Abstract
The Pectobacterium genus comprises pectolytic enterobacteria defined as the causal agents of soft rot, blackleg, and aerial stem rot diseases of potato and economically important crops. In this study, we undertook extensive genome-wide comparative analyses of twelve species that conform the Pectobacterium genus. Bioinformatics approaches outlined a low nucleotide identity of P. parmentieri and P. wasabiae with other species, while P. carotovorum subsp. odoriferum was shown to harbor numerous pseudogenes, which suggests low coding capacity and genomic degradation. The genome atlases allowed for distinguishing distinct DNA structures and highlighted suspicious high transcription zones. The analyses unveiled a noteworthy heterogeneity in the pathogenicity determinants. Specifically, phytotoxins, polysaccharides, iron uptake systems, and the type secretion systems III–V were observed in just some species. Likewise, a comparison of gene clusters encoding antimicrobial compounds put in evidence for high conservation of carotovoricin, whereas a few species possessed the phenazine, carbapenem, and carocins. Moreover, three clustered regularly interspaced short palindromic repeats-Cas (CRISPR-Cas) systems: I-E, I-F, and III-A were identified. Surrounding some CRISPR-Cas regions, different toxin and antitoxin systems were found, which suggests bacterial suicide in the case of an immune system failure. Multiple whole-genome alignments shed light on to the presence of a novel cellobiose phosphotransferase system (PTS) exclusive to P. parmenteri, and an unreported T5SS conserved in almost all species. Several regions that were associated with virulence, microbe antagonism, and adaptive immune systems were predicted within genomic islands, which underscored the essential role that horizontal gene transfer has imparted in the dynamic evolution and speciation of Pectobacterium species. Overall, the results decipher the different strategies that each species has developed to infect their hosts, outcompete for food resources, and defend against bacteriophages. Our investigation provides novel genetic insights that will assist in understanding the pathogenic lifestyle of Pectobacterium, a genus that jeopardizes the agriculture sustainability of important crops worldwide.
1. Introduction
Members of Pectobacterium genus are defined as plant pathogenic enterobacteria that are responsible for the most devastating diseases, including common soft rot, blackleg, wilt, and aerial stem rot [1,2]. Strains of this genus cause severe losses worldwide in field crops, diverse vegetables, fruits, and ornamental plants [3,4]. Pectobacterium ranks among the top 10 of the most hazardous plant pathogenic bacteria due to its destructiveness impact that is exerted in agriculture fields as well as during storage and transportation [5], especially on potato, the third main staple food crop [6] and the major host of several Pectobacterium species [4,7]. Species of this genus have been isolated from infected tissues, soil, ground water, and can be associated with fruit flies, which aids in their rapid dissemination [8,9].
In regards to its taxonomy, the Pectobacterium genus has been restructured and subjected to constant revisions. First characterized at the beginning of the 20th century, Pectobacterium was once considered to be part of the Erwinia genus. Subsequent analyses that were based upon 16S rDNA revealed three major phylogenetic clusters; as a result, Brenneria and Pectobacterium were proposed and conceived as new genera independent from Erwinia [10]. At that time, Pectobacterium harbored eight species: P. carotovorum subsp. atrosepticum, P. c. subsp. betavasculorum, P. c. subsp. carotovorum, P. c. subsp. odoriferum, P. c. subsp. wasabiae, P. cacticidum, P. chrysanthemi, and P. cypripedii [10]. In 2003, three subspecies were elevated to the species level giving rise to P. atrosepticum, P. betavasculorum, and P. wasabiae [11]. Concurrently, highly virulent Brazilian strains were isolated from potato and named P. c. subsp. brasiliense [12]. Later, P. chrysanthemi was reclassified as Dickeya chrysanthemi [13], whereas P. cypripedii was transferred to the genus Pantoea [14]. Years later, the causal agent of canker in Actinidia chinensis was isolated and named P. c. subsp. actinidiae [15]. Furthermore, some isolates with preference for monocotyledonous hosts were designated as P. aroidearum [16]. In a similar way, P. wasabiae was split and P. parmentieri was established [17]. Recently, strains that were previously confirmed as P. carotovorum were re-classified and established as P. polaris [18], P. peruviense [19], and Candidatus P. maceratum [20]. At the time of writing, Pectobacterium is comprised of thirteen species, although there was not even a draft genome of P. cacticidum in any databases.
The host range of these thirteen species is quite divergent. P. betavasculorum, P. cacticidum, P. wasabiae, and P. c. subsp. actinidiae exhibit a narrow host range, as they are restricted to sugar beet, cacti, horseradish, and kiwifruit, respectively [11,15,17]. P. aroidearum shows a preference for aroid hosts [16]. P. atrosepticum and P. parmentieri seem to be limited to potato [17,21]. Likewise, P. peruviense, P. polaris, and C. P. maceratum so far have only been found in potato. In contrast, P. c. subsp. carotovorum and P. c. subsp. brasiliense have been isolated from a great variety of vegetables (tomato, pepper, cucumber, eggplant, carrot, lettuce, onion, and Chinese cabbage), as well as other solanaceous hosts, including potato and tobacco [22,23,24]. P. c. subsp. odoriferum was thorough to affect solely chicory, but posterior studies confirmed this pathogen in different vegetables [25].
Each Pectobacterium species exerts their characteristic symptoms, such as slimy, wilting, black rot, and gummosis through the deployment of pathogenicity determinants, which suppress the plant defense mechanisms. The secretion systems, proteases, the plant cell wall degrading enzymes (PCWDE), effector molecules, flagella-based motility, iron acquisition systems, polysaccharides, cell attachment proteins, the type IV pilus, phytotoxins, 3-hydroxy-2-butanone pathway (3H2B), and other virulent elements that are encoded by individual genes comprise these virulence-associated promoters [2,26,27]. Pectobacterium sp. produce specific compounds that display antimicrobial activity, either with a broad antibiosis spectrum, like carbapenem and phenazine, or with a narrow effect, including carotovoricin and carocins [21,28,29,30,31,32]. The transcription of pathogenicity factors and antimicrobial components is controlled by regulators that operate jointly as a complex network.
High throughput sequencing has unveiled a set of genomic data to conduct bacterial comparative genomic analyses. P. atrosepticum SCRI1043 was the first soft rot bacterium being sequenced [21]. Later, the complete genomes of P. c. subsp. carotovorum PCC21 [33], P. parmentieri SCC3193 [26], P. wasabiae CFBP 33304 [17], P. carotovorum SCC1 [34], P. polaris [18], and P. c. subsp. brasiliense BZA12 [24] were analyzed. At the time of writing this article (September 2018), 18 complete and 90 high quality draft genomes of the Pectobacterium species have been published in the GenBank genome database. The easy access of several published genomes illuminates new strategies for exploring the genetic datasets, giving rise to comparative genomics [35,36]. Bacterial genome comparison is a powerful tool for studying evolutionary biology, taxonomy, phylogeny, and it can provide insights to distinct niches adaptation [35,37]. Furthermore, comparing genomes has allowed for identifying the core repertoire of virulence genes shared by related organisms and, likewise, to locate gene clusters or genome islands exclusive of species or even unique of a strain [38]. Putative genes that promote specific interactions between bacteria and their cognate plant hosts have also been uncovered by using comparative genomics approaches [35]. To date, few comparison studies have been conducted regarding Pectobacterium [8,24,27,39]. However, none of them have compared the virulence mechanisms, antimicrobial gene clusters, and the diversity of the immune system clustered regularly interspaced short palindromic repeats-Cas (CRISPR-Cas) across all species that currently comprise the Pectobacterium genus in detail.
When considering this background, the primary purpose of the present investigation was to develop an extensive genome-wide comparative analysis among the 12 species of the genus Pectobacterium. First, we evaluated the main genomic features and DNA structural properties of genome atlases in all species. Second, we assessed the current taxonomy within the Pectobacterium genus based upon the average nucleotide identity (ANI), the DNA-DNA hybridization (dDDH), phylogenetic tree, proteome comparison, codon and amino acid usage profiles, and core-genome dendrogram. Third, we compared the sequence organization and arrangements of gene clusters defined as pathogenicity determinants and those clusters that are implicated in the synthesis of antimicrobial compounds. Fourth, we in silico identified the distinct CRISPR-Cas systems harbored in 34 Pectobacterium genomes. Our data aim to shed light on novel genetic regions and elucidate remarkable divergences that are involved in pathogenesis, antimicrobial components, and immune adaptative system (CRISPR-Cas), which will contribute to understanding the evolutionary dynamics and virulent lifestyle of Pectobacterium, a complex and heterogeneous genus that is responsible for disastrous economic losses in important agricultural crops.
2. Materials and Methods
2.1. Strains Information, Genome Sequencing, Assembly and Annotation
All of the genome assemblies used in this study were retrieved from NCBI GenBank genome database, and Table S1 provides details. The strains that were used for genome sequencing originated worldwide, including Scotland, China, Belarus, South Korea, Denmark, Peru, Finland, Canada, France, Israel, Pakistan, Russia, Japan, Norway, the Netherlands, and the United States of America (Table S1); the strains were isolated from Solanum tuberosum, Brassica rapa, Cucumis sativus, Apium graveolens, Cichorium intybus, Actinidia deliciosa, Ornithogalum dubium, Eutrema wasabi, Beta vulgaris, and Brassica oleracea (Table S1). The genomes were sequenced while using different sequencing platforms from Illumina MiSeq (Illumina Inc., San Diego, CA, USA) to PacBio (Pacific Biosciences of California, Inc., Menlo Park, CA, USA); PHRAP, SOAPdenovo, SPAdes, Newbler, Velvet and HGAP (Hierarchical Genome Assembly Process) were used to assemble the genomes (Table S1). The genomes of two neighbor species, Dickeya zeae and D. solani, were also included for phylogenomic and pairwise nucleotide identity. Geneious 10.2.4 was used to visualize and analyse the genome sequences and feature annotations. The Reference Sequence (RefSeq) numbers for either the International Nucleotide Sequence Database Collaboration (INSDC) or Whole-genome-sequence (WGS) projects were used to download the genomes.
2.2. Phylogenomic Analyses
With the aim of assessing the level of discrepancy among the Pectobacterium species, the Average Nucleotide Identity (ANI) based on the Nucleotide MUMmer algorithm (ANIm) was computed and then determined in the JSpecies Web Server with default parameters [40]. Additionally, the in silico DNA-DNA hybridization (dDDH) or genome-to-genome distance between the genomes were calculated while using the Genome-to-Genome Distance Calculator (GGDC) version 2.1 with the recommended formula 2 and the BLAST + alignment criteria [41,42]. ANI and dDDH data were both compiled in a single matrix and visualized as a color-coded heatmap while using MORPHEUS (https://software.broadinstitute.org/morpheus). The cut-off values of 95% and 70% were assigned as a species delineation framework for ANI and dDDH, respectively.
