Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells
Abstract
1. Introduction
2. Results
2.1. Generation of Primary Kidney Cells from Miniopterus fuliginosus
2.2. Generation of Pseudoviruses
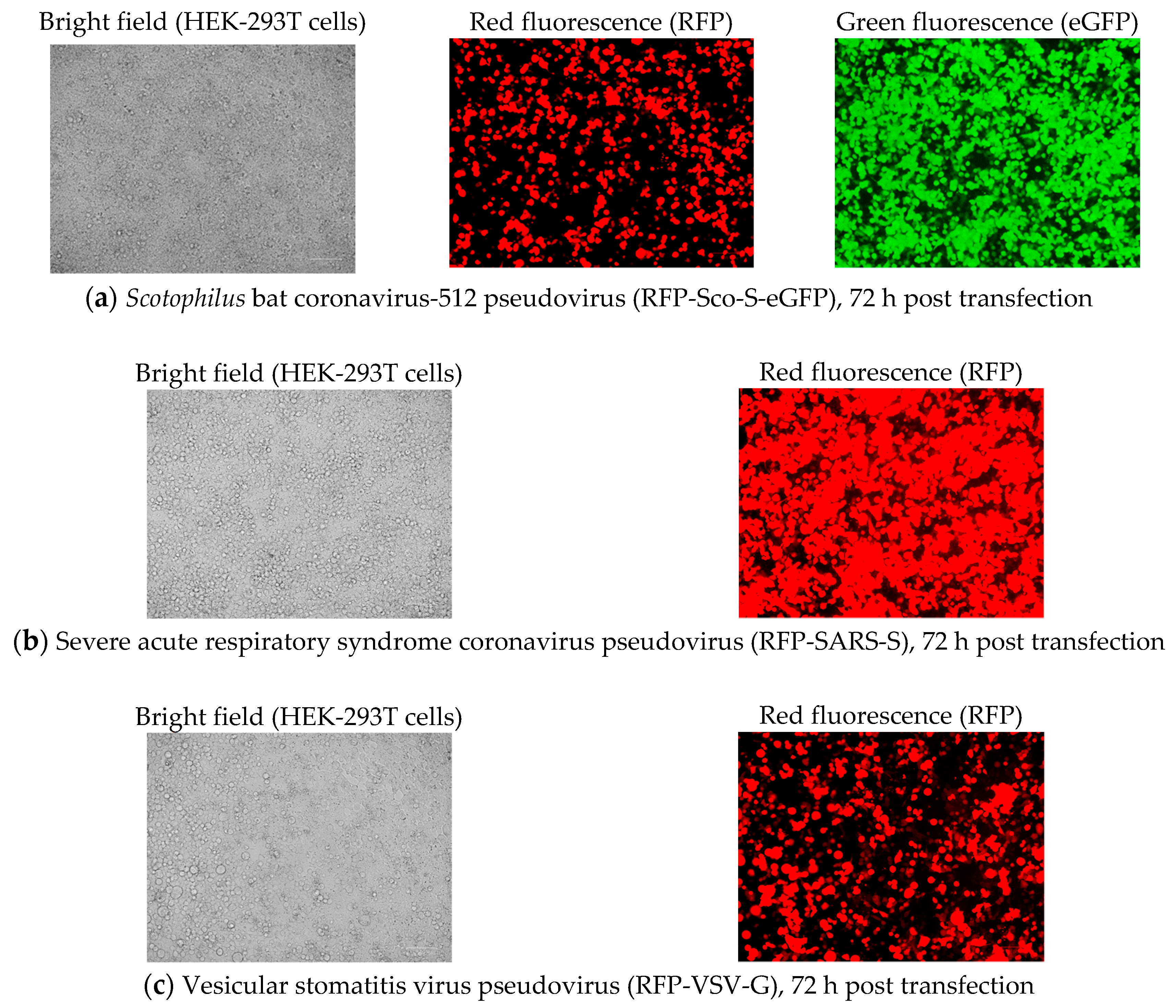
2.2.1. Pseudovirus Production after Co-Transfection
2.2.2. Pseudovirus Titration
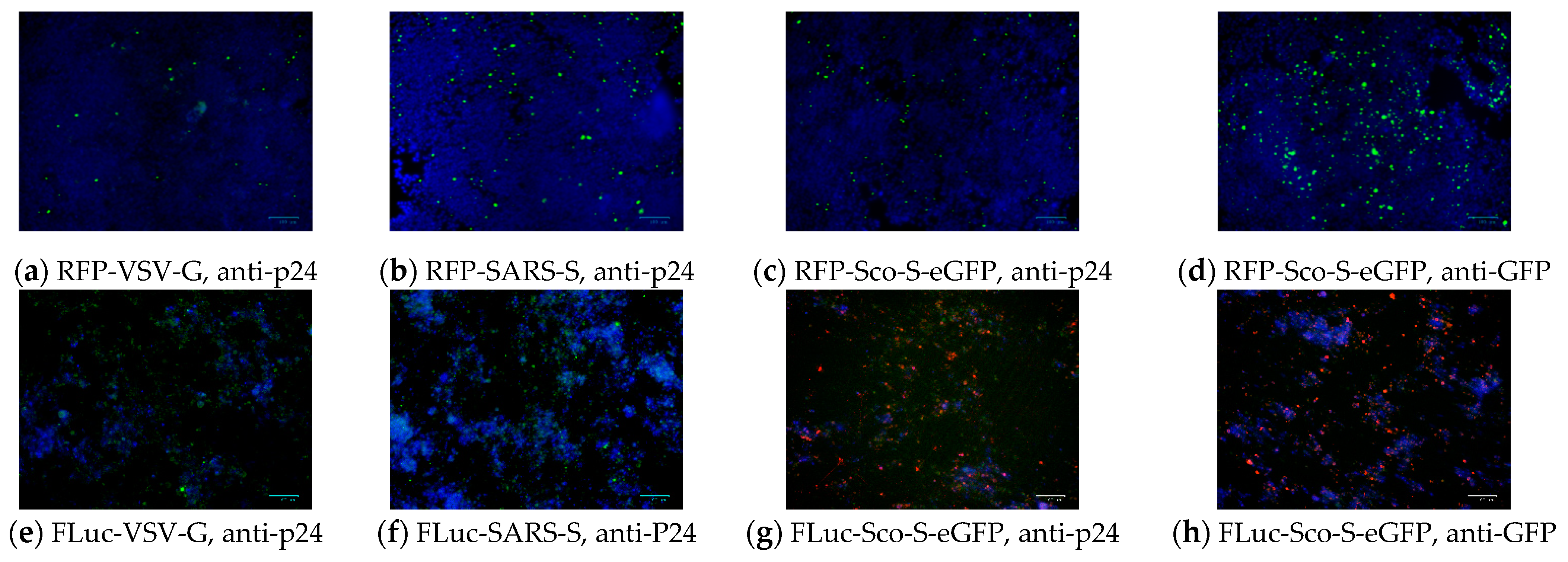
2.2.3. Detection of Pseudoviral Proteins after Infection
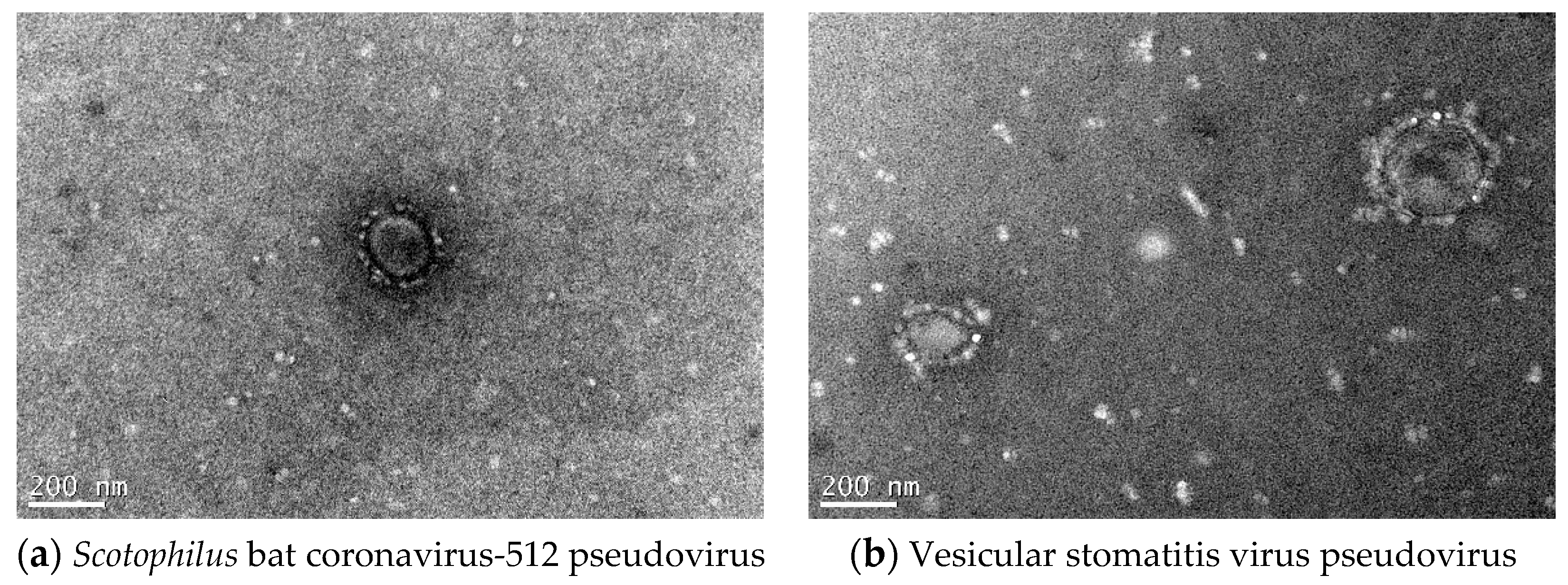
2.2.4. Electron Microscopic Morphology of Pseudoviruses
2.3. Cell Entries of RFP-Pseudoviruses
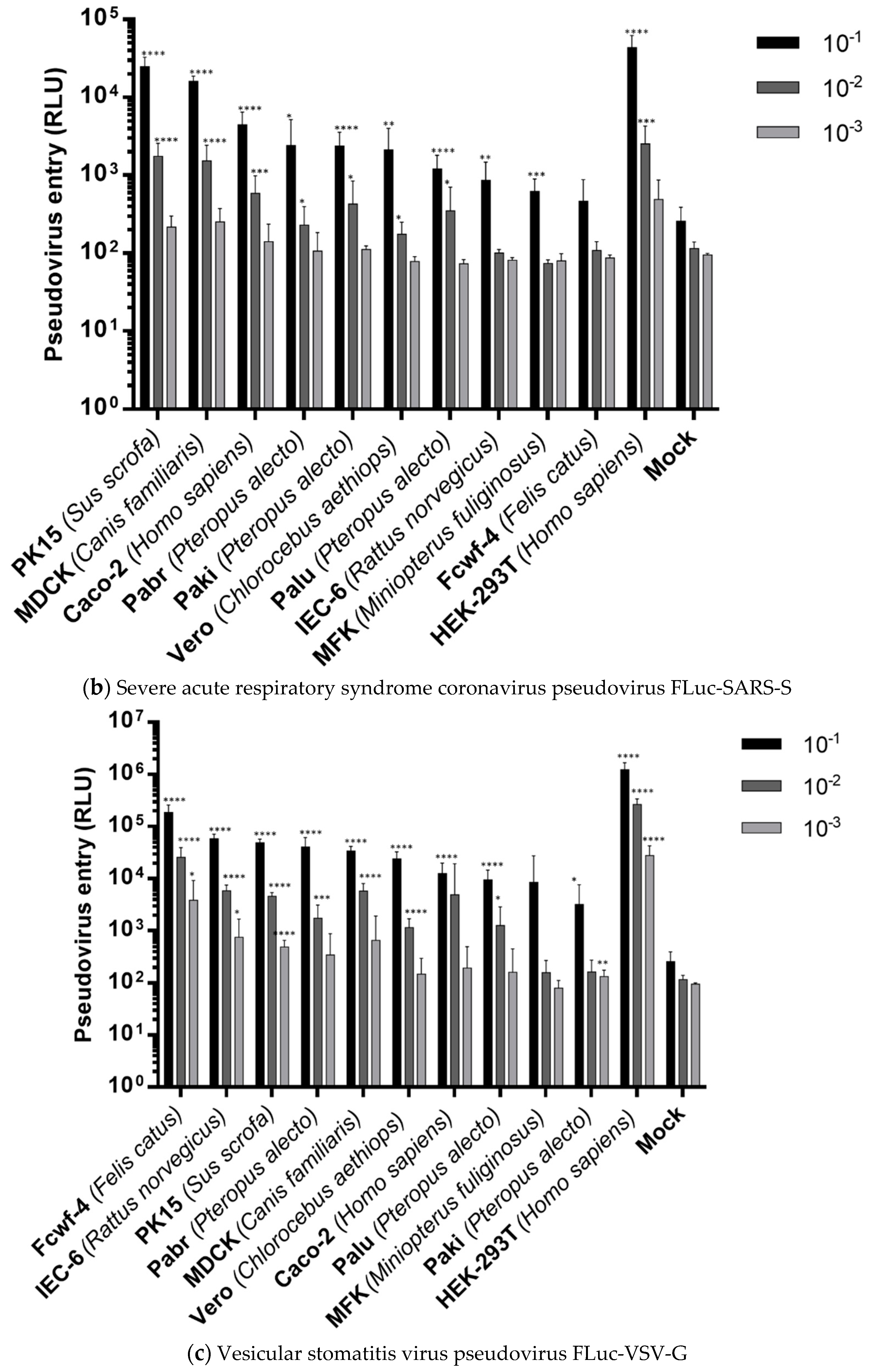
2.4. Cell Entries of FLuc-Pseudoviruses
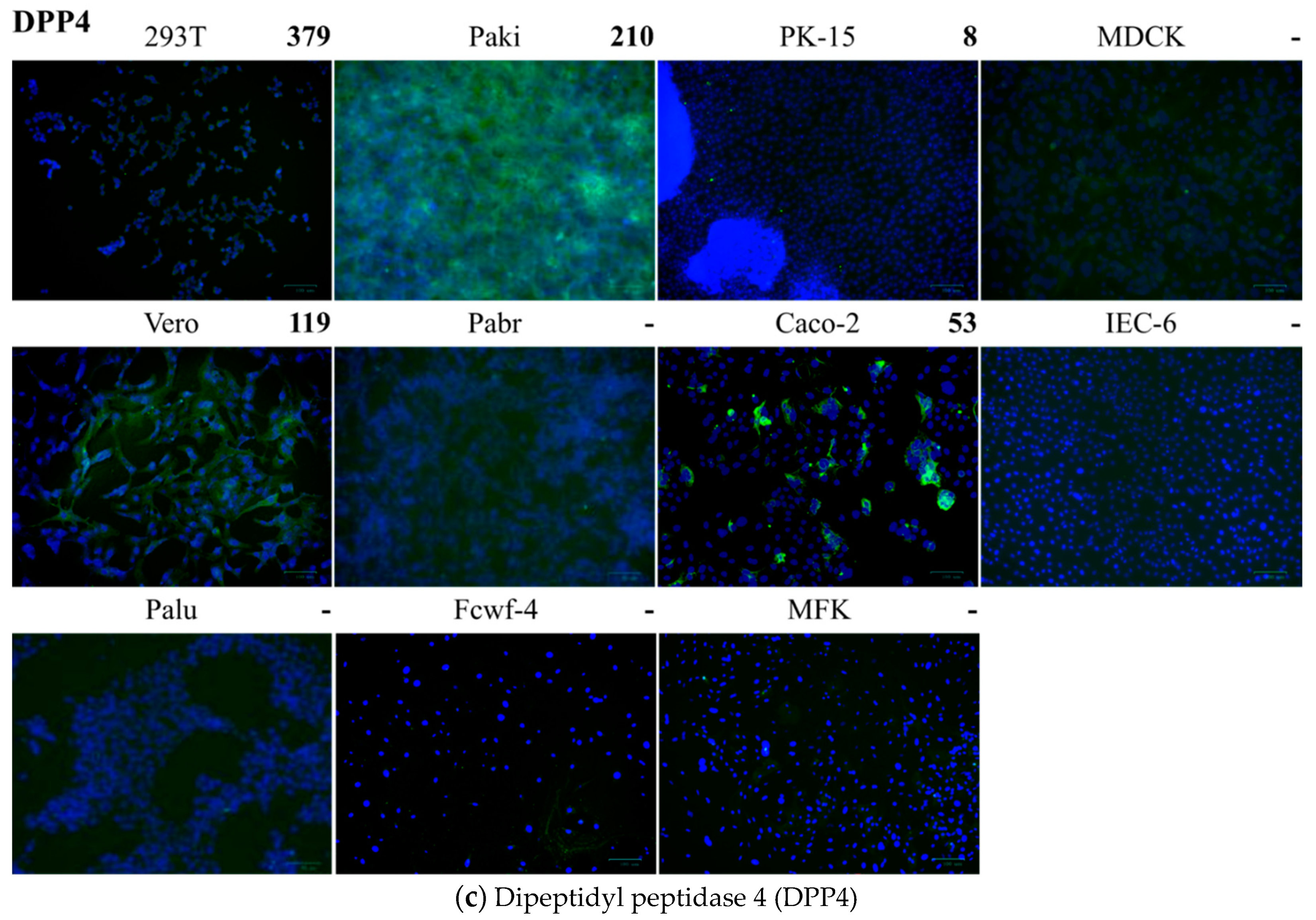
2.5. Detection of Cell Receptors on Different Cells
3. Discussion
4. Materials and Methods
4.1. Animal Experiment Ethics
4.2. Primary Bat Cell Culture
4.3. Cell Line Culture
4.4. Pseudovirus Generation
4.5. Pseudovirus Entry Assay
4.6. Immunofluorescent Antibody Assay
4.7. Detection of Cell Receptor
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease and Prevention. Update: Outbreak of severe acute respiratory syndrome-worldwide. MMWR 2003, 52, 269–272. [Google Scholar]
- World Health Organization. Disease Outbreak news: Middle East Respiratory Syndrome Coronavirus (MERS-CoV)-Saudi Arabia, 31 August 2019. Available online: https://www.who.int/csr/don/26-august-2019-mers-saudi-arabia/en/ (accessed on 31 August 2019).
