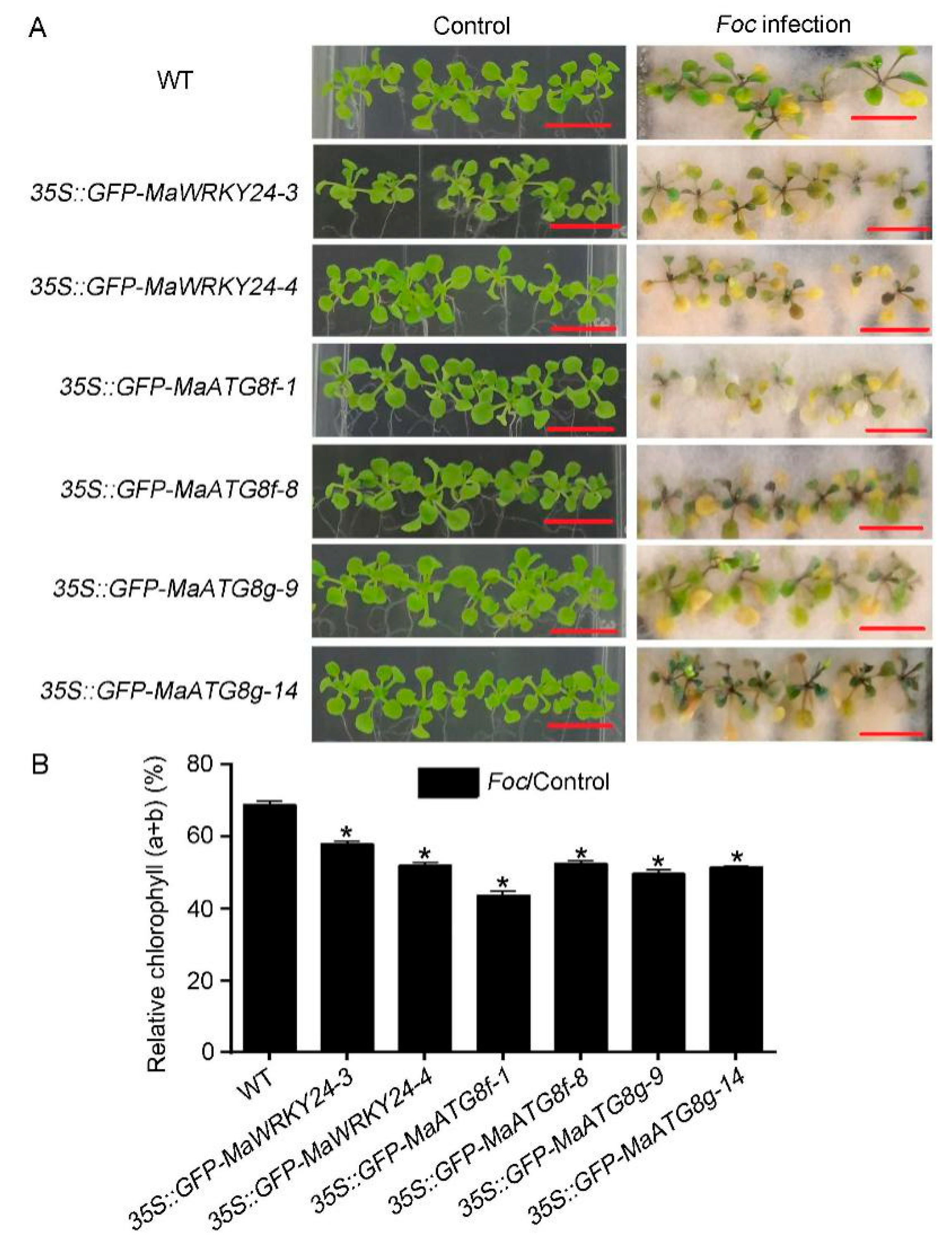
3.5. Overexpression of MaWRKY24 and MaATG8f/g Negatively Regulate Plant Disease Susceptibility to Foc 4
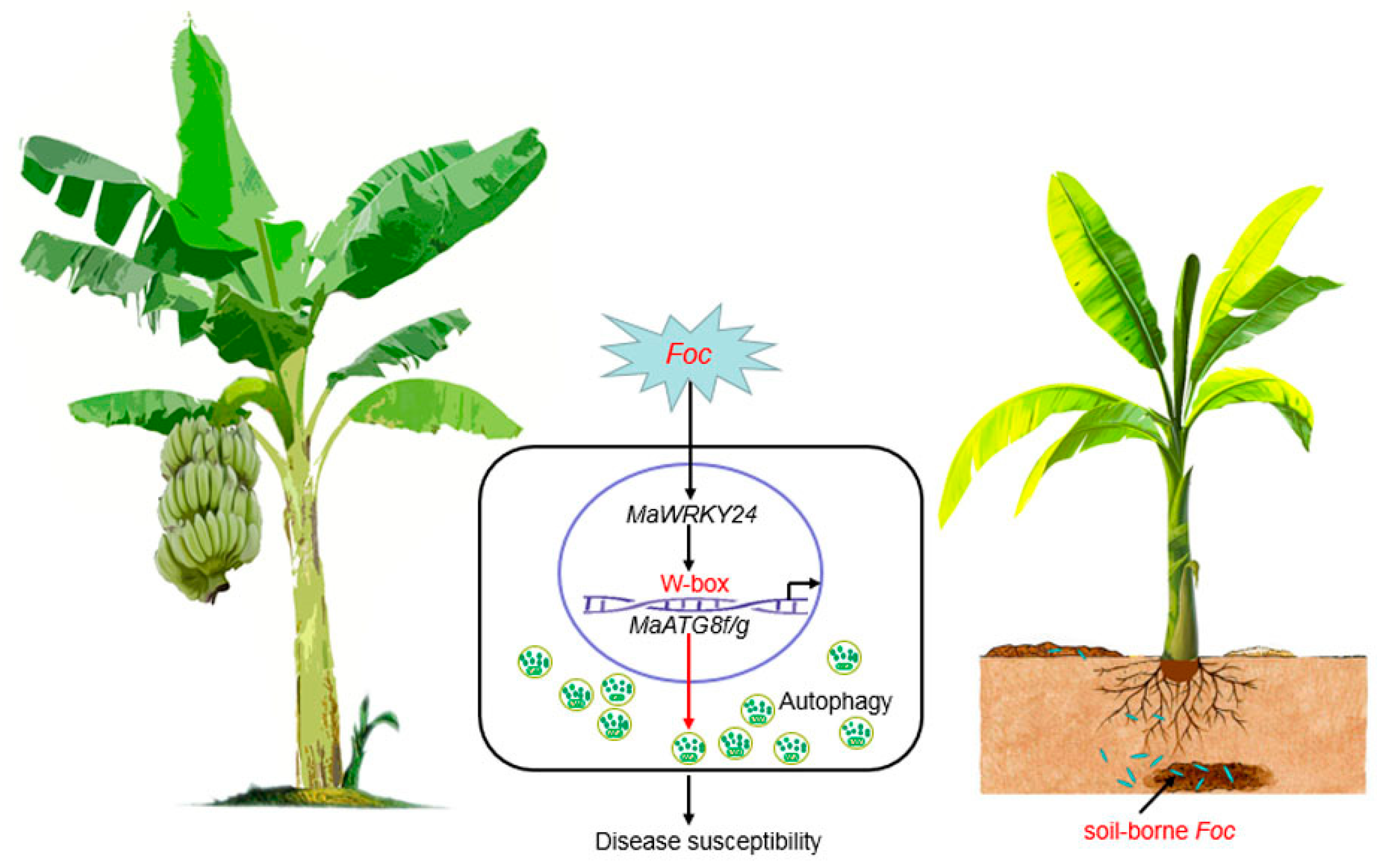
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Di, X.; Takken, F.; Tintor, N. How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front. Plant Sci. 2016, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shao, J.; Wang, Y.; Li, W.; Guo, D.; Yan, B.; Xia, Y.; Peng, M. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genom. 2013, 14, 851. [Google Scholar] [CrossRef]
- Berg, N.; Berger, D.; Hein, I.; Birch, P.; Wingfield, M.; Viljoen, A. Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol. Plant Pathol. 2007, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, N.; Seidl, M.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.; Ploetz, R.; Kema, G. Worse comes to worst: Bananas and panama disease-when plant and pathogen clones meet. PLoS Pathog. 2015, 11, e1005197. [Google Scholar] [CrossRef]
- Ploetz, R. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Cheng, C.; Liu, F.; Sun, X.; Tian, N.; Mensah, R.; Li, D.; Lai, Z. Identification of Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) responsive miRNAs in banana root. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Thangavelu, R.; Palaniswami, A.; Velazhahan, R. Mass production of Trichoderma harzianum for managing Fusarium wilt of banana. Agric. Ecosyst. Environ. 2004, 103, 259–263. [Google Scholar] [CrossRef]
- Dong, X.; Ling, N.; Wang, M.; Shen, Q.; Guo, S. Fusaric acid is a crucial factor in the disturbance of leaf water imbalance in Fusarium-infected banana plants. Plant Physiol. Biochem. 2012, 60, 171–179. [Google Scholar] [CrossRef]
- Goel, R.; Pandey, A.; Trivedi, P.; Asif, M. Genome-wide analysis of the Musa WRKY gene family: Evolution and differential expression during development and stress. Front. Plant Sci. 2016, 7, 299. [Google Scholar] [CrossRef]
- Rushton, P.; Somssich, I.; Ringler, P.; Shen, Q. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-Wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yu, S.; Li, J.; Li, Q.; Wang, K.; Huang, J.; Liu, Z. Molecular characterization of WRKY transcription factors that act as negative regulators of O-methylated catechins biosynthesis in tea plant (Camellia Sinensis L.). J. Agric. Food Chem. 2018, 66, 11234–11243. [Google Scholar] [CrossRef] [PubMed]
- Akagi, A.; Fukushima, S.; Okada, K.; Jiang, C.; Yoshida, R.; Nakayama, A.; Shimono, M.; Sugano, S.; Yamane, H.; Takatsuji, H. WRKY45-Dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol. Biol. 2014, 86, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Deng, X.; Qu, A.; Zhang, M.; Tao, Y.; Yang, L.; Liu, Y.; Xu, J.; Zhang, S. Regulation of pollen lipid body biogenesis by MAP kinases and downstream WRKY transcription factors in Arabidopsis. PLoS Genet. 2018, 14, e1007880. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Mitogen-Activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef]
- Guan, Y.; Meng, X.; Khanna, R.; LaMontagne, E.; Liu, Y.; Zhang, S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 2014, 10, e1004384. [Google Scholar] [CrossRef]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirau, K.; Yoshioka, H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 2007, 19, 819–830. [Google Scholar] [CrossRef]
- Wang, X.; Guo, R.; Tu, M.; Wang, D.; Guo, C.; Wan, R.; Li, Z.; Wang, X. Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the Necrotrophic Pathogen Botrytis cinerea. Front. Plant Sci. 2017, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, J.; Yang, F.; Zhang, G.; Wang, D.; Zhang, L.; Ou, Y.; Yao, Y. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant 2019. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, L.; Ma, X.; Zhao, X.; Jiao, B.; Li, C.; Luo, K. The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 2017, 37, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Kaliyappan, R.; Viswanathan, S.; Suthanthiram, B.; Subbaraya, U.; Marimuthu, S.; Muthu, M. Evolutionary expansion of WRKY gene family in banana and its expression profile during the infection of root lesion nematode, Pratylenchus coffeae. PLoS ONE 2016, 11, e0162013. [Google Scholar] [CrossRef]
- Shekhawat, U.; Ganapathi, T. MusaWRKY71 overexpression in banana plants leads to altered abiotic and biotic stress responses. PLoS ONE 2013, 8, e75506. [Google Scholar] [CrossRef]
- Tang, Y.; Kuang, J.; Wang, F.; Chen, L.; Hong, K.; Xiao, Y.; Xie, H.; Lu, W.; Chen, J. Molecular characterization of PR and WRKY genes during SA- and MeJA-induced resistance against Colletotrichum musae in banana fruit. Postharvest Biol. Technol. 2013, 79, 62–68. [Google Scholar] [CrossRef]
- Luo, D.; Ba, L.; Shan, W.; Kuang, J.; Lu, W.; Chen, J. Involvement of WRKY transcription factors in abscisic-acid-induced cold tolerance of banana fruit. J. Agric. Food Chem. 2017, 65, 3627–3635. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, W.; Hu, W.; Liu, G.; Wu, C.; Liu, W.; Zeng, H.; He, C.; Shi, H. Genome-Wide analysis of autophagy-related genes (ATGs) in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017, 36, 1237–1250. [Google Scholar] [CrossRef]
- Cutler, S.; Ehrhardt, D.; Griffitts, J.; Somerville, C. Random GFP: cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 2000, 97, 3718–3723. [Google Scholar] [CrossRef]
- Sparkes, I.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Sagi, L.; Remy, S.; Panis, B.; Swennen, R.; Volckaert, G. Transient gene expression in electroporated banana (Musa spp., cv. ‘Bluggoe’, ABB group) protoplasts isolated from regenerable embryogenetic cell suspensions. Plant Cell Rep. 1994, 13, 262–266. [Google Scholar] [CrossRef]
- Ream, J.; Lewis, L.; Lewis, K. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions. Anal. Biochem. 2016, 511, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.; Bent, A. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, G. Construction of overexpressing MaATG8g Arabidopsis and analysis of the promoter. 2020. Available online: https://kns.cnki.net/KCMS/detail/46.1068.S.20191008.1121.008.html (accessed on 22 November 2019).