A comprehensive phylogenetic relationship across the genus Pectobacterium was established by aligning ~18 kb of genome region comprising nucleotide sequences from 12 housekeeping genes, namely chromosomal replication initiator protein DnaA (dnaA—1382 bp), DNA polymerase III subunit beta (dnaN—1101 bp), DNA polymerase III subunit gamma/tau (dnaX—1101 bp), elongation factor G (fusA—2102 bp), glyceraldehyde-3-phosphate dehydrogenase A (gapA—996 bp), DNA gyrase subunit A (gyrA—2633 bp), DNA gyrase subunit B (gyrB—2411 bp), DNA recombination and repair protein RecA (recA—1074 bp), DNA repair protein RecN (recN—1661 bp), DNA-directed RNA polymerase subunit alpha (rpoA—990 bp), RNA polymerase sigma factor RpoD (rpoD—1826 bp), and RNA polymerase sigma factor RpoS (rpoS—990 bp). The alignment was individually performed for each gene while using the MUSCLE algorithm [43]. Afterward, all of the individual alignments from 12 genes were concatenated using Geneious. Lastly, the phylogenetic analysis based on the concatenated sequences was performed while using MEGA X [44]; evolutionary history along the Pectobacterium genus was inferred using the Neighbor-Joining clustering algorithm [45]. The Maximum Composite Likelihood method was applied for determining the evolutionary distances among the isolates [46]. The bootstrap test based on 1000 replicates was computed to estimate reliability [47], and the associated taxa clustered were expressed as percentage units next to the branches of outputted tree. In addition, two closest taxon D. solani IPO2222, as well as D. zeae EC1, were included in this study as an outgroup for obtaining a comprehensive taxonomic position within the Pectobacterium species.
2.3. Basic Statistics and Genomic Profile Features
The basic parameters for each genome, such as GC content, number of coding sequence regions (CDS), tRNA genes, pseudogenes, and others features (Table 1), were fetched from the data that were available in both the NCBI GenBank database and the Bioinformatic Resource Center PATRIC [48,49]. Additionally, genome atlases were only constructed for complete genomes to illustrate the different structural components of each genome sequence, such as GC skew, stacking energy, intrinsic curvature, position preference, global direct, and indirect repeats. The previous parameters were visualized and drawn as a circle plot by using the script atlas_createConfig while using CMG-Biotools [50].

Table 1.
General genome characteristics described for Pectobacterium species.
2.4. General Genomic and Proteome Analyses
A comparative genomic ring plot was executed using BLAST Ring Image Generator (BRIG) [51] to illustrate the degree of homology among the twelve Pectobacterium species. P. atrosepticum SCRI1043 genome was taken as reference for the nucleotide BLAST genome comparison. In proteomics analyses, an overall pairwise proteome comparison that was based on BLAST [52] was performed to construct a BLAST matrix, which is a visual graphic showing the total number of proteins common within and between a given set of proteomes [53]. The entire analyses were performed while using the CMG-Biotool package. The script matrix_createConfig was used to create a BLAST matrix set-up file in an XML format; this file was used as an input data file to plot the BLAST matrix using the program ‘matrix’. Finally, the generated matrix plot was visualized as a color scale heatmap that represented the percentage wise numerical homology values across all compared proteomes. BLAST hits with at least 50% of identical matches and 50% of coverage were considered to be significant.
2.5. Codon and Amino Acid Usage and Pan-Core Genome Analyses
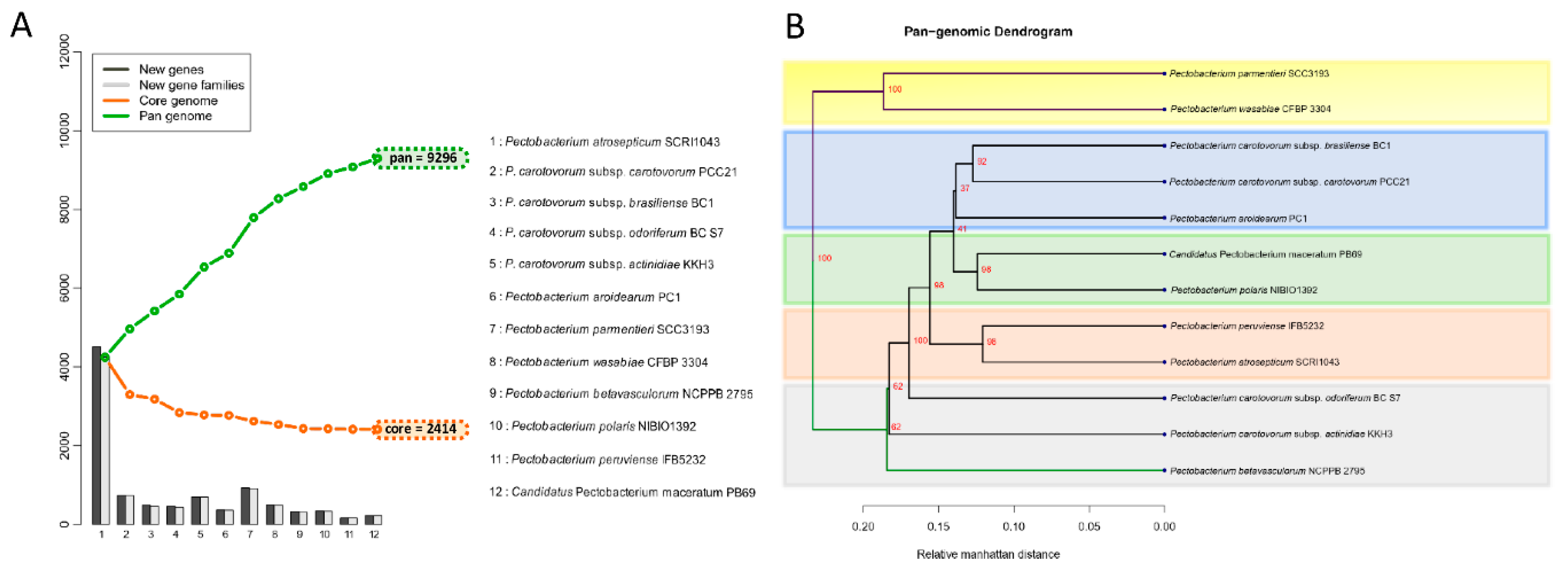
The comparative analyses that were based on the amino acid and codon usage between the Pectobacterium species were determined using BioPerl modules and implemented with the script stats_usage using CMG-Biotools package. Two heatmaps depicted the different clustering patterns across the Pectobacterium genomes, in terms of amino acid and codon usage, were generated in R: The R Project for Statistical Computing (https://www.r-project.org/). The number of Pan and Core genes across the Pectobacterium species were analyzed and calculated while using the BLAST algorithm [52] with 50% of cutoff values for either cover or identity percentage parameters. The output plot was constructed using CMG-Biotools pipeline package and then implemented in the program ‘pancoreplot’. Afterward, a pan-genome phylogenetic tree that was based upon those gene families shared between all Pectobacterium species was created and plotted using the script pancoreplot_tree [50].
2.6. Genomic Islands (GI) Prediction
The Integrative Conjugative Elements (ICE), such as Horizontal Acquired Islands (HAI), were studied and predicted using the IslandViewer 4 webserver [54], which allowed for an interactive visualization of genomic islands (GI) as either in circular or linear plots by integrating four different and accurate predictor tools IslandPath-DIMOB [55], SIGI-HMM, IslandPick [56], and Islander [57].
2.7. Analyses of Pathogenicity, Virulence, Antimicrobial Gene Clusters and CRISPR-Cas
The clusters that were involved in diverse virulence and pathogenicity events, including type secretion systems (TSS), phytotoxins, iron uptake, polysaccharides biosynthesis, flagella encoding genes, cell attachment, and agglutination were screened and compared across all the Pectobacterium species. The clusters comparison was performed while using Proteome Comparison tool of the Pathosystems Resource Integration Center (PATRIC) web server [49]. Two separate genome comparison tables were generated for P. atrosepticum SCRI1043 and P. parmentieri SCC3193, since both species were used as reference genomes. On the other hand, the biosynthetic gene clusters encoding secondary metabolites related to the synthesis of phytotoxins, antibiotics and antimicrobial compounds were predicted while using antiSMASH 4.0 [58]. In addition, BAGEL4 was applied as a genome mining tool for identifying putative gene clusters involved in bacteriocin biosynthesis [59]. Finally, the synteny and different rearrangements between the genomic clusters were visualized as linear arrow comparison plots, generated while using Easyfig v2.2.3 [60]. Circa was used to illustrate a summary of different clusters involved in different functions of Pectobacterium species, their shared homology, and connections (http://omgenomics.com/circa). The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) arrays, as well as the type of CRISPR associated proteins (Cas) systems, were predicted while using CRISPRCasFinder [61]. CRISPRs loci features, such as total size, number of spacers and direct repeats, length, and consensus sequence of direct repeats were also analyzed and corroborated by using CRISPRDetect [62] and the NCBI GenBank database. The different CRISPR-Cas systems that were analyzed in this study were illustrated and manually drawn.
3. Results
3.1. General Genomic Features and DNA Structure Properties
Overall genomic features, such as genome size, GC (guanine-cytosine) content, conserved coding sequences, number of RNA genes, pseudo-genes, proteins with assigned function in metabolic pathways, as well as virulence factors and antibiotic resistance genes, were retrieved from either NCBI GenBank database or PATRIC [49]. The corresponding data set of the twelve species: Pectobacterium atrosepticum SCRI1043, P. carotovorum subsp. carotovorum PCC21, P. carotovorum subsp. brasiliense BC1, P. carotovorum subsp. odoriferum BC S7, P. carotovorum subsp. actinidiae KKH3, P. arodiearum PC1, P. parmentieri SCC3193, P. wasabiae CFBP 3304, P. betavasculorum NCPPB 2795, P. polaris NIBIO 1392, P. peruviense IFB5232, and Candidatus Pectobacterium maceratum PB69, was compiled in a single table (Table 1). The length of the genomes ranged between 5.16 Mb to 4.68 Mb, with P. parmentieri and P. betavasculorum, possessing the largest and smallest genomes, respectively. The GC content percent was almost same among Pectobacterium species (around 51%); P. c. subsp. carotovorum showed maximum GC content of 52.2%. The highest numbers of conserved coding sequence (CDS) were observed in P. pamentieri, with 4,449 confirmed gene sequences, whereas the lowest number of CDS were found in P. c. subsp. odoriferum with 3,855 confirmed gene sequences. However, P. c. subsp. odoriferum was found to maintain the highest number of pseudogenes (385) among the Pectobacterium species (Table 1). The rRNA and tRNA genes were similar in all species, except for P. peruviense, which expressed the lowest number of 5S RNA, 16S RNA, and 23S RNA genes (2, 1, 2, respectively). P. parmentieri and P. peruviense showed the highest number of proteins with an assigned metabolic function (835 CDS), followed by P. atrosepticum (829 CDS). The highest known virulence and predicted antibiotic resistance CDSs were found in P. parmentieri and P. atrosepticum, each possessing 53 and 60 genes, respectively. The recently identified species C. P. maceratum possesses the highest number of 35 CDS for drug targets.
Genome atlases were generated for eight complete genomes to contrast relevant characteristics based upon structural DNA properties (Figure S1). Atlases were not created for P. c. subsp. actinidiae, P. betavasculorum, P. peruviense, and C. P. maceratum due to the unavailability of the complete genomes. GC skew pointed out the start and the end point of replication and displayed as blue and pink for G and C content, respectively; generally, Gs represent the leading strand. Among the atlases, P. atrosepticum harbored the most global direct and indirect repeats (24 blue and 18 red lines), whereas the atlas of P. aroidearum only presented 13 direct and seven invert global repeats. The position reference parameter is a value that associates DNA regions of greater rigidity (0.15 and featured in pink in the atlas) or flexibility (0.14 and featured with a green line in the atlas), according to the position preference of the DNA within the nucleosome [50]. The DNA atlas of P. aroidearum extends over 59 regions with high positional preference, as indicated by characteristic rigidity that appears pink on the atlas.