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Tsoi, H.W.; Wong, B.H.; Wong, S.S.; Leung, S.Y.; Chan, K.H.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Li, K.S.; Tsang, A.K.; Yuen, K.Y. Genetic relatedness of the novel human group C Betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg. Microbes. Infect. 2012, 1, e35. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Huynh, J.; Li, S.; Yount, B.; Smith, A.; Sturges, L.; Olsen, J.C.; Nagel, J.; Johnson, J.B.; Agnihothram, S.; Gates, J.E.; et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012, 86, 12816–12825. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–256. [Google Scholar] [CrossRef]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Caì, Y.; Yú, S.; Postnikova, E.N.; Mazur, S.; Bernbaum, J.G.; Burk, R.; Zhāng, T.; Radoshitzky, S.R.; Müller, M.A.; Jordan, I. CD26/DPP4 cell-surface expression in bat cells correlates with bat cell susceptibility to middle east respiratory syndrome coronavirus (MERS-CoV) infection and evolution of persistent infection. PLoS ONE 2014, 9, e112060. [Google Scholar] [CrossRef] [PubMed]
- Eckerle, I.; Corman, V.M.; Müller, M.A.; Lenk, M.; Ulrich, R.G.; Drosten, C. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg. Infec. Dis. 2014, 20, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-N.; Phuong, V.N.; Chen, H.C.; Chou, C.-H.; Cheng, H.-C.; Wu, C.-H. Detection of the severe acute respiratory syndrome-related coronavirus and Alphacoronavirus in the bat population of Taiwan. Zoonoses Public Health 2016, 63, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-N.; Su, B.-G.; Chen, H.-C.; Chou, C.-H.; Cheng, H.-C. Detection of specific antibodies to the nucleocapsid protein fragments of severe acute respiratory syndrome-coronavirus and Scotophilus bat coronavirus-512 in three insectivorous bat species. Taiwan Vet. J. 2018, 44, 179–188. [Google Scholar] [CrossRef]
- Huang, Y.W.; Dickerman, A.W.; Pineyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef]
- Liu, C.; Tang, J.; Ma, Y.; Liang, X.; Yang, Y.; Peng, G.; Qi, Q.; Jiang, S.; Li, J.; Du, L.; et al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015, 89, 6121–6125. [Google Scholar] [CrossRef]
- Lin, S.-C.; Kappes, M.A.; Chen, M.-C.; Lin, C.-C.; Wang, T.T. Distinct susceptibility and applicability of MDCK derivatives for influenza virus research. PLoS ONE 2017, 12, e0172299. [Google Scholar] [CrossRef]
- O’Brien, A.; Mettelman, R.C.; Volk, A.; André, N.M.; Whittaker, G.R.; Baker, S.C. Characterizing replication kinetics and plaque production of type I feline infectious peritonitis virus in three feline cell lines. Virology 2018, 525, 1–9. [Google Scholar] [CrossRef]
- Li, B.X.; Ge, J.W.; Li, Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 355, 166–172. [Google Scholar] [CrossRef]
- Li, W.; Luo, R.; He, Q.; van Kuppeveld, F.J.M.; Rottier, P.J.M.; Bosch, B.J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017, 235, 6–13. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Z.; Yang, H. Aminopeptidase N knockout pigs are not resistant to porcine epidemic diarrhea virus infection. Virol. Sin. 2019. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Maejima, M.; Islam, M.T.; Miyazaki, A.; Kawase, M.; Matsuyama, S.; Taguchi, F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, bur promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016, 97, 2528–2539. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, W.; Begeman, L.; Schipper, D.; Run, P.R.V.; Cunningham, A.A.; Kley, N.; Reusken, C.B.; Haagmans, B.L.; van den Brand, J.M.A. Tissue distribution of the MERS-Coronavirus receptor in bats. Sci. Rep. 2017, 7, 1193. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, W.; Raj, V.S.; Schipper, D.; Kolijn, K.; van Leenders, G.; Bosch, B.J.; Bensaid, A.; Segales, J.; Baumgartner, W.; Osterhaus, A.; et al. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J. Virol. 2016, 90, 4838–4842. [Google Scholar] [CrossRef] [PubMed]
- Meyerholz, D.K.; Lambertz, A.M.; McCray, P.B., Jr. Dipeptidyl peotidase 4 distribution in the human respiratory tract: Implications for the Middle East respiratory syndrome. Am. J. Pathol. 2016, 186, 78–86. [Google Scholar] [CrossRef]
- Li, W.; Sui, J.; Huang, I.C.; Kuhn, J.H.; Radoshitzky, S.R.; Marasco, W.A.; Choe, H.; Farzan, M. The S protein of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology 2007, 367, 367–374. [Google Scholar] [CrossRef]
- Liu, C.; Ma, Y.; Yang, Y.; Zheng, Y.; Shang, J.; Zhou, Y.; Jiang, S.; Du, L.; Li, J.; Li, F. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 2016, 291, 24779–24789. [Google Scholar] [CrossRef]
