Construction of A New Dose–Response Model for Staphylococcus aureus Considering Growth and Decay Kinetics on Skin
Abstract
1. Introduction
2. Results and Discussion
2.1. Review of the Previous Model
2.2. Gompertz Growth and Decay Model
2.3. Development of a New Dose–Response Model
3. Method
3.1. Development of the S. aureus Growth Model
3.1.1. Data Source
3.1.2. Review of the Previous Model
3.1.3. Gompertz Growth and Decay Models
3.1.4. Parameter Estimation Methods
Ordinary Least Squares Estimation (OLS).
Scaled Sensitivity Coefficients
3.2. Development of the S. aureus Dose-Response Model
3.2.1. Data Source
3.2.2. Revised S. aureus Dose
3.2.3. Fitting Dose–Response Model and Uncertainty Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.W.; Biancotti, J.C.; Berg, B.L.; Gate, D.; Kolar, S.L.; Müller, S.; Rodriguez, M.D.; Rezai-Zadeh, K.; Fan, X.; Beenhouwer, D.O.; et al. Increased susceptibility of humanized NSG mice to Panton-Valentine leukocidin and Staphylococcus aureus skin infection. PLoS Pathog. 2015, 11, e1005292. [Google Scholar] [CrossRef] [PubMed]
- Verbrugh, H.A. Colonization with Staphylococcus aureus and the role of colonization in causing infection. In Staphylocci in Human Disease; Crossley, K.B., Jefferson, K.K., Archer, G.L., Fowler, V.G., Jr., Eds.; Wiley-Blackwell: Chichester, UK, 2009; pp. 255–271. [Google Scholar]
- Ryan, M.O.; Haas, C.N.; Gurian, P.L.; Gerba, C.P.; Panzl, B.M.; Rose, J.B. Application of quantitative microbial risk assessment for selection of microbial reduction targets for hard surface disinfectants. Am. J. Infect. Control 2014, 42, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Krebes, J.; Al-Ghusein, H.; Feasey, N.; Breathnach, A.; Lindsay, J.A. Are nasal carriers of Staphylococcus aureus more likely to become colonized or infected with methicillin-resistant Staphylococcus aureus on admission to a hospital? J. Clin. Microbiol. 2011, 49, 430–432. [Google Scholar] [CrossRef]