The stacking energy, on the other hand, is a -3.82 kcal/mol to 14.59 kcal/mol based parameter, which allows for distinguishing between the regions that were susceptible to de-stacking (less negative values or closer to 0) and regions of greater stability (more negative values) [50]. The intrinsic curvature is a parameter that quantifies the degree of DNA wrapping around a histone core and it ranges between the values of 0 and 1: P. c. subsp. carotovorum and P. parmentieri exhibited the strongest and weakest wrapping intensity, respectively (as indicated with the prevalence of blue lines). Pink, red, and dark blue colors in the atlases highlight specific regions with high positions preference, stacking energy, and high intrinsic curvature characteristics.
3.2. Phylogenetic Relationships within Pectobacterium Genus
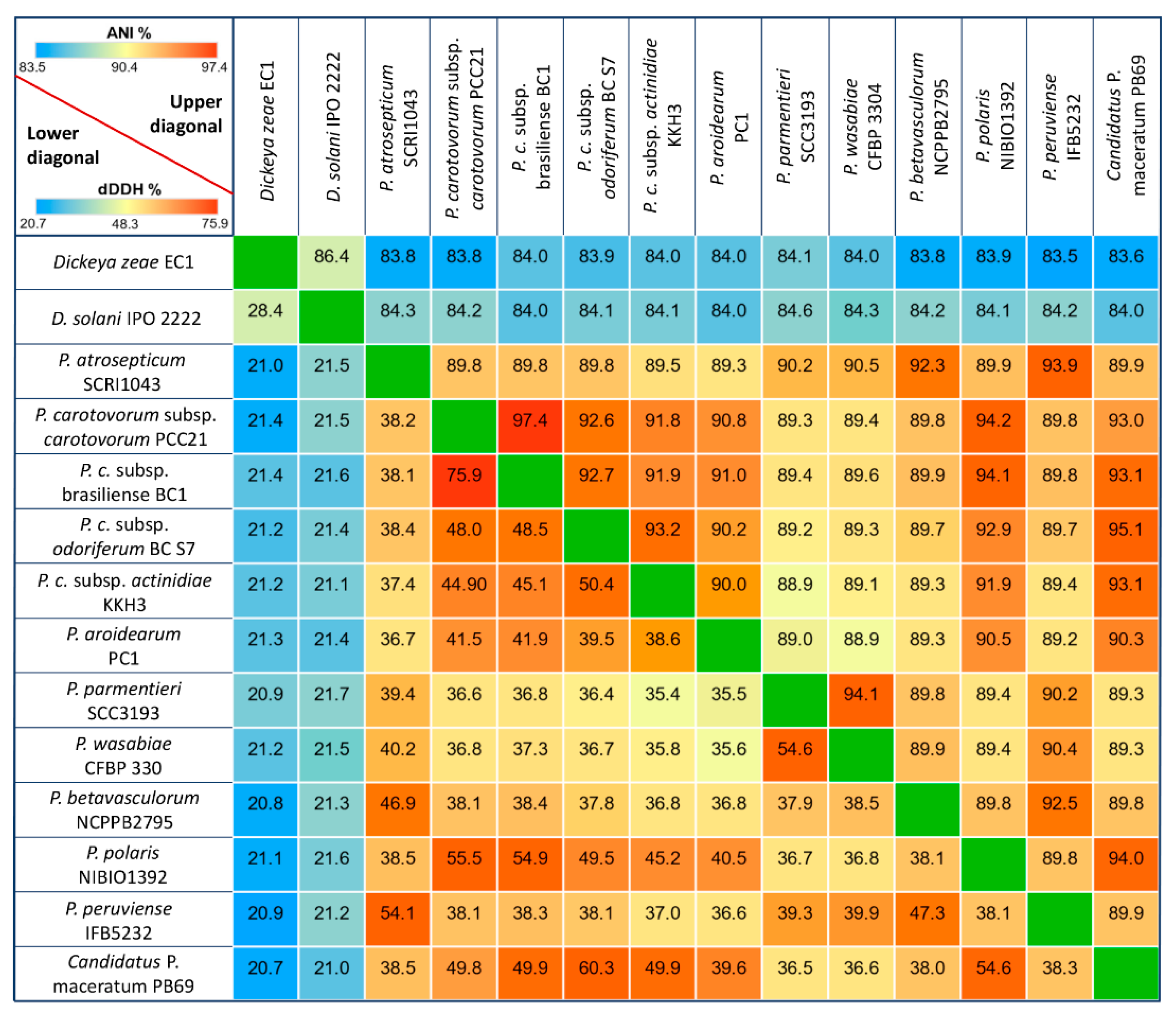
The genome homology between the Pectobacterium and Dickeya taxa oscillated from 83.5 to 97.4% based on ANI analysis effectuated with the MUMmer algorithm (Figure 1) in the JSpecies software. The highest degree of genome similarity was observed between P. carotovorum subsp. carotovorum PCC21 and P. carotovorum subsp. brasiliense with an ANI value of 97.4%, and related species P. parmentieri SCC3193 and P. wasabiae CFBP 3304 exhibited a high homology of 94.1%. On the other hand, P. parmentieri presented the highest divergence with subspecies P. c subsp. actinidiae KKH3, showing a low ANI value of 88.9%.
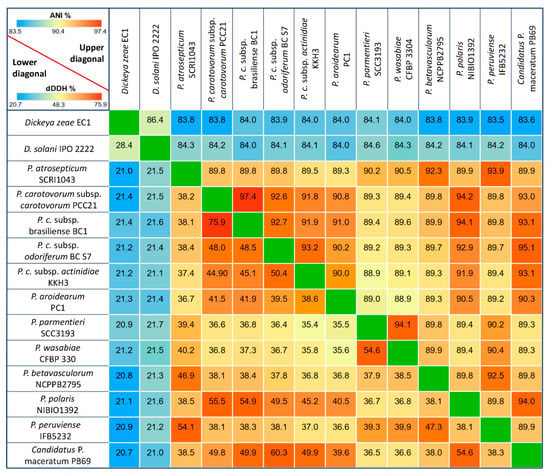
Figure 1.
Pairwise heatmap of Pectobacterium species based on average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH). Both ANI and dDDH are represented as percentage values among the twelve Pectobacterium species and two Dickeya species (D. zeae EC1 and D. solani IPO 2222). The upper diagonal displays ANI data, whereas the lower diagonal depicts the in silico dDDH data. The cut-off values for species delineation are 95 and 70% for ANI and dDDH, respectively.
The digital DNA-DNA hybridization (dDDH) analysis allows for inferences to be drawn between the sequenced genomes relative to genome distances and provides analogues data within the range of 20.7 (with Dickeya species) to 75.9%, a value that is consistent with the ANI results (Figure 1). In general, all of the dDDH values were below 70%. Additionally, the lowest dDDH values were observed by comparing the genome distances between the Dickeya and Pectobacterium isolates. Likewise, a 60.3% genome-to-genome distance was estimated between C. P. maceratum and P. c susbp. odoriferum. A divergence trend was also observed for P. parmentieri and P. wasabie with P. c. subsp. actinidiae and P. aroidearum, respectively.
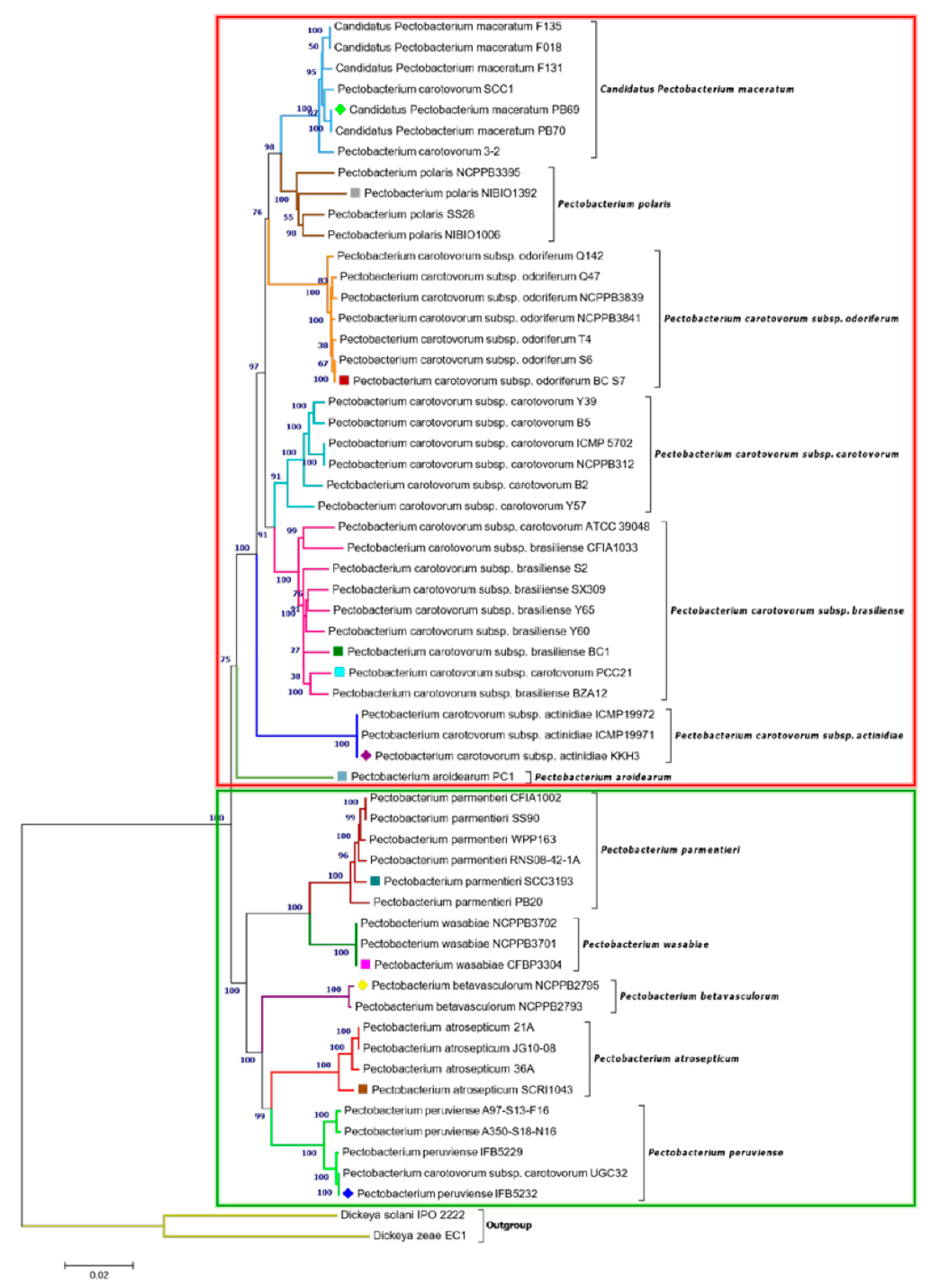
In addition, a phylogenetic tree that was based on concatenated sequences of twelve housekeeping genes (dnaA, dnaN, dnaX, fusA, gapA, gyrA, gyrB, recA, recN, rpoA, rpoD, and rpoS) was constructed, as in Figure 1. with MEGA X using the Neighbor-Joining method (Figure 2) to evaluate the current evolutionary relationships among all Pectobacterium species described to date. The 12 gene sequences of 57 Pectobacterium strains and 2 Dickeya species (used as out-group) were extracted from the respective species genomes, retrieved from NCBI GenBank genome database (Table S1), and aligned using MUSCLE algorithm. The generated dendogram revealed two main clades: the first clade (red dashed square) featured an aggregate of the P. carotovorum subspecies as well as C. P. maceratum, P. polaris, and the aroids plant pathogen, P. aroidearum; the second clade (green dashed square) featured the species P. parmentieri, P. wasabiae, P. betavasculorum, P atrosepticum, and P. peruviense (Figure 2). In clade I, a major cluster was formed from two sub-clusters, while P. aroidearum PC1 was isolated. The first sub-cluster allocated the genomespcies C. P. maceratum as a close relative of P. polaris. In the phylogenetic tree below, the node of the P. polaris and C. P. maceratum strains lineage with P. c. subsp. odoriferum; it can be concluded that the three species evolved from a common ancestor.