- Tang, X.C.; Zhang, J.X.; Zhang, S.Y.; Wang, P.; Fan, X.H.; Li, L.F.; Li, G.; Dong, B.Q.; Liu, W.; Cheung, C.L.; et al. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006, 80, 7481–7490. [Google Scholar] [CrossRef]
- Kaye, M.; Druce, J.; Tran, T.; Kostecki, R.; Chibo, D.; Morris, J.; Catton, M.; Birch, C. SARS–associated coronavirus replication in cell lines. Emerg. Infect. Dis. 2006, 12, 128–133. [Google Scholar] [CrossRef]
- Mossel, E.C.; Huang, C.; Narayanan, K.; Makino, S.; Tesh, R.B.; Peters, C. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J. Virol. 2005, 79, 3846–3850. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE 2011, 6, e23710. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Dominguez, S.R.; Holmes, K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 2013, 8, e76469. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.M.; Scobey, T.; Swanstrom, J.; Jensen, K.L.; Burch, C.L.; Baric, R.S.; Heise, M.T. Permissively of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017, 19, e00534-17. [Google Scholar] [CrossRef]
- Wentworth, D.E.; Holmes, K.V. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): Influence of N-linked glycosylation. J. Virol. 2001, 75, 9741–9752. [Google Scholar] [CrossRef] [PubMed]
- Tusell, S.M.; Schittone, S.A.; Holmes, K.V. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants o viral host range. J. Virol. 2007, 81, 1261–1273. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.; Huang, I.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef]