- Xing, D.; Lai, Z.; Zheng, Z.; Vinod, K.; Fan, B.; Chen, Z. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 2008, 1, 459–470. [Google Scholar] [CrossRef]
- Li, J.; Zhong, R.; Palva, E. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE 2017, 12, e0183731. [Google Scholar] [CrossRef]
- García-Bastidas, F.; Van der Veen, A.; Nakasato-Tagami, G.; Meijer, H.; Arango-Isaza, R.; Kema, G. An improved phenotyping protocol for panama disease in banana. Front. Plant Sci. 2019. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, D. WRKY transcription factors: Links between phytohormones and plant processes. Sci. China Life Sci. 2015, 58, 501–502. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Li, Y.; Wang, F.; Cheng, Y.; Fan, B.; Yu, J.; Chen, Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011, 23, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.; Kracher, B.; Somssich, I. Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Pandey, S.; Somssich, I. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Yu, D. Wounding-Induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe. Interact. 2010, 23, 558–565. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, H.; Zhang, L.; Wang, F.; Yu, D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013, 74, 730–745. [Google Scholar] [CrossRef]
- Pandey, S.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef]
- Karim, A.; Jiang, Y.; Guo, L.; Ling, Z.; Ye, S.; Duan, Y.; Li, C.; Luo, K. Isolation and characterization of a subgroup iia WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 2015, 35, 1129–1139. [Google Scholar] [CrossRef]
- Liu, X.; Song, Y.; Xing, F.; Wang, N.; Wen, F.; Zhu, C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana Benthamiana. Protoplasma 2016, 253, 1265–1281. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.; Robatzek, S.; Somssich, I. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Eulgem, T. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Marè, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Stanca, A.; Cattivelli, L. Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef]
- Marshall, R.; Vierstra, R. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Thompson, A.; Vierstra, R. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef]
- Klionsky, D.; Abdalla, F.; Abeliovich, H.; Abraham, R.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef]
- Dagdas, Y.; Beihaj, K.; Maqbool, A.; Chaparro-Garcia, A.; Pandey, P.; Petre, B.; Tabassum, N.; Cruz-Mireles, N.; Hughes, R.; Sklenar, J.; et al. An effector of the irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 2016, 5, e10856. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Baluška, F.; Bozhkov, P.; Elander, P.; Fernie, A.; Galili, G.; Hassan, A.; Hofius, D.; Isono, E.; Bars, R.; et al. Autophagy-Related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 2018, 69, 1335–1353. [Google Scholar] [CrossRef]
- Minina, E.; Moschou, P.; Bozhkov, P. Limited and digestive proteolysis: Crosstalk between evolutionary conserved pathways. New Phytol. 2017, 215, 958–964. [Google Scholar] [CrossRef]
- Ustun, S.; Hafren, A.; Liu, Q.; Marshall, R.; Minina, E.; Bozhkov, P.; Vierstra, R.; Hofius, D. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 2018, 30, 668–685. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.; Vierstra, R.; Nürnberger, T.; Gust, A. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal. Behav. 2011, 6, 1040–1042. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y.; Bassham, D. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, P.; He, C.; Shi, H. MeWRKY20 and its interacting and activating autophagy-related protein 8 (MeATG8) regulate plant disease resistance in cassava. Biochem. Biophys. Res. Commun. 2017, 494, 20–26. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, J.; Chen, Z. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 2014, 15, 637–645. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Zeng, H.; Li, X.; Wei, Y.; Shi, H. Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense. Pathogens 2019, 8, 264. https://doi.org/10.3390/pathogens8040264
Liu G, Zeng H, Li X, Wei Y, Shi H. Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense. Pathogens. 2019; 8(4):264. https://doi.org/10.3390/pathogens8040264
Chicago/Turabian StyleLiu, Guoyin, Hongqiu Zeng, Xiang Li, Yunxie Wei, and Haitao Shi. 2019. "Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense" Pathogens 8, no. 4: 264. https://doi.org/10.3390/pathogens8040264
APA StyleLiu, G., Zeng, H., Li, X., Wei, Y., & Shi, H. (2019). Functional Analysis of MaWRKY24 in Transcriptional Activation of Autophagy-Related Gene 8f/g and Plant Disease Susceptibility to Soil-Borne Fusarium oxysporum f. sp. cubense. Pathogens, 8(4), 264. https://doi.org/10.3390/pathogens8040264