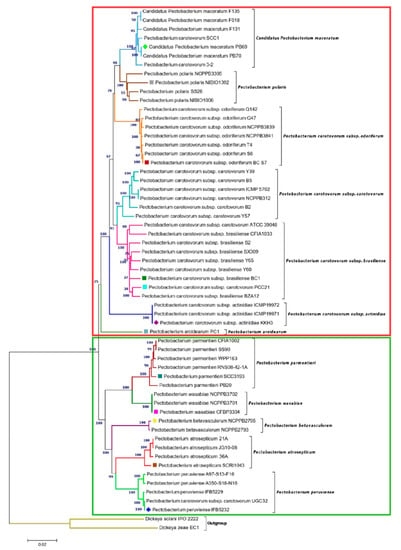
Figure 2.
Concatenated phylogenetic tree illustrating the relationships among all Pectobacterium species. A total of 59 strains from all known Pectobacterium species were selected to generate Neighbor-Joining tree based on 12 housekeeping genes: dnaA, dnaN, dnaX, fusA, gapA, gyrA, gyrB, recA, recN, rpoA, rpoD, and rpoS. The alignment of individual genes was performed using MUSCLE, and a concatenated tree was subsequently generated after aligning all 12 genes (total length ~ 18,269 bp). The Maximum Composite Likelihood method was applied to determine the evolutionary distances, with each node being supported by a bootstrap of 1,000 replicates to assess reliability. Bootstrap values are displayed next to the branches. The branches were colored to highlight the distinct strains clustered in each species. The dendrogram was drawn to scale, and all positions containing gaps and missing data were eliminated. DNA sequences of D. zeae EC1 and D. solani IPO 2222 were included as out-groups to root the tree. The evolutionary analysis was developed in MEGA X. Colored square and diamond shapes indicate the complete and draft genomes included in the study, respectively. Two main clades were identified and marked with dark red and green rectangles.
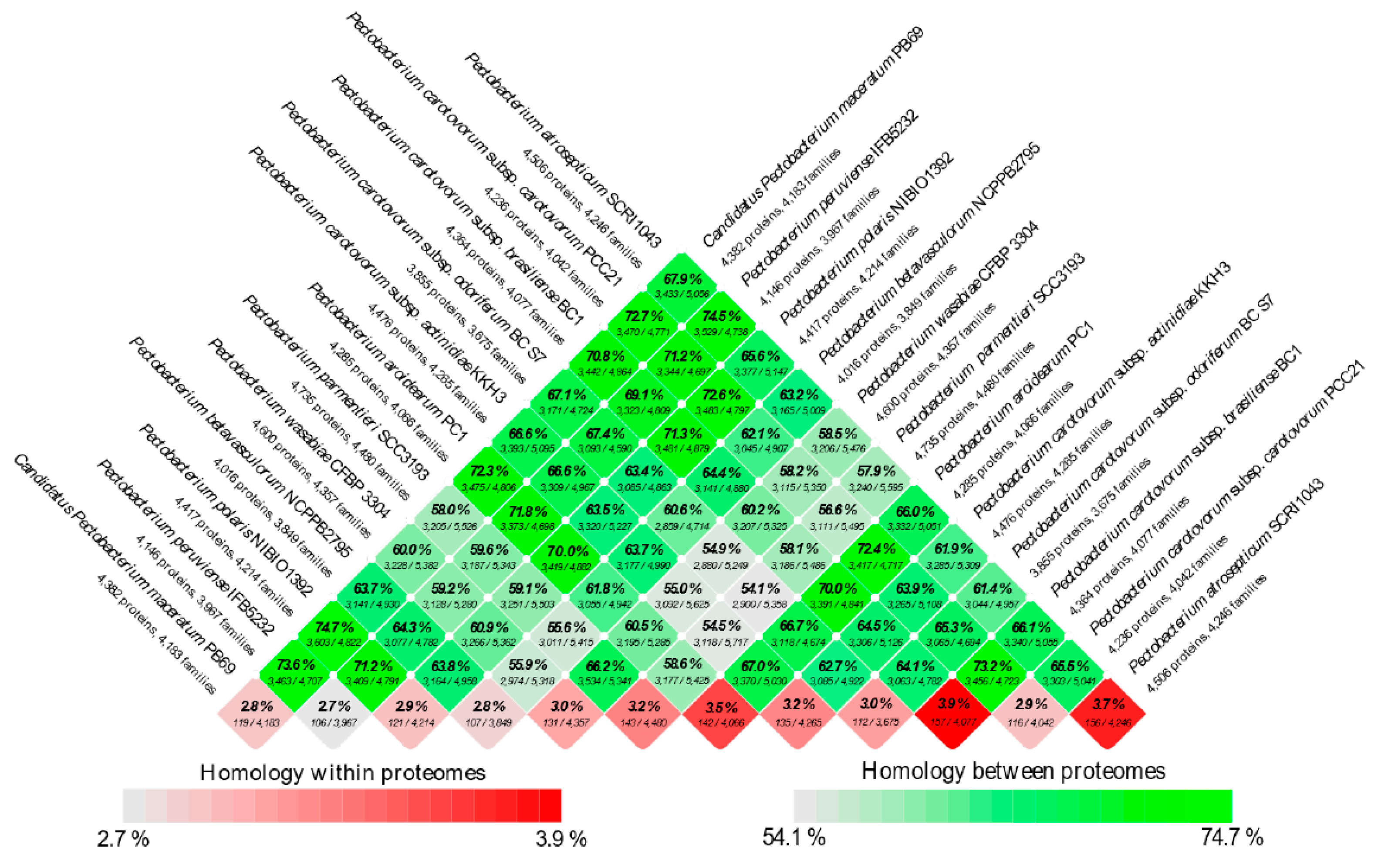
The second sub-cluster of the clade I grouped P. c. subsp. carotovorum and P. c. subsp. brasiliense. Interestingly, two strains, PCC21 and ATCC 39048, available in the NCBI as P. c. subsp. carotovorum, clustered within the group of P. c. subsp. brasiliense strains, suggesting a misidentification of these both strains. The three isolates of kiwi P. c. subsp. actinidiae clustered in a separate node of the two sub-clusters mentioned above. Likewise, two distinct clusters can be observed in clade II. The first cluster comprised P. parmentieri and P. wasabiae, as expected. The second cluster included the strains from P. betavasculorum, P. atrosepticum and P. peruviense. In this cluster, the newly categorized species, P. peruviense formed a sub-cluster along with P. atrosepticum, whereas P. betavasculorum split into a different lineage. This outcome is concordant with that of ANI and dDDH. Based on analyses, it can be concluded that P. peruviense and P. atrosepticum both co-evolved, but eventually diverged into two separate species. The pairwise comparison of total protein coding genes among Pectobacterium species demonstrated a range of 54.1% to 74.7% shared proteins, with the lowest value being represented between P. c. subsp. odoriferum and P. parmentieri, and the highest between P. polaris and C. P. maceratum (Figure 3). Likewise, the proteins (3,855) and protein families (3,675) that were used for the comparison were the lowest for P. c. subsp. odoriferum (Figure 3). In agreement with ANI, dDDH, and phylogenetic analysis outcomes, P. peruviense shared 74.5% proteins in common with P. atrosepticum. The sugar beet pathogen, P. betavasculorum, showed the least similarity (55.6% to 64.4%) when compared to the rest of the species.
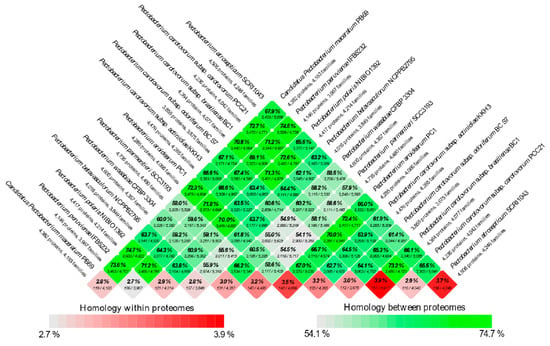
Figure 3.
BLAST matrix between and within total proteomes of Pectobacterium genus. Pairwise protein comparison was performed using BLAST and protein coding sequences were compared among the genomes. A BLAST hit was considered significant when 50% of the alignment revealed identical matches and covered at least 50% Alignment. If two protein sequences were similar based on cut-off value, they were grouped within the same protein family. The color scale saturation changes from dark green to light green indicated the degree of homology between the proteomes, whereas the color scale from dark red to light red showed the homologous hits within the proteome itself (internal paralogs at the bottom row of the matrix). Twelve genomes of different Pectobacterium species were compared in the analysis.
3.3. Genomic Evolution of Pectobacterium Species—Analysis of Codon and Amino Acid Usage
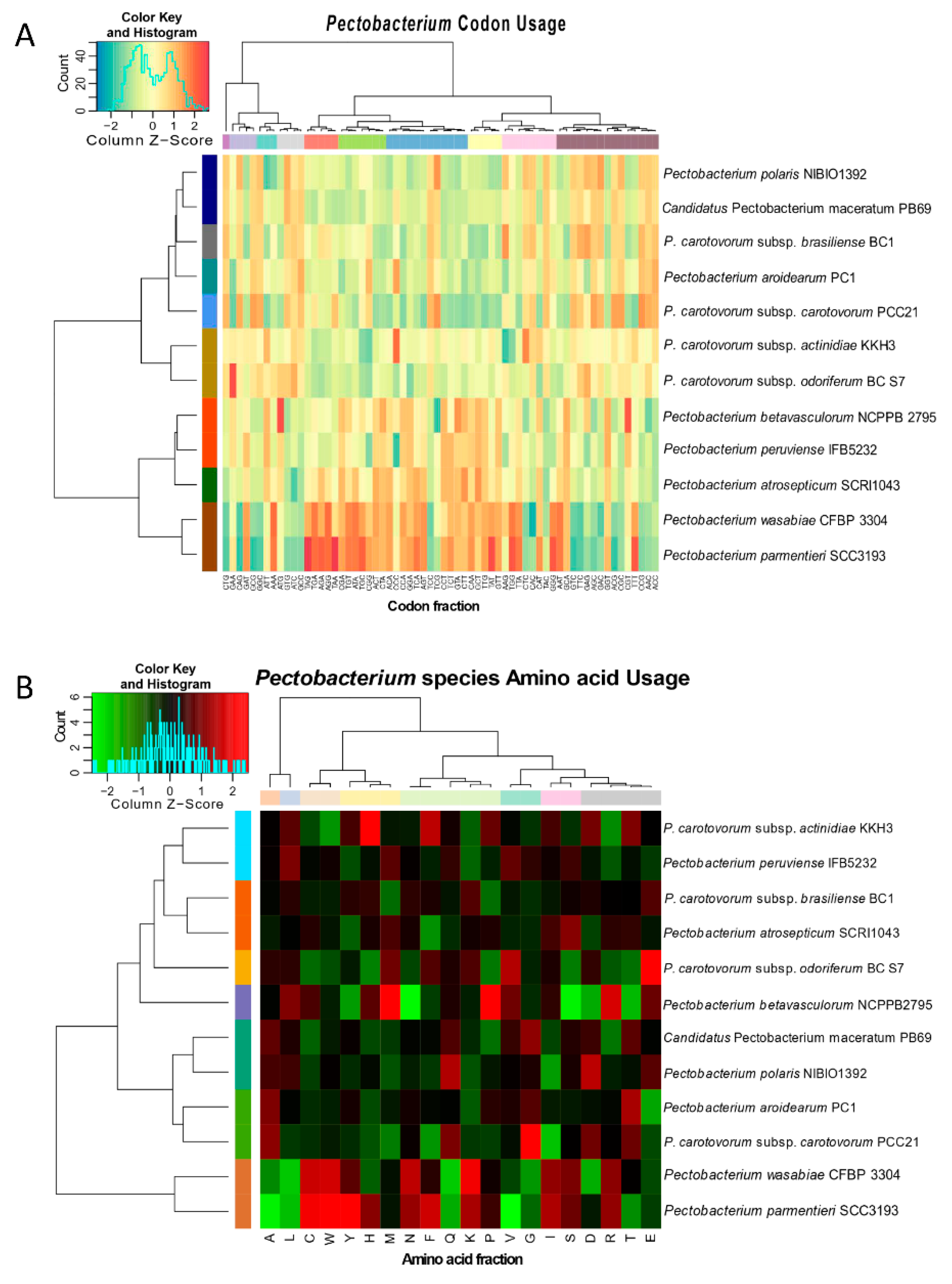
The preference of codon usage varies between organisms and even between species. Therefore, the codon usage pattern constitutes an exclusive property for each species and genome [63]. Studying the divergences of codon usage bias contributes to understanding the basis of pathogen evolution and their adaptation to specific niche [63]. The bias score was evaluated at +1 if 100% in the third position corresponds to G or C, or −1 if the bias score was 100% of A or T in the third position. Overall, all Pectobacterium sp. genomes exhibited bias towards either G or C nucleotides (Figure S2); however, the value ranges were low (0.1133 to 0.1894). The AT content for all species was low, and it ranged from 46.50 % (P. c. subsp. carotovorum) to 48.34% (P. parmentieri). P. c. subsp. carotovorum, P. polaris, and P. c. subsp. brasiliense showed a slightly higher bias score towards G usage when compared with the other bacteria (0.1894, 0.1827, and 0.1809, respectively). The calculated percentages of the codon and amino acid profiles, among the Pectobacterium sp., were compiled and visualized in two dimensional heatmaps that were created in R (Figure 4). Besides, rose plots on amino acid and codon usage of each genome were also created (Figures S3 and S4).