- Jarvis, W.R.; Jarvis, A.A.; Chinn, R.Y. National prevalence of methicillin-resistant Staphylococcus aureus in inpatients at United States health care facilities, 2010. Am. J. Infect. Control 2012, 40, 194–200. [Google Scholar] [CrossRef]
- Lindsay, J.A. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 2014, 304, 103–109. [Google Scholar] [CrossRef]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, J.M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef]
- See, I.; Mu, Y.; Albrecht, V.; Karlsson, M.; Dumyati, G.; Hardy, D.J.; Koeck, M.; Lynfield, R.; Nadle, J.; Ray, S.M.; et al. Trends in incidence of methicillin-resistant Staphylococcus aureus bloodstream infections differ by strain type and healthcare exposure, United States, 2005–2013. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Grigg, C.; Palms, D.; Stone, N.D.; Gualandi, N.; Bamberg, W.; Dumyati, G.; Harrison, L.H.; Lynfield, R.; Nadle, J.; Petit, S.; et al. Burden of Invasive Methicillin-Resistant Staphylococcus Aureus Infections in Nursing Home Residents. J. Am. Geriatr. Soc. 2018. [Google Scholar] [CrossRef]
- Kluytmans, J.; Van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.F. Pathogenesis of MRSA infections. Dis. Mon. 2008, 54, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Van Wamel, W.J. Staphylococcus aureus infections, some second thoughts. Curr. Opin. Infect. Dis. 2017, 30, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Van