- Amman, B.R.; Carroll, S.A.; Reed, Z.D.; Sealy, T.K.; Balinandi, S.; Swanepoel, R.; Kemp, A.; Erickson, B.R.; Comer, J.A.; Campbell, S.; et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012, 8, e1002877. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Madan, V.; Garcia, M.J.; Sanz, M.A.; Carrasco, L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005, 579, 3607–3612. [Google Scholar] [CrossRef]
- Yount, B.; Roberts, R.S.; Sims, A.C.; Deming, D.; Frieman, M.B.; Sparks, J.; Denison, M.R.; Davis, N.; Baric, R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005, 79, 14909–14922. [Google Scholar] [CrossRef] [PubMed]
- De Haan, C.A.; Masters, P.S.; Shen, X.; Weiss, S.; Rottier, P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 2002, 296, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Narayan, K.; Huang, C.; Lokugamage, K.; Kamitani, W.; Ikegami, T.; Tseng, C.T.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008, 82, 4471–4479. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.G.; Wang, N.; Chen, Z.; Chen, Z.; Tseng, M.; Barretto, N.; Lin, R.; Peters, C.J.; Tseng, C.T.; Baker, S.C.; et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007, 282, 32208–32221. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, D.; Zillinger, T.; Nuth, D.; Zielecki, F.; Horvath, G.; Suliman, T.; Barchet, W.; Weber, F.; Drosten, C.; Muller, M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonists. J. Virol. 2013, 87, 12489–12495. [Google Scholar] [CrossRef]