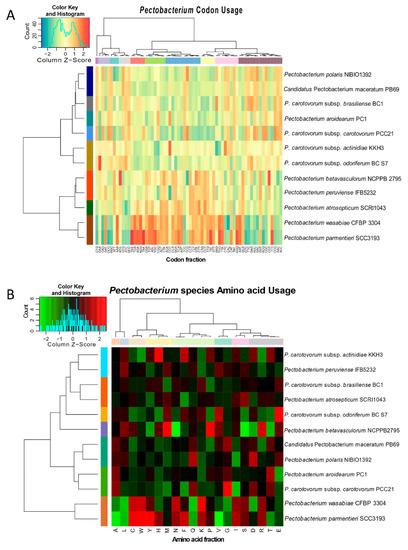
Figure 4.
Heat maps featuring the relationships across the twelve Pectobacterium species based on (A) codon usage and (B) amino acid usage. The identified gene data was plotted and used to construct heat maps in R with heatmap.2 function. The species were reorganized according to the profiles of codon and amino acid usage. Dendrograms at the top and the left side of both heat maps were drawn to illustrate the species clustering pattern. The row and column colors next to both dendrograms highlighted the different clusters observed after analysis. Histograms indicated the scale colors based upon the percentages of codon usage and amino acid frequencies.
The CTG, GAA, and CAG were frequently used by all species, while the CCG, AAC, and ACC codons exhibited the least usage (Figure S3). Furthermore, amino acid composition revealed that the Leucine (L) and Alanine (A) were used in higher frequencies in all the Pectobacterium species, while the amino acids Tryptophan (W) and Cytosine (C) displayed the minimum values (Figure S4). The heat map that was generated based on codon compositions (Figure 4A) demonstrated concordant clustering, as revealed by phylogenetic analysis (Figure 2). Thus, for instance, P. betavasculorum, P. peruviense, P. atrosepticum, P. wasbiae, and P. parmentieri grouped in the same clade, while the remaining species formed a distinct lineage. In amino acid usage heat map (Figure 4B), two main clades were formed; P. wasabiae and P. parmentieri grouped separately. In disagreement with the previous phylogenic analysis, the amino acid tree displayed a close relationship between P. c. subsp. actinidiae and P. peruviense.
3.4. Analysis of the Pangenome and Coregenome
The proteomes of the twelve species were extracted, and a BLASTp was conducted against each genome. The genes were assigned as conserved between the species only if they fulfilled the criteria of at least 50% of sequence similarity over no less than 50% of the length of the alignment. The 12 Pectobacterium species showed an accumulative number of 9296 gene families (Figure 5A).
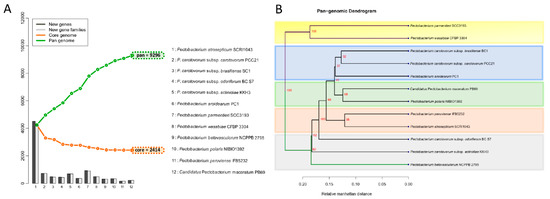
Figure 5.
Pan and core genome analysis among the Pectobacterium species. (A) Pan-Core genome plot. The pan and core genome were calculated using BLAST with a cutoff of 50% identity and 50% coverage. The total number of genes, conformed for pan and core genomes, are represented with green and orange squares, respectively. The bars in the plot represent the number of new genes (dark grey bars) or new gene families (light grey bars) found after the addition of each new genome. Therefore, the size of the pan-genome increased with each new genome. While the number of core genome decrease with the addition of each new genome. (B) Pan-genome tree. The dendrogram illustrates the grouping among the Pectobacterium species based upon shared gene families (core genome) defined in the pan and core genome analysis. Two main clades were defined, and the main branches of each clade are highlighted in green and violet colors. Shaded color rectangles were added manually to emphasize how the species were clustered according to the core genome.
It is important to clarify that either the new gene(s) or new gene families were determined based on the BLASTp hit against the previously included genomes in the list (x-axis in the plot). Hence, the order in which each species was added influenced the addition of novel gene families, as depicted in the bar graphs. However, the total number of genes in core and pan genomes were always the same, regardless of the species order. In contrast with the pan-genome, after the addition of each genome in the analysis, the total number of genes decreased—12 Pectobacterium genomes depicted a total of 2414 core genes (Figure 5A); this number corresponds to the genes conserved or orthologous across all the genomes. The size of the pan-genome was more than three times when compared to the core genome, indicating great genetic heterogeneity and evolutionary dynamics among the Pecotbacterium species. In addition, a pan-genome phylogenetic tree (Figure 5B) based on the reported 2414 common genes was constructed. Two main clades were found, the first cluster included P. parmentieri and P. wasabiae, with a 100% bootstrap value (Figure 5B). P. c. subsp. carotovorum (Pcc), P. c. subsp. brasiliense (Pcb), and P. aroidearum formed a group together, while C. P. maceratum with P. polaris were closer to each other. P. peruviense with P. atrosepticum formed their group. The remaining species did not constitute a defined cluster.
3.5. Versatile Pathogenicity Mechanisms have been Acquired Through Integration of Genomic Islands (GIs) Across the Pectobacterium Species
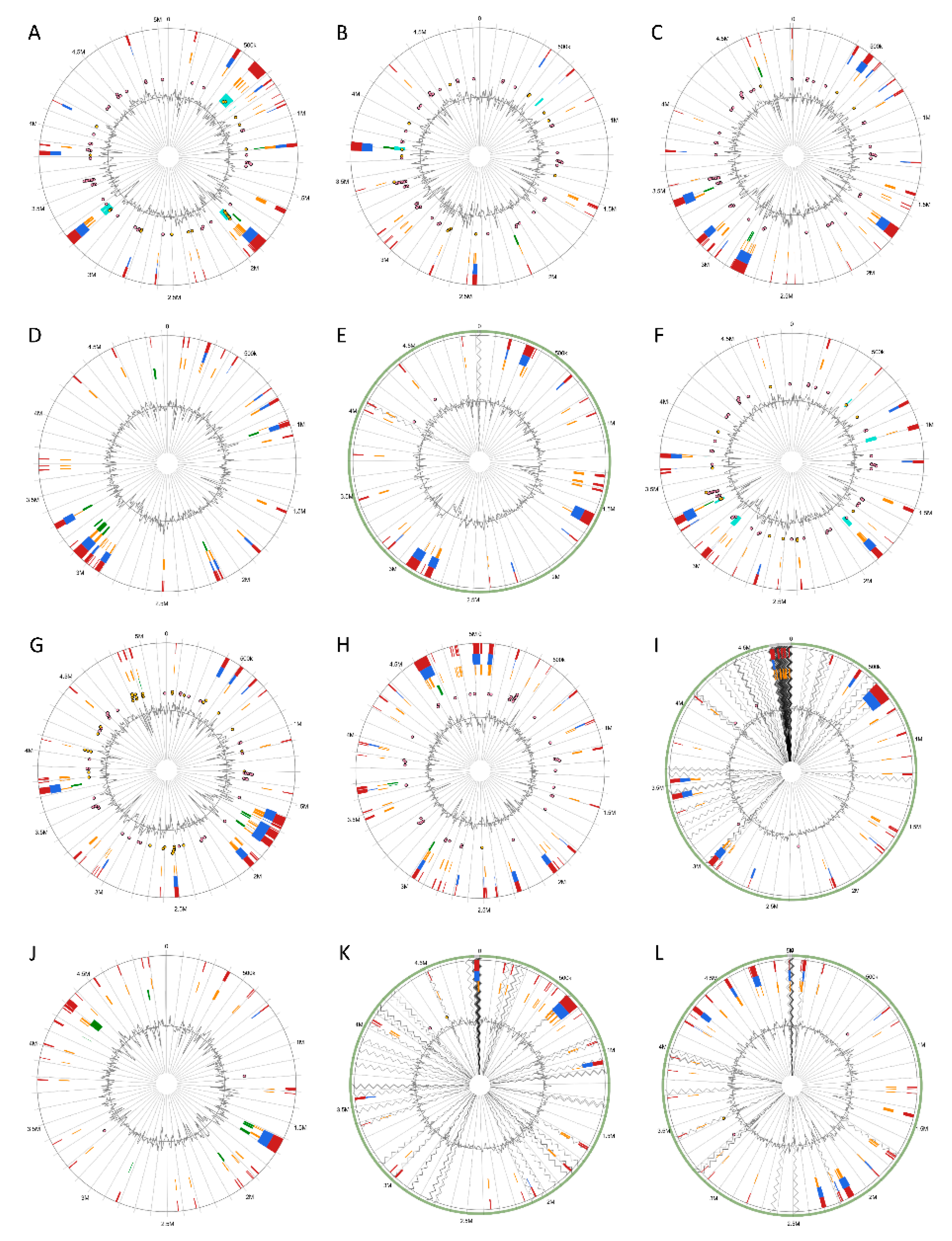
Genomic Islands were predicted and visualized the genomic islands across the 12 Pectobacterium species; different circle plots depicting different GIs in each of the species (Figure 6). A total of 22 GIs was found for P. atrosepticum (Table S2). However, no method, which integrates IslandViewer 4, could identify HAI6, HAI10, HAI11, HAI14, and HAI15 documented by Bell et al. [21]. For the rest of the species, the following number of GIs were found: P. c. subsp. carotovorum—25 GIs (Table S3), P. c. subsp. brasiliense—33 GIs (Table S4), P. c. subsp. odoriferum—29 GIs (Table S5), P. c. subsp. actinidiae—23 GIs (Table S6), P. aroidearum—24 GIs (Table S7), P. parmentieri—40 GIs (Table S8), P. wasabiae—45 GIs (Table S9), P. betavasculorum—32 GIs (Table S10), P. polaris—30 GIs (Table S11), P. peruviense—20 GIs (Table S12), and C. P. maceratum—28 GIs (Table S13). In P. c. subsp. carotovorum, GIs ranging from 4.11 kb with 6 CDS (GI-23) to 50.30 kb with 36 CDS (GI-24) were predicted (Table S3). In P. c. subsp. brasiliense, the GI-18 with a length of 94.09 kb harboring 75 CDS was found as being the largest, while the shortest was the GI-23 of 4.12 kb in size with just three CDS (Table S4). The GIs of P. c. subsp. odoriferum (Table S5) ranged between 4.13 kb with 3 CDS (GI-17) and 72.62 with 39 CDS (GI-21). The GIs of the kiwi phytopathogen P. c. subsp. actinidiae (Table S6) ranged from 70.58 kb with 57 CDS (GI-16) to 4.23 kb with six CDS (GI-12). P. aroidearum exhibited the GIs with the smallest sizes among the species (Table S7), being the largest GI-20 with 43 CDS and 46.07 kb, whereas the shortest was GI-7 of 4.01 kb with just three CDS. The genome of P. parmentieri was predicted to harbor GIs from 4.03 kb (GI-28 with six CDS) to 82.93 kb (GI-15 with 81 CDS) (Table S8). P. wasabiae, with the highest number of predicted GIs, has the largest GI of 91.27 kb with 84 CDS (GI-39) and the smallest GI-6 with five CDS of 4.22 kb in size (Table S9). The sugar beet bacterial pathogen, P. betavasculorum, displayed a large GI of 100.94 kb with 41 CDS, and the shortest GI of 4.18 kb with only eight CDS (Table S10). In the case of P. polaris, the shortest GI found was the GI-23 with six CDS with 4 kb length, whereas the GI-9 was identified as the largest with 94 CDS with a size of 103.55 kb (Table S11). The largest GI of 100.96 kb (GI-6) containing 91 CDS and the shortest GI with only two CDS with a length of 4.23 kb (GI-17) were identified for P. peruviense (Table S12). C. P. maceratum showed GIs that vary in sizes, shortest GI-17 of 4.244 kb with four CDS, and the largest GI-13 of 53.43 kb with 42 CDS (Table S13).