Kleef, M.; Vos, M.C.; Ott, A.; Verbrugh, H.A.; Fokkens, W. Nose picking and nasal carriage of Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 2006, 27, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Byrne, F.M.; Wilcox, M.H. MRSA prevention strategies and current guidelines. Injury 2011, 42, S3–S6. [Google Scholar] [CrossRef]
- Sollid, J.U.E.; Furberg, A.S.; Hanssen, A.M.; Johannessen, M. Staphylococcus aureus: Determinants of human carriage. Infect. Genet. Evol. 2014, 21, 531–541. [Google Scholar] [CrossRef]
- Kazakova, S.V.; Hageman, J.C.; Matava, M.; Srinivasan, A.; Phelan, L.; Garfinkel, B.; Boo, T.; McAllister, S.; Anderson, J.; Jensen, B. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 2005, 352, 468–475. [Google Scholar] [CrossRef]
- Scott, E.; Bloomfield, S.F. Investigations of the effectiveness of detergent washing, drying and chemical disinfection on contamination of cleaning cloths. J. Appl. Bacteriol. 1990, 68, 279–283. [Google Scholar] [CrossRef]
- Marples, R.R.; Towers, A.G. A laboratory model for the investigation of contact transfer of micro-organisms. Epidemiol. Infect. 1979, 82, 237–248. [Google Scholar] [CrossRef]
- Sattar, S.A.; Springthorpe, S.; Mani, S.; Gallant, M.; Nair, R.C.; Scott, E.; Kain, J. Transfer of bacteria from fabrics to hands and other fabrics: Development and application of a quantitative method using Staphylococcus aureus as a model. J. Appl. Microbiol. 2001, 90, 962–970. [Google Scholar] [CrossRef]
- Kusumaningrum, H.D.; Paltinaite, R.; Koomen, A.J.; Hazeleger, W.C.; Rombouts, F.M.; Beumer, R.R. Tolerance of Salmonella enteritidis and Staphylococcus aureus to surface cleaning and household bleach. J. Food Prot. 2003, 66, 2289–2295. [Google Scholar] [CrossRef] [PubMed]