- Kopecky-Bromberg, S.A.; Martinez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. SARS coronavirus protein Orf3b, Orf6, and nucleocapsid function as interferon antagonists. J. Virol. 2006, 81, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Zhang, L.; Luk, H.K.H.; Xiong, L.; Peng, X.; Li, K.S.M.; He, X.; Zhao, P.S.-H.; Fan, R.Y.Y.; Wong, A.C.P.; et al. Receptor usage of a novel bat lineage C Betacoronavirus reveals evolution of Middle East respiratory syndrome-related coronavirus spike proteins for human dipeptidyl peptidase 4 binding. J. Infect. Dis. 2018, 218, 197–207. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Fan, R.Y.Y.; Luk, H.K.H.; Zhu, L.; Fung, J.; Li, K.S.M.; Wong, E.Y.M.; Ahmed, S.S.; Chan, J.F.W.; Kok, R.K.H.; et al. Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins. Emerg. Microbe Infect. 2018, 7, 209–219. [Google Scholar] [CrossRef]
- Müller, M.A.; Raj, V.S.; Muth, D.; Meyer, B.; Kallies, S.; Smits, S.L.; Wollny, R.; Bestebroer, T.M.; Specht, S.; Suliman, T.; et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio 2012, 3, e00515-12. [Google Scholar] [CrossRef]
- Letko, M.; Miazgowicz, K.; McMinn, R.; Seifert, S.N.; Sola, I.; Enjuanes, L.; Carmody, A.; van Doremalen, N.; Munster, V. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018, 24, 1730–1737. [Google Scholar] [CrossRef]

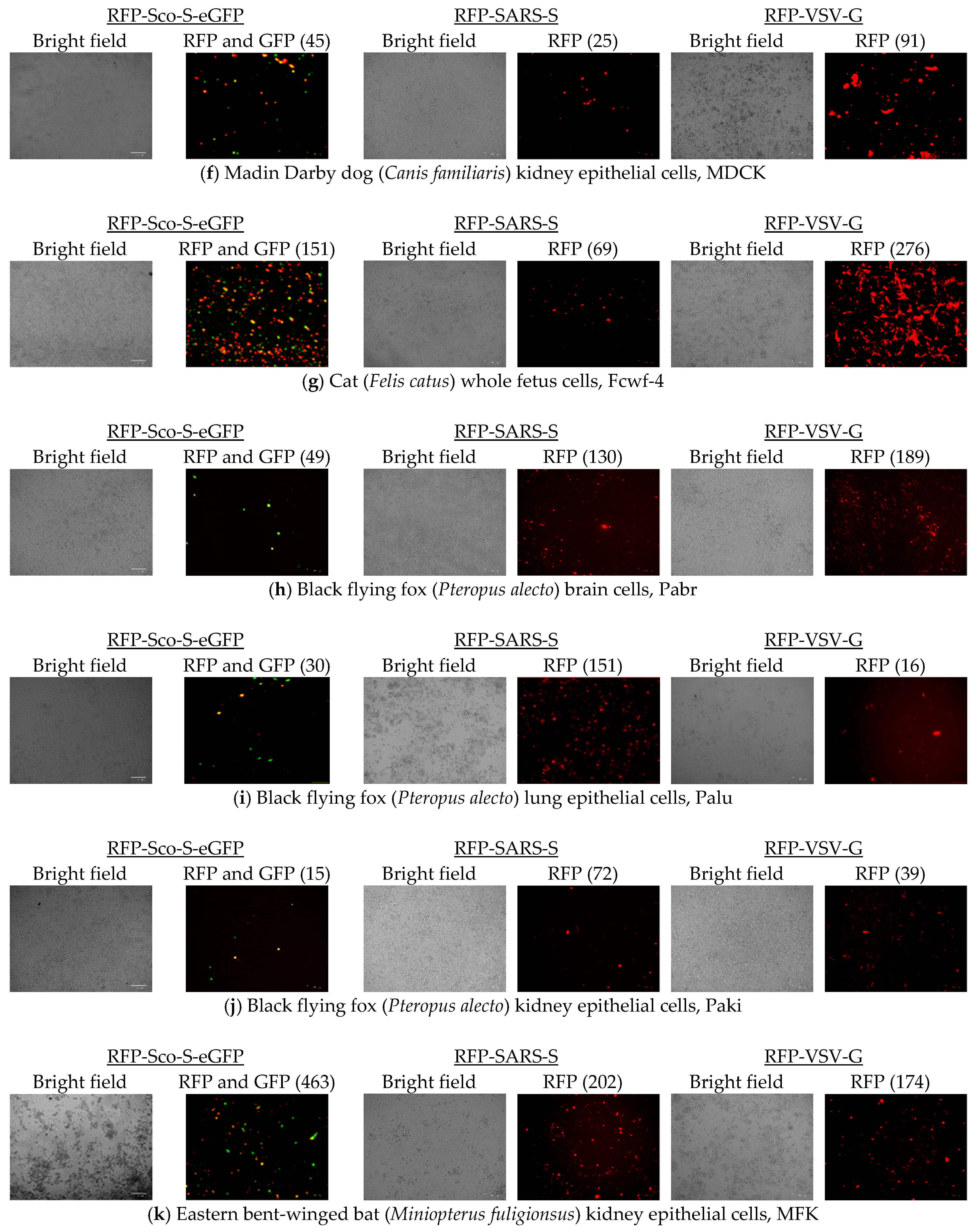
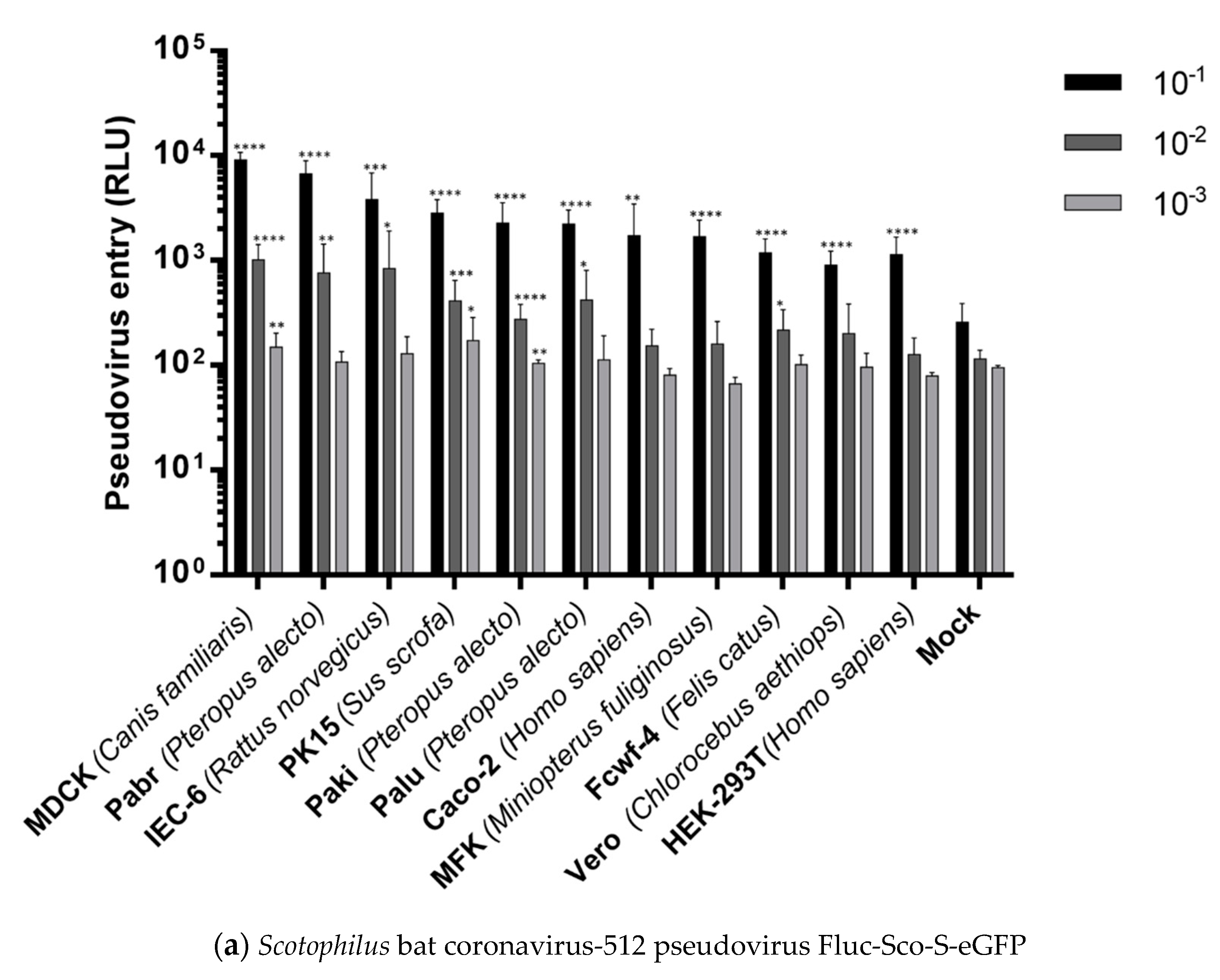
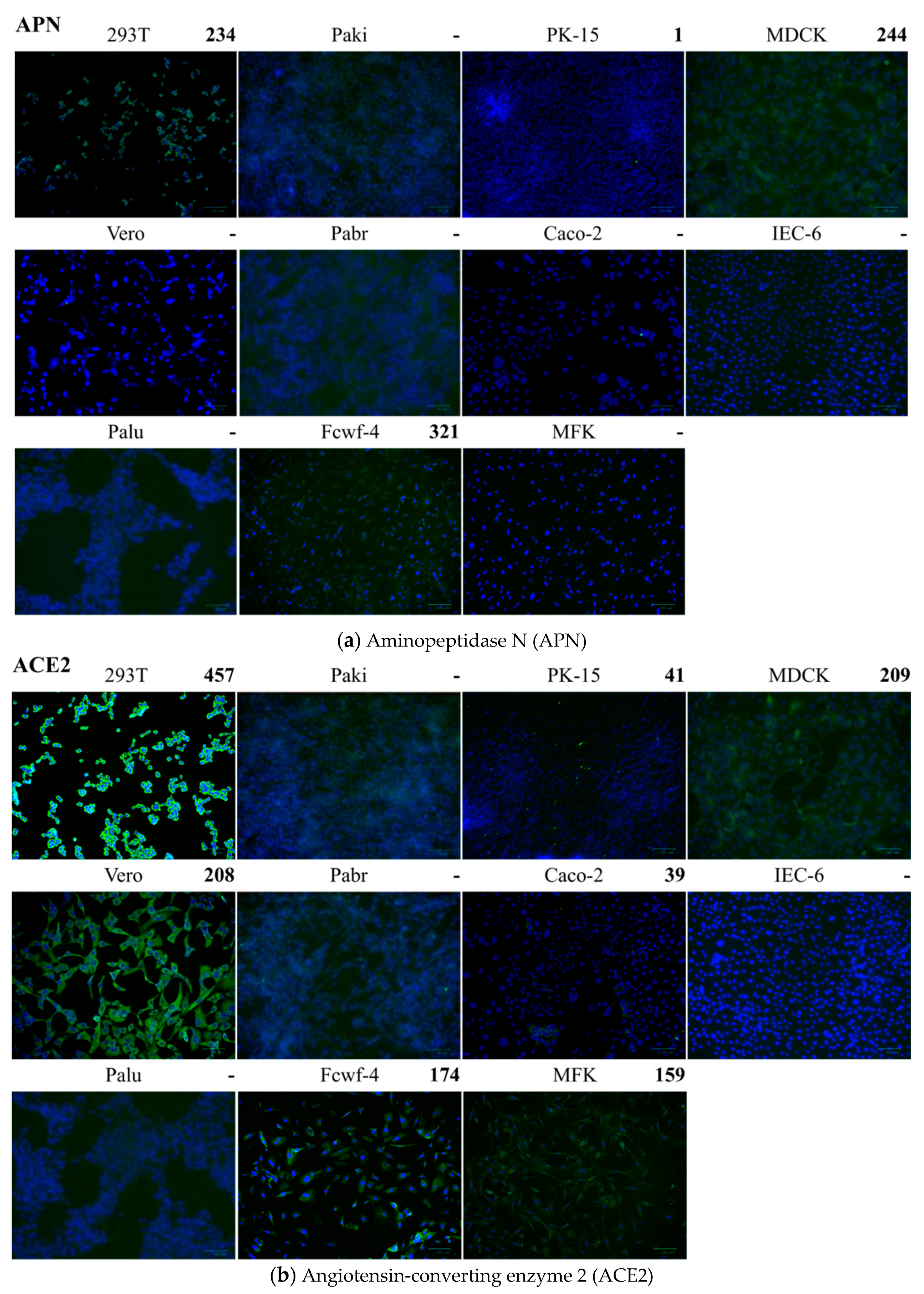

| Cell 1 | Source | Tissue | APN 2 | ACE2 3 | DPP4 4 | FLuc-Sco-S-eGFP 5 | FLuc-SARS-S 6 |
|---|---|---|---|---|---|---|---|