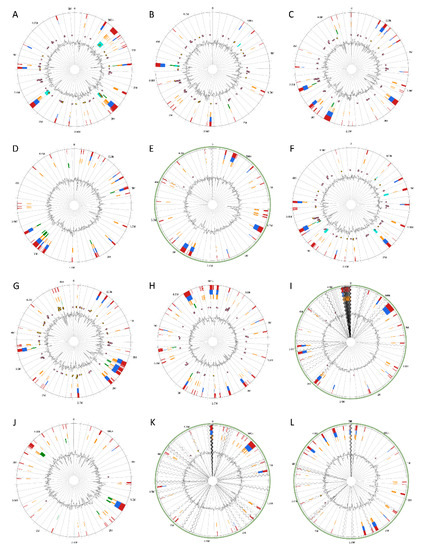
Figure 6.
Circular visualization of predicted Genomic Islands (GIs) among the 12 Pectobacterium species. Starting from the top left row to the bottom right—illustrated the circular GIs plots for the species: (A) P. atrosepticum SCRI1043, (B) P. carotovorum subsp. carotovorum PCC21, (C) P. carotovorum subsp. brasiliense BC1 (D) P. carotovorum subsp. odoriferum BC S7, (E) P. carotovorum subsp. actinidiae KKH3, (F) P. aroidearum PC1, (G) P. parmentieri SCC3193, (H) P. wasabiae CFBP 3304, (I) P. betavasculorum NCPPB 2795, (J) P. polaris NIBIO1392, (K) P. peruviense IFB5232 and (L) C. Pectobacterium maceratum PB69. The interactive visualization of distinct islands across the genomes are shown with colored blocks according to the description provided in the predictor tool: IslandPick, based on genome comparison (green); IslandPath-DIMOB, based on associated GIs features such as tRNAs, transposon elements, integrases and sequence bias (blue); SIGI-HMM, based on the codon usage bias with a Hiddden Markov model criterion (orange); Islander, based on the mapping of GIs in regards with the recurrent use of tRNA and tmRNA genes (turquoise); and, the integrated results of four tools (dark red). Antimicrobial resistance genes as well as pathogen-associated genes are also displayed as pink and yellow circular glyphs, respectively. Species with incomplete genomes, including P. c. subsp. actinidiae, P. betavasculorum, P. peruviense, and C. P. maceratum, the contigs were ordered against the closest relative reference genome (light green outer circle). The references were selected based upon the ANI and dDDH analysis; consequently, P. atrosepticum was used as the reference for P. betavasculorum and P. peruviense, P. c. subsp. brasiliense as the reference for P. c. subsp. actinidiae, and P. polaris as the reference for C. P. maceratum. Gray areas represented unaligned contigs.
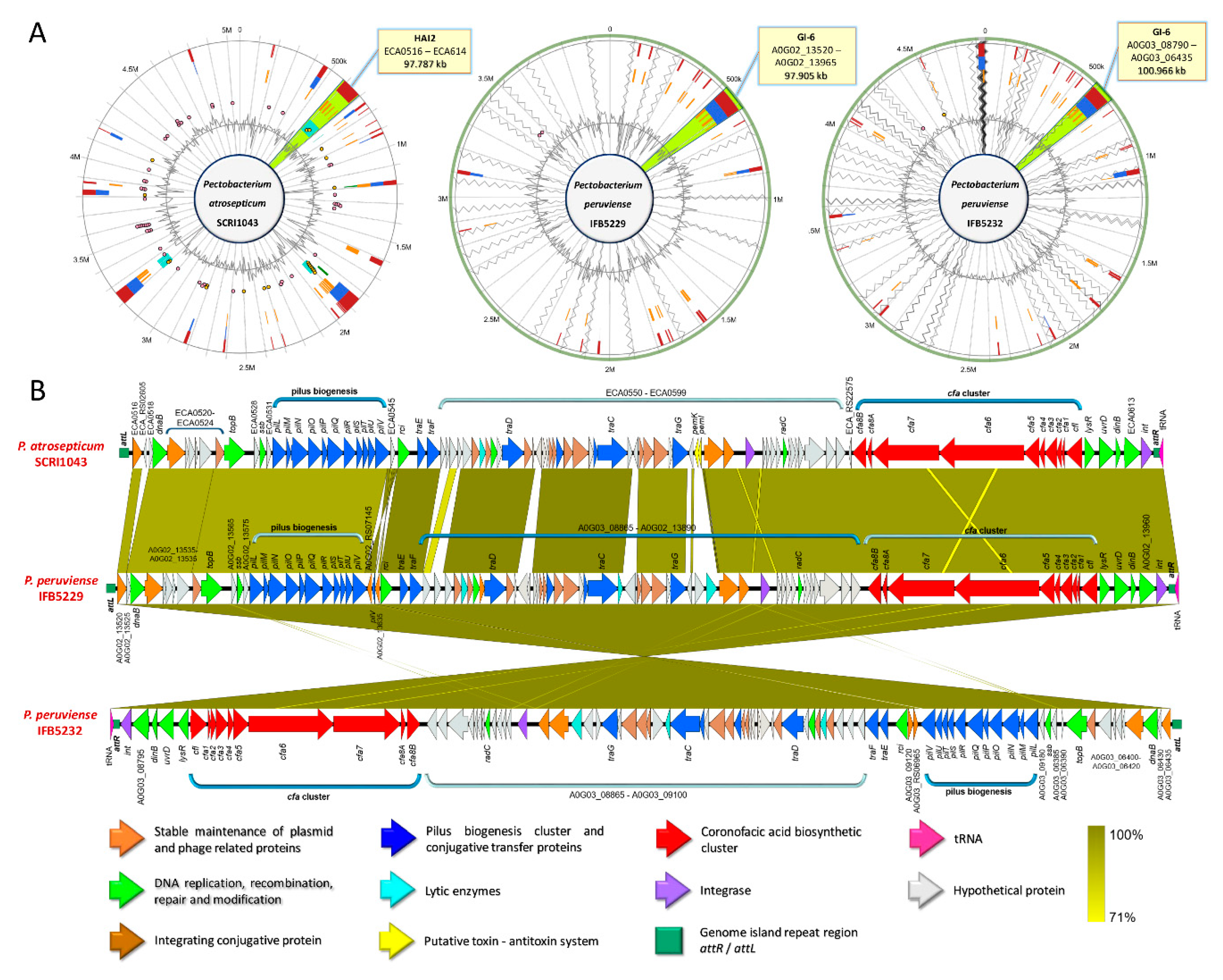
By comparing the GIs across the Pectobacterium species, high synteny between the HAI2 of P. atrosepitucm and the GI-6 of P. peruviense was observed (Figure 7). The HAI2 in P. peruviense was delineated, like P. atrosepticum, by the direct repeats attR and attL (green squares in Figure 7B). The organization between both GIs was also highly conserved (Figure 7B). The pilus biogenesis cluster (blue arrows in the figure), as well as the coronofacic acid (cfa) cluster (red arrows in the figure), exhibited high homology between both species. Other similarities that were observed in both islands were: genes related to the stable maintenance of plasmids and phage proteins (orange arrows in Figure 7B), a tRNA gene (pink arrow in Figure 7B) located next to attR, an integrase (violet arrow), genes that are involved in conjugal integration (brown arrow) and coding regions related to DNA replication, repair, recombination, and modification (green arrows). The pemK and pemI genes (yellow arrows in the graphic), which encode for a toxin and an antitoxin protein, were only found in P. atrosepticum. Additionally, we distinguished some genes encoding lytic enzymes (light blue arrows), as well as some hypothetical proteins, were absent in P. atrosepticum. The entire GI-6 of P. peruviense IFB5232 was arranged in reverse orientation. We extended the comparison of HAI2 of P. atrosepticum to the rest of P. peruviense strains available in NCBI GenBank database—a highly similar HAI2 was found in P. peruviense strain IFB5929, which displayed a positive orientation contrast to strain IFB5232—since our previous analysis (ANI, dDDH, phylogenetic, codon usage) indicated that P. atrospeticum and P. peruviense are closely related species (Figure 7B). However, the other two P. peruviense strains A97-S13-F16 and A350-S18-N16, isolated from the Durance river stream in France [64], lacked the entire HAI2.
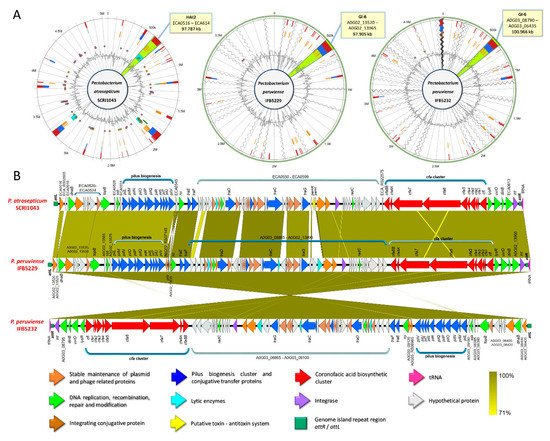
Figure 7.