- Neely, A.N.; Maley, M.P. Survival of enterococci and staphylococci on hospital fabrics and plastic. J. Clin. Microbiol. 2000, 38, 724–726. [Google Scholar] [PubMed]
- Archer, G.L. Staphylococcus aureus: A well-armed pathogen. Rev. Infect. Dis. 1998, 26, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- CDC. Staphylococcus aureus in Healthcare Settings; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011. Available online: https://www.cdc.gov/hai/organisms/staph.html (accessed on 19 September 2017).
- Cohen, J.; Pinching, A.J.; Rees, A.J.; Peters, D.K. Infection and immunosuppression: A study of the infective complications of 75 patients with immunologically-mediated disease. QJM: Int. J. Med. 1982, 51, 1–15. [Google Scholar]
- Finkey, M.B.; Corbin, N.C.; Aust, L.B.; Aly, R.; Maibach, H.I. In vivo effect of antimicrobial soap bars containing 1.5% and 0.8% trichlorocarbanilide against two strains of pathogenic bacteria. J. Soc. Cosmet. Chem. 1984, 35, 351–355. [Google Scholar]
- Rose, J.B.; Haas, C.N. A risk assessment framework for the evaluation of skin infections and the potential impact of antibacterial soap washing. Am. J. Infect. Control 1999, 27, S26–S33. [Google Scholar] [CrossRef]
- Haas, C.; Gerba, C.; Rose, J.B. Quantitative Microbial Risk Assessment; Wiley: New York, NY, USA, 2014. [Google Scholar]
- Haas, C.N. Microbial dose response modeling: Past, present, and future. Environ. Sci. Technol. 2015, 49, 1245–1259. [Google Scholar] [CrossRef]
- Lee, H.; Kim, K.; Lee, S.; Han, M.; Yoon, Y. Growth kinetics of Staphylococcus aureus on Brie and Camembert cheeses. J. Dairy Res. 2014, 81, 252–256. [Google Scholar] [CrossRef]