| HEK-293T Caco-2 Vero | Human Human Monkey | Kidney Colon Kidney | >200 Neg.7 Neg. | >200 <200 >200 | >200 <200 <200 | 2804 efg 1715 def 909 fg | a 4463 d 2118 d |
| IEC-6 PK15 MDCK Fcwf-4 Pabr Palu Paki MFK | Rat Pig Dog Cat Fruit Bat Fruit Bat Fruit Bat Bat | Intestine Kidney Kidney Fetus Brain Lung Kidney Kidney | Neg. <200 >200 >200 Neg. Neg. Neg. Neg. | Neg. <200 >200 <200 Neg. Neg. Neg. <200 | Neg. <200 Neg. Neg. Neg. Neg. >200 Neg. | 3822 c 2835 cd 9126 a 1188 efg 6708 b 2225 de 2270 de 1683 def | 859 d 24,723 b 16,024 c 466 d 2398 d 1201 d 2382 d 623 d |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-N.; Hsu, H.-C.; Wang, S.-W.; Lien, H.-C.; Lu, H.-T.; Peng, S.-K. Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells. Pathogens 2019, 8, 259. https://doi.org/10.3390/pathogens8040259
Chen Y-N, Hsu H-C, Wang S-W, Lien H-C, Lu H-T, Peng S-K. Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells. Pathogens. 2019; 8(4):259. https://doi.org/10.3390/pathogens8040259
Chicago/Turabian StyleChen, Yi-Ning, Hsiao-Chin Hsu, Sheng-Wei Wang, Hao-Chiang Lien, Hsin-Ti Lu, and Sheng-Kai Peng. 2019. "Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells" Pathogens 8, no. 4: 259. https://doi.org/10.3390/pathogens8040259
APA StyleChen, Y.-N., Hsu, H.-C., Wang, S.-W., Lien, H.-C., Lu, H.-T., & Peng, S.-K. (2019). Entry of Scotophilus Bat Coronavirus-512 and Severe Acute Respiratory Syndrome Coronavirus in Human and Multiple Animal Cells. Pathogens, 8(4), 259. https://doi.org/10.3390/pathogens8040259