Comparison between the horizontal acquired island 2 (HAI2) of Pectobacterium atrosepticum SCRI043 with the genomic island 6 (GI-6) of the newly reclassified species P. peruviense. (A) Genomic Islands (GIs) plots illustrating the location of HAI2 in P. atrosepticum and P. peruviense. The plots displayed the predicted GIs based on five approaches (from the inner to the outermost): Islander (turquoise), IslandPick (green), SIGI-HMM (orange), IslandPath-DIMOB (blue), and the integrated methods of the four tools (red). Pink and yellow dots indicated antimicrobial resistance and pathogen-associated genes. The olive green color in each island was manually added to highlight the location of the HAI2 among the three genomes. Moreover, the callout lines in each genome point out the approximate length size of HAI2. (B) Linear genomic representation of HAI2 in the three genomes. The arrows depict the genome organization within each island. In addition, the orientation of the arrows indicated forward or reversed the position of each gene. Arrows Color highlighted different genetic functions within the island as well as gene clusters. Dark yellow shaded regions represent areas with high identity between the analyzed genomes with cut-off values of 100% and 71%.
3.6. Genome-Wide Comparison of Gene Clusters Associated with Pathogenesis
Several pathogenicity determinants have been described in some species of Pectobacterium, including the six type secretion systems (TSS), the plant cell wall degrading enzymes (PCWDE), synthesis of polysaccarides (enterobacterial common antigen, capsular polysaccharide, lipopolysaccharides, exopolysaccharides, and O-antigen), iron acquisition systems, secretion of phytotoxins (coronofacic acid and syringomycin), bacterial attachment operons (type IV pili or fimbriae), secretion of effector proteins (DspE, HrpK, Hrp, Vgr), a necrosis/inducing protein (Nip), an avirulence protein (Svx) similar to Xanthomonas, and the 3-hydroxy-2-butanone pathway (3H2B) [2,26,27,65]. We have compared the similarities, divergences, or absence of the virulent determinants among all Pectobacterium species.
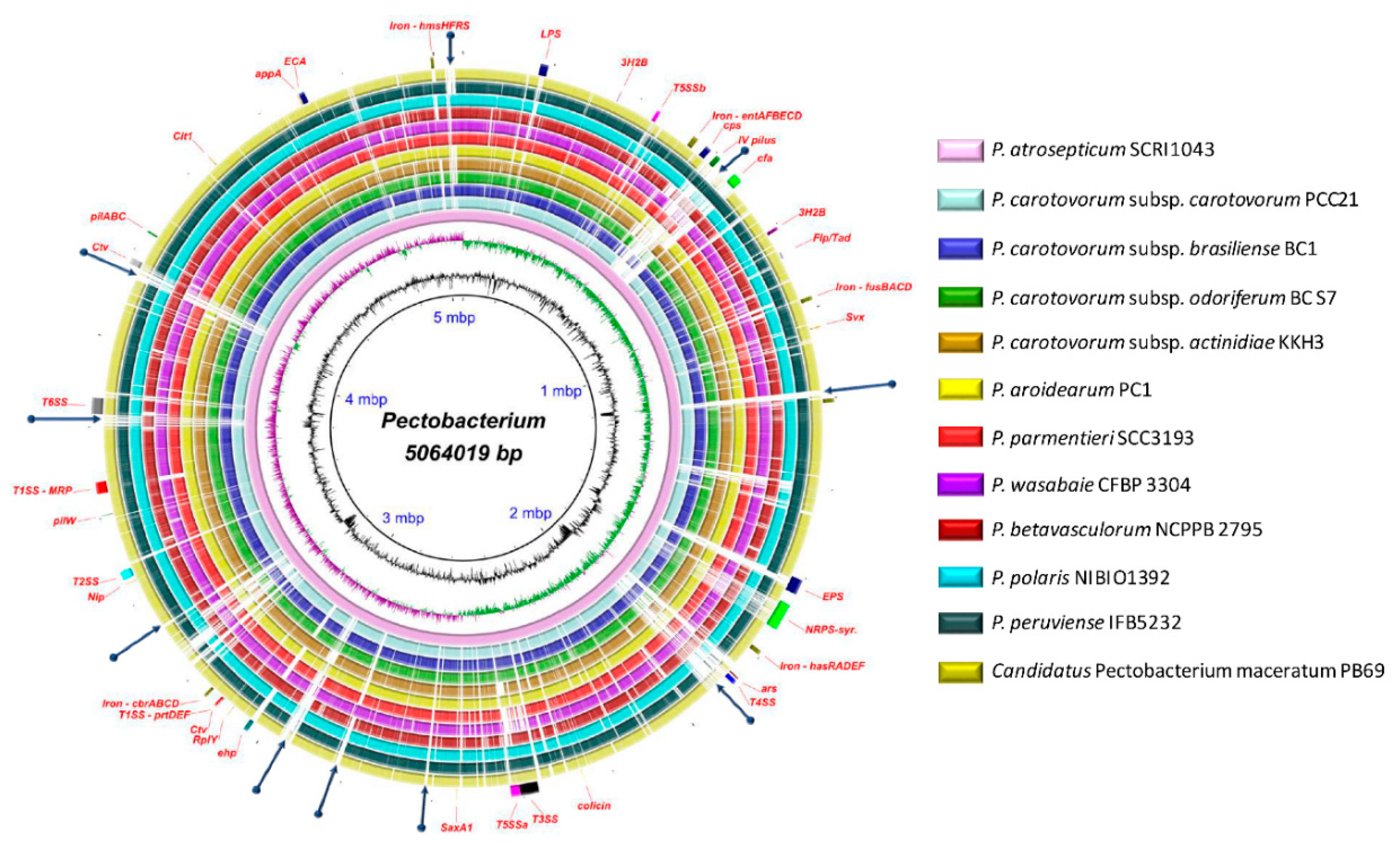
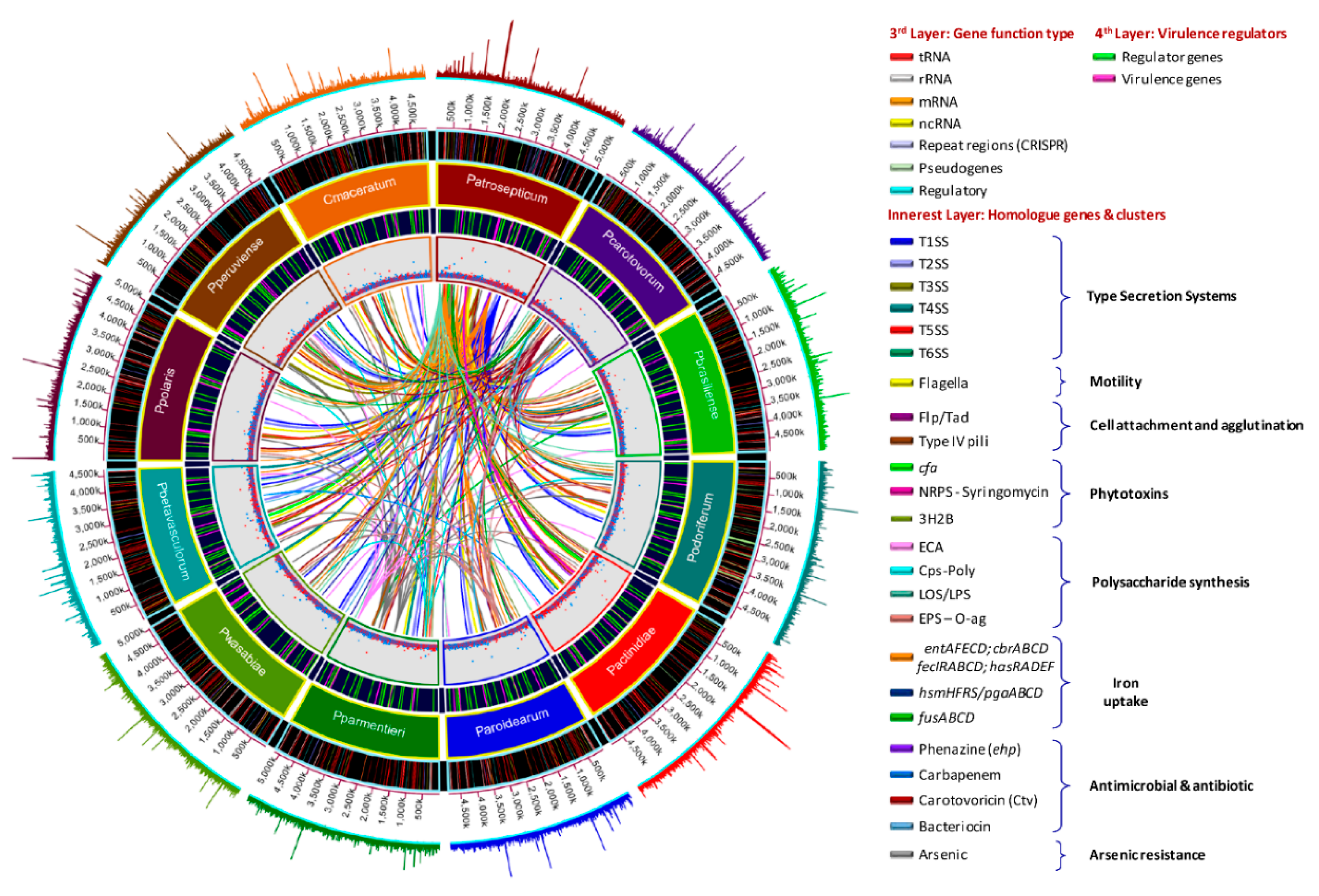
The antimicrobial compounds, such as carotovoricin [31,66,67], carbapenem [28], phenazine [21], and colicins-like bacteriocins [29,30,68,69], which were identified in some Pectobacterium species to out-compete and attack the other microbes for food and specific niche, were also analyzed. A BLAST ring image (Figure 8) displayed nucleotide comparison among 12 species—the outer arrows highlight the locus of the above-described virulent determinants and the antimicrobial components. P. atrosepticum SCRI1043 was used as a reference.
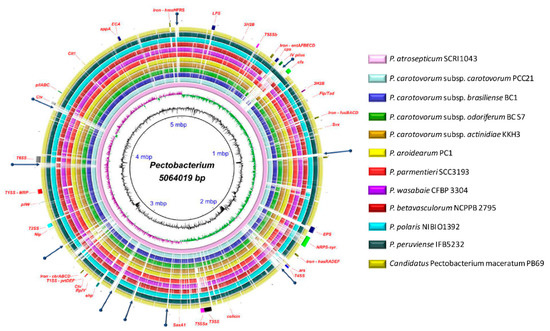
Figure 8.