- Mansur, A.R.; Park, J.H.; Oh, D.H. Predictive model for growth of staphylococcus aureus on raw pork, ham, and sausage. J. Food Prot. 2016, 79, 132–137. [Google Scholar] [CrossRef]
- Fujikawa, H.; Morozumi, S. Modeling Staphylococcus aureus growth and enterotoxin production in milk. Food Microbiol. 2006, 23, 260–267. [Google Scholar] [CrossRef]
- Huang, L. Growth of Staphylococcus aureus in Cooked Potato and Potato Salad—A One-Step Kinetic Analysis. J. Food Sci. 2015, 80, M2837–M2844. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Koseki, S.; Chung, M.J.; Oh, D.H. A Novel Approach to Predict the Growth of Staphylococcus aureus on Rice Cake. Front. Microbiol. 2017, 8, 1140. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Shim, Y.H.; Choi, N.J.; Ha, S.D.; Chung, M.S.; Hwang, I.G.; Oh, D.H. Mathematical modeling on the growth of Staphylococcus aureus in sandwich. Food Sci. Biotechnol. 2010, 19, 763–768. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Pearl, R. The growth of populations. Quart. Rev. Biol. II 1927, 4, 532–548. [Google Scholar] [CrossRef]
- Singh, G.; Marples, R.R.; Kligman, A.M. Experimental Staphylococcus aureus infections in humans. J. Investig. Dermatol. 1971, 57, 149–162. [Google Scholar] [CrossRef]
- Gil, M.M.; Miller, F.A.; Brandao, T.R.; Silva, C.L. On the use of the Gompertz model to predict microbial thermal inactivation under isothermal and non-isothermal conditions. Food Eng. Rev. 2011, 3, 17–25. [Google Scholar] [CrossRef]
- Dolan, K.D.; Mishra, D.K. Parameter estimation in food science. Annu. Rev. Food Sci. Technol. 2013, 4, 401–422. [Google Scholar] [CrossRef]
- Huang, L. Thermal inactivation of Listeria monocytogenes in ground beef under isothermal and dynamic temperature conditions. J. Food Eng. 2009, 90, 380–387. [Google Scholar] [CrossRef]
- McKellar, R.C.; Lu, X. (Eds.) Modeling Microbial Responses in Food; CRC Press: Boca Ranto, FL, USA, 2003. [Google Scholar]
- Mishra, D.K.; Dolan, K.D.; Yang, L. Confidence intervals for modeling anthocyanin retention in grape pomace during nonisothermal heating. J. Food Sci. 2008, 73, E9–E15. [Google Scholar] [CrossRef]
- Dolan, K.D.; Yang, L.; Trampel, C.P. Nonlinear regression technique to estimate kinetic parameters and confidence intervals in unsteady-state conduction-heated foods. J. Food Eng. 2007, 80, 581–593. [Google Scholar] [CrossRef]
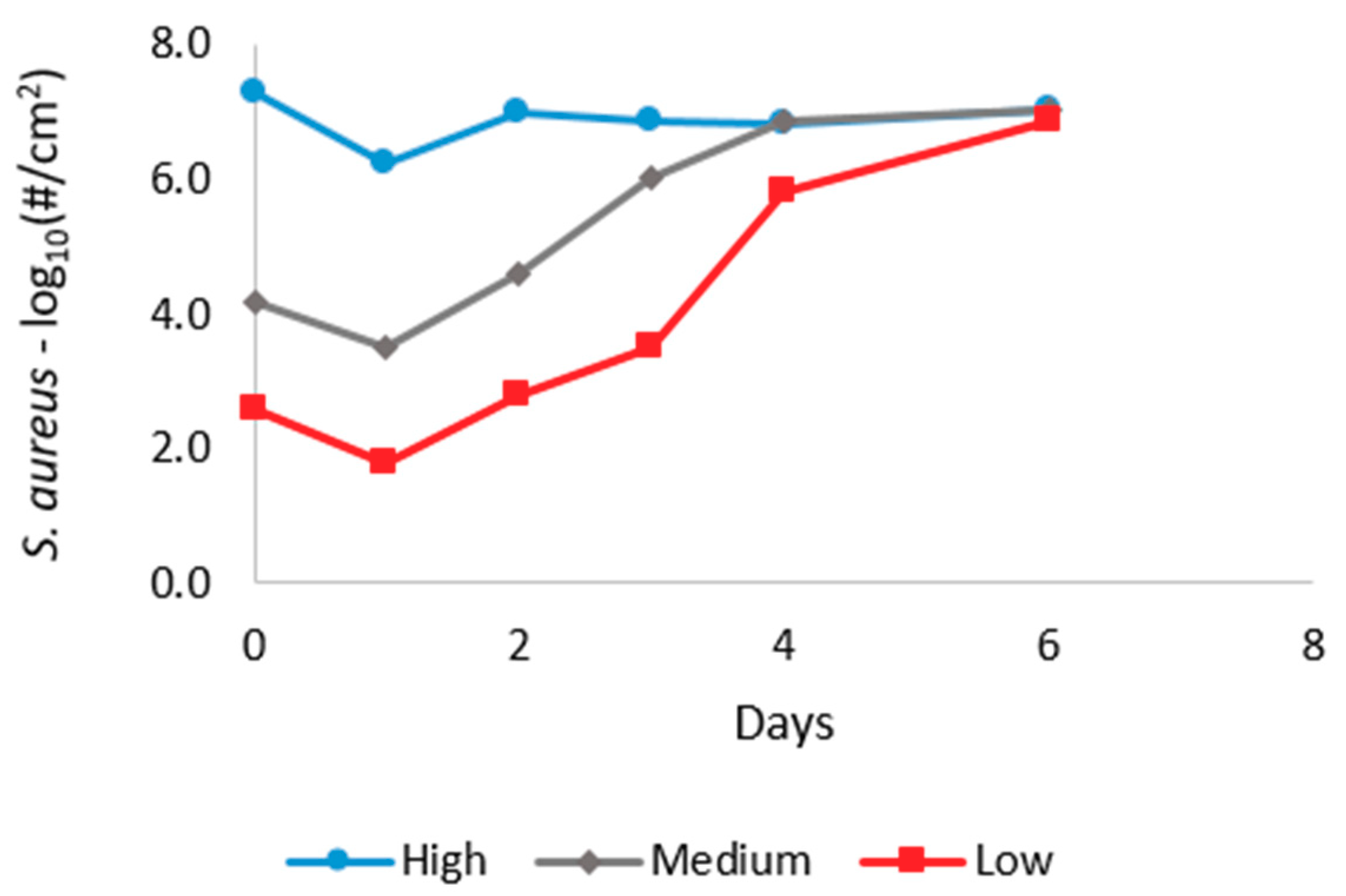
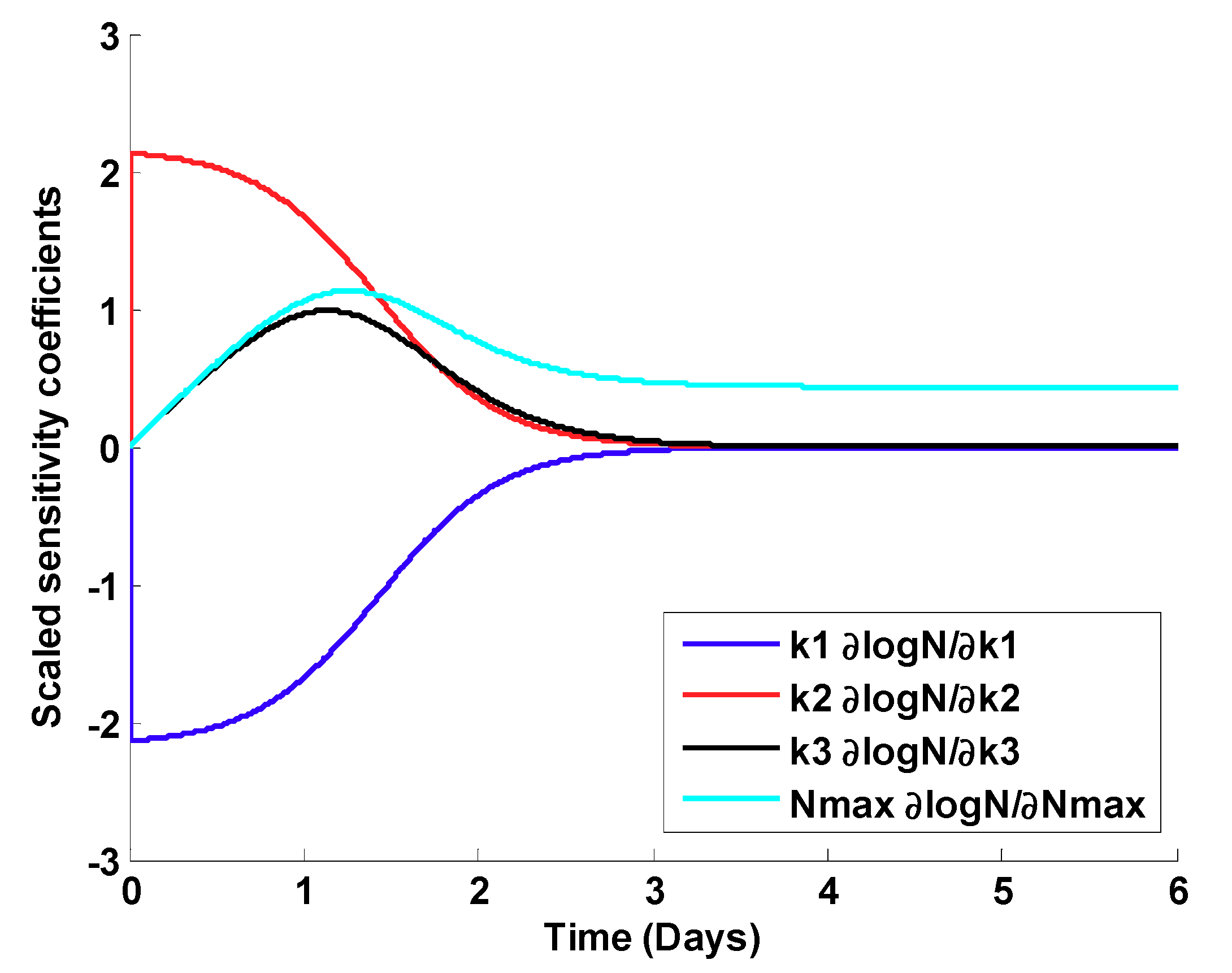
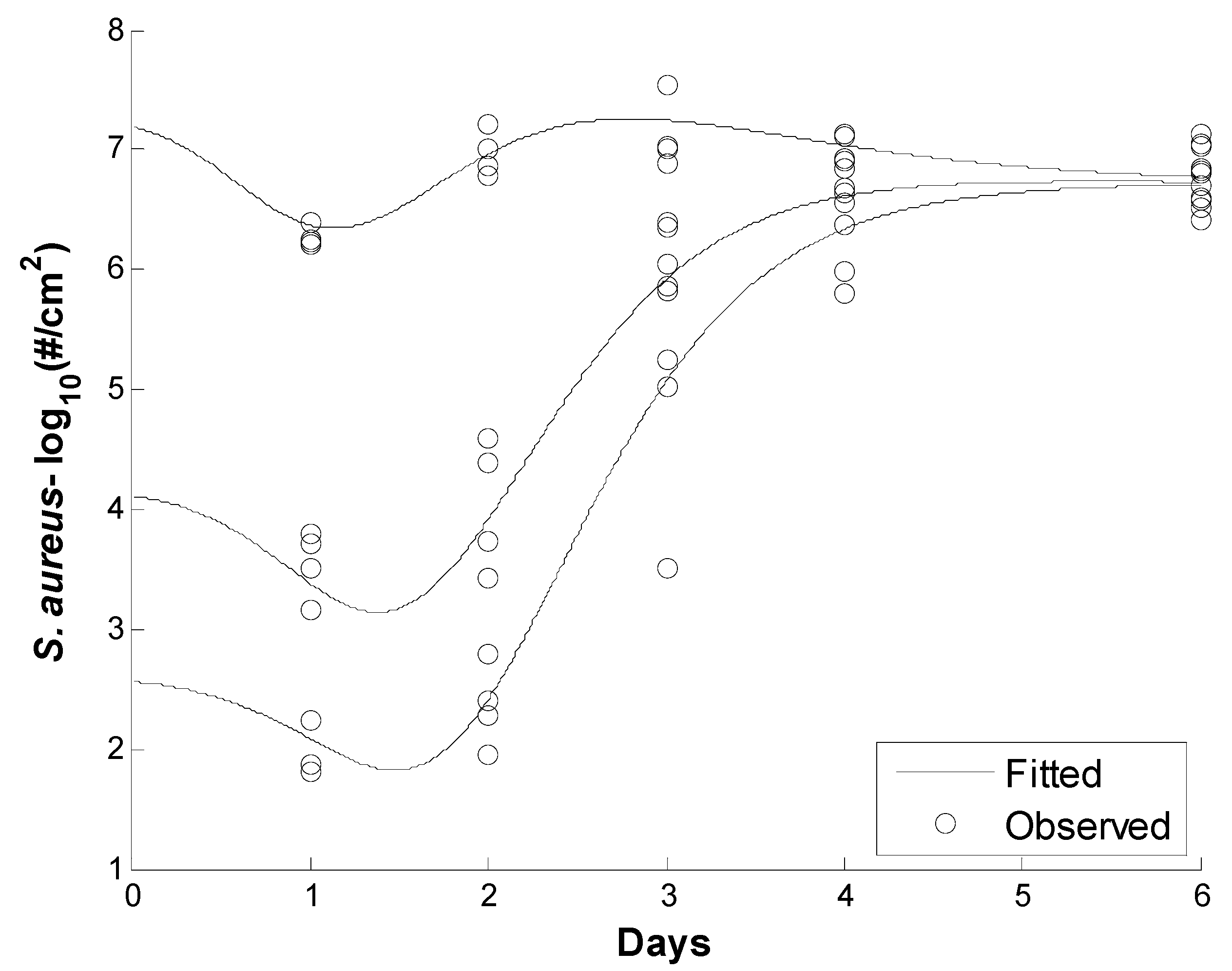

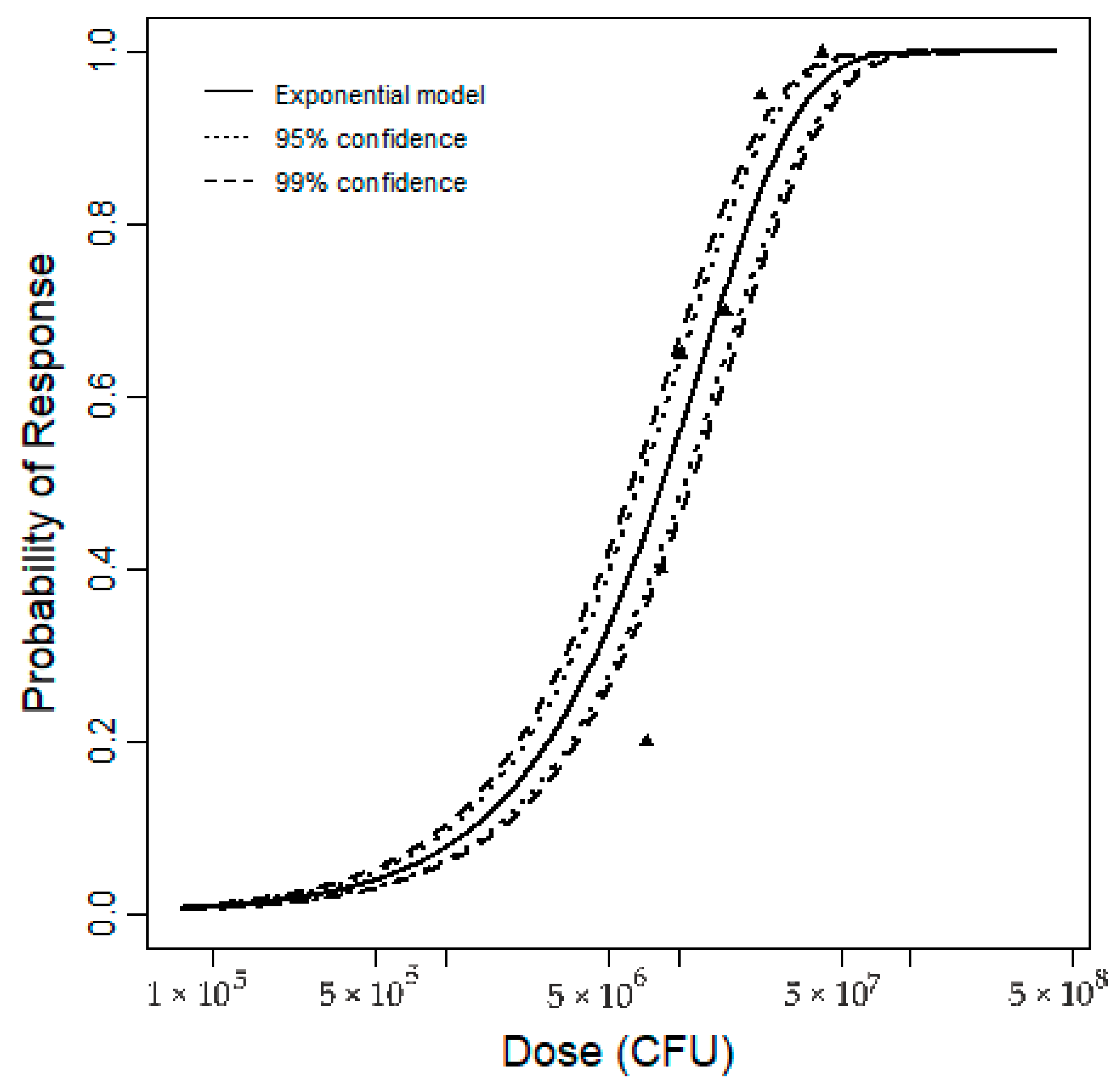
| K1 | K2 | K3 | Nmax | |
|---|---|---|---|---|
| K1 | 1 | Symmetric | ||
| K2 | 0.9932 | 1 | ||
| K3 | −0.0392 | −0.0943 | 1 | |
| Nmax | −0.2894 | −0.2788 | −0.7364 | 1 |
| Parameters | Estimate | 95% Confidence Interval | Relative Error (%) | |
|---|---|---|---|---|
| A | 1.00 | 0.94 | 1.05 | 2.63 |
| µ | 1.02 | 0.80 | 1.25 | 10.95 |
| M | 1.48 | 1.20 | 1.76 | 9.45 |
| C | 6.71 | 6.51 | 6.91 | 1.52 |
| B | 1.47 | 1.25 | 1.69 | 7.48 |
| β6 | 2.35 | 2.25 | 2.45 | 2.1 |
| β7 | 1.63 | 1.40 | 1.87 | 7.33 |
| A | µ | M | C | B | β6 | β7 | |
|---|---|---|---|---|---|---|---|
| a | 1.00 | ||||||
| µ | −0.42 | 1.00 | Symmetric | ||||
| M | −0.56 | −0.07 | 1.00 | ||||
| C | −0.08 | 0.37 | −0.26 | 1.00 | |||
| B | −0.24 | 0.57 | 0.18 | −0.28 | 1.00 | ||
| β6 | −0.07 | −0.37 | 0.51 | −0.04 | −0.13 | 1.00 | |
| β7 | −0.19 | −0.34 | 0.85 | −0.26 | 0.04 | 0.53 | 1.00 |