Genomes comparison across the Pectobacterium genus. The BLAST ring image generated using BRIGS displayed the genome comparison of 12 genomes within genus Pectobacterium. From the innermost origin of each lane, the image features were characterized: the axis with the size of genome (kbp), the GC content (black line), the GC skew (purple color indicates G’s), and afterward each color ring depicting each Pectobacterium species genome, namely, P. atrosepticum SCRI1043 (NC_004547), P. carotovorum subsp. carotovorum PCC21 (NC_018525), P. c. subs. brasiliense BC1 (NZ_CP009769), P. c. subsp. odoriferum BC S7 (CP009678), P. c. subsp. actinidiae KKH3 (NZ_JRMH00000000), P. aroidearum PC1 (NC_012917), P. parmentieri SCC3193 (NC_017845), P. wasabiae CFBP 3304 (NZ_CP015750), P. betavasculorum NCPPB2795 (NZ_JQHM00000000), P. polaris NIBIO1392 (NZ_CP017482), P. peruviense IFB5232 (NZ_LXFV00000000), and Candidatus Pectobacterium maceratum (NZ_PDVY00000000). The outermost ring identified the different loci of most relevant pathogenicity determinants and antimicrobial biosynthetic clusters. The complete genome of P. atrospeticum served as a template for the generation of the BRIG image based upon the nucleotide BLAST analysis. Blue arrows highlighted genomic regions that exhibited divergences when compared to the reference genome. Key acronyms (red legends) used to name the clusters or single virulence genes in the outermost ring are detailed: T1SS-MRP (Type I secretion system—multi-repeat protein), T1SS-prtDEF (Type I secretion system—protein transporters PrtDEF), T2SS (Type II secretion system), T3SS (Type III secretion system), T4SS (Type IV secretion system), T5SSa (Type V secretion system-hecAB), T5SSb (Unreported type V secretion system—described in this study), LPS (lipopolysaccharide), cps (capsular polysaccharide), ECA (Enterobacteria Common Antigen), EPS (Exopolysaccharide O-antigen), Iron-hmsHFRS (hemin storage), Iron- entAFECD (enterobactin), Iron-fusBACD (ferredoxin uptake), Iron-hasRADEF (heme acquisition), Iron-cbrABCD (achromobactin), 3H2B (3-hydroxy-2-butanone pathway), Flp/Tad (Fimbrial low-molecular-weight protein/Tight adherence protein), IV pilus (Type IV pilus), pilW (Pilus assembly protein PilW), pilABC (pilus production proteins PilABC), cfa (coronofacic acid), NRPS-syr (Non-Ribosomal Peptide-Synthetase like syringomycin), ars (arsenic resistance), Ctv (carotovoricin), ehp (phenazine antibiotic biosynthesis), colicin (colicin-like bacteriocin), Svx (virulence protein homologue to Xanthomonas campestris), SaxA1 (aliphatic isothiocyanate resistance protein SaxA), RplY (ribosomal protein RplY), Cit1 (citrate transporter), and Nip (Necrosis-inducing protein), and appA (phytase).
3.7. Type Secretion Systems
The type I secretion system (T1SS) secrets hydrolases, toxins, nodulation factors, adhesins, and hemophores from the cytoplasm to the extracellular media, and it consists of three elemental proteins, including a specific outer membrane protein (OMP) or TolC, an ATP-binding protein, and a membrane fusion protein (MFP) [70]. In Pectobacterium, three different T1SS have been described [65]. The first one is formed by the operon prtD, prtE, and prtF, and it allows the secretion of metalloproteases (prtW) [27,65]. In our analysis, the prtDEF operon shown to be highly conserved in all the species (Table S14). Likewise, the metalloprotease prtW was also observed in all genomes (Table S16). The second T1SS, involved in secretion of hemophore HasA homologue to T1SS-HasA of Serratia marcescens [21,65], was absent in P. betavasculorum and closely related P. parmentieri and P. wasabiae (Table S30). In the analysis, the entire cluster of the third T1SS, involved in secretion of a proteinaceous multi-repeat adhesin protein (MRP) [71], was observed among all species and exhibited high similarity (Table S14 and Figure S5). However, the gene encoding the multi-repeat adhesin protein (MRP) displayed a huge divergence in size among the Pectobacterium species (Figure S5). For instance, the lengths of the MRP in P. parmentieri and P. peruviense were 4,173- and 3,829-bp, respectively; however, the largest MRP-encoding gene was observed in P. polaris (15,240 bp) and P. c. subsp. carotovorum (14,613 bp) (Table S14). Moreover, P. c. subsp. odoriferum harbor MRP and LapG proteins as pseudogenes—indicating that this species might lack the functionality of MRP/T1SS.
The type II secretion system (T2SS) or Out system constitutes a two-step process secretion machinery, which secretes the proteins from cytoplasm to extracellular medium, composed with a set of 12 to 16 genes denominated as the out gene cluster [72,73]. The out operon was highly conserved across all Pectobacterium species (Table S15 and Figure S6). However, the outN gene was missing in P. parmentieri and P. wasabiae (Figure S5). Likewise, neither P. parmentieri nor P. wasabiae harbored a pectate lyase located close to the T2SS cluster (Figure S6). Based on the literature [8,21,23,26], we have predicted that all of the Plant cell wall degrading enzymes (PCWDE) present in Pectobacterium species (Table S16). In the category of pectate lyases, 11 enzymes were found; among these, the HrpK protein with a pectate lyase domain was absent in both P. parmentieri and P. wasabiae. Moreover, the periplasmic pectate lyase was not found in P. betavasculorum. In regards to pectin lyase (pnl), pectin acetylesterases (paeX and paeY) and pectin methylesterases (pemA and pemB) were present in all species. In contrast, a putative polygalacturonase, annotated as “glycoside hydrolase family 28”, was only present in P. aroidearum. The polygalacturonase PehK was not observed in P. parmentieri and P. wasabiae, while the rest of polygalacturonases (pehA, pehN, and pehX) were present in all 12 species. In the case of cellulases, the glycoside hydrolase (celB) was not present in P. c. subsp. brasiliense and P. polaris, whereas the celV and bcsZ genes were highly conserved across the Pectobacterium species. The oligogalacturonide liase (ogl) and Rhamnogalacturonate lyase (rhiE) were also present in all Pectobacterium species. Proteases are fundamental pieces in bacterial virulence and they are secreted through the prtDEF operon; these group of proteins exhibited the highest difference when compared with PCWDE (Table S16). While some proteases, such as metalloproteases PrtW and Prt1, peptidases M3, M20, and S58, deacetylase, D-aminopeptidase, tripeptide aminopeptidase, and zinc dependent protease, were common among the 12 species, the other proteases, such as peptidase M50 and peptidase S9 prolyl oligopeptidase, were solely restricted to P. aroidearum. Similarly, Omptin family outer membrane protease 7 and serine protease were only found in P. parmentieri and P. wasabiae. Other proteins, including a peptidase S53, a putative zinc protease (pqqL), and an acetylornithine deacetylase were only absent in P. betavasculorum. The staphylolytic protease, LasA, was not present in P. parmentieri and P. wasabiae.
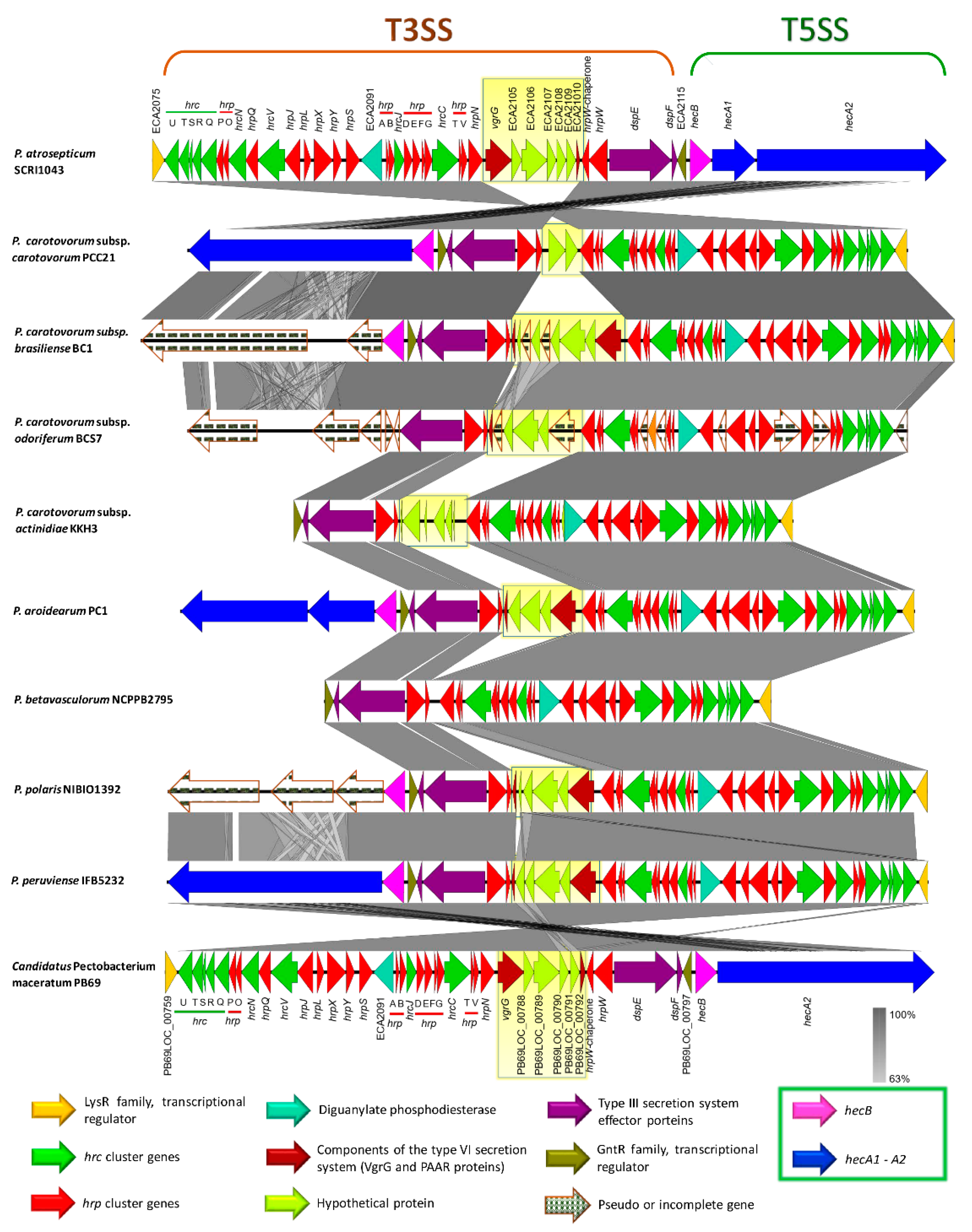
The type III secretion system (T3SS) is the most extensively studied bacterial pathogen system. This system forms injection machinery that allows close interaction of bacteria with the eukaryotic hosts [2,27]. It is reported that the T3SS cluster is present in P. atrosepticum SCRI1043 and harbored within the genomic island HAI7. Within the T3SS, a genetic region between the hrpN and hrpW genes (Table S17, marked in red bold) displayed a high discrepancy among the species (yellow blur rectangle in Figure 9), we named this section as a variable region. P. betavasculorum, for instance, did not have this region. This variable region harbored two components of type VI secretion system: a valine glycine repeats G protein (VgrG) and a proline-alanine-alanine-arginine repeat protein (PAAR), as presented in some species. No remnant of T3SS was found in P. parmentieri and P. wasabiae.
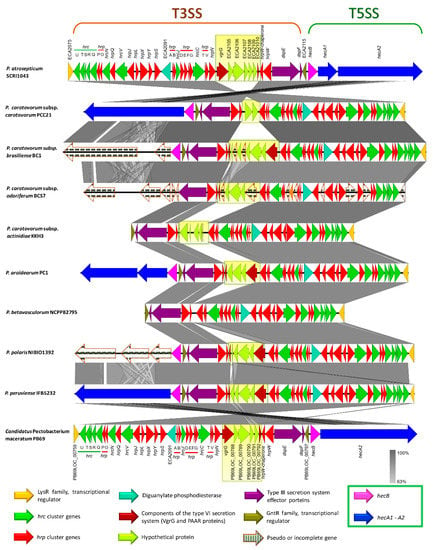
Figure 9.
Comparison of the genetic organization of type III secretion system (T3SS) among the Pectobacterium species. The arrow position represented forward/reverse gene orientation. Arrow color signified specific gene composition within the T3SS. Gene names were provided at the top and bottom of the linear graph; the locus-tag was used for hypothetical or