| Initial Dose (No./cm2) | Subjects with Infection | Total Subjects |
|---|---|---|
| 40 | 4 | 20 |
| 220 | 8 | 20 |
| 2000 | 13 | 20 |
| 105,000 | 14 | 20 |
| 1,600,000 | 19 | 20 |
| 10,000,000 | 20 | 20 |
| Integrated Dose (AUC) (Days × No./cm2) | Subjects with Infection | Total Subjects |
|---|---|---|
| 7.32 × 106 | 4 | 20 |
| 8.45 × 106 | 8 | 20 |
| 1.03 × 107 | 13 | 20 |
| 1.59 × 107 | 14 | 20 |
| 2.26 × 107 | 19 | 20 |
| 4.15 × 107 | 20 | 20 |
| Parameter | MLE Estimate | Percentiles | |||||
|---|---|---|---|---|---|---|---|
| 0.5% | 2.5% | 5% | 95% | 97.5% | 99.5% | ||
| k | 8.05 × 10−8 | 6.06 × 10−8 | 6.46 × 10−8 | 6.70 × 10−8 | 9.69 × 10−8 | 1.00 × 10−7 | 1.08 × 10−7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esfahanian, E.; Adhikari, U.; Dolan, K.; Mitchell, J. Construction of A New Dose–Response Model for Staphylococcus aureus Considering Growth and Decay Kinetics on Skin. Pathogens 2019, 8, 253. https://doi.org/10.3390/pathogens8040253
Esfahanian E, Adhikari U, Dolan K, Mitchell J. Construction of A New Dose–Response Model for Staphylococcus aureus Considering Growth and Decay Kinetics on Skin. Pathogens. 2019; 8(4):253. https://doi.org/10.3390/pathogens8040253
Chicago/Turabian StyleEsfahanian, Elaheh, Umesh Adhikari, Kirk Dolan, and Jade Mitchell. 2019. "Construction of A New Dose–Response Model for Staphylococcus aureus Considering Growth and Decay Kinetics on Skin" Pathogens 8, no. 4: 253. https://doi.org/10.3390/pathogens8040253
APA StyleEsfahanian, E., Adhikari, U., Dolan, K., & Mitchell, J. (2019). Construction of A New Dose–Response Model for Staphylococcus aureus Considering Growth and Decay Kinetics on Skin. Pathogens, 8(4), 253. https://doi.org/10.3390/pathogens8040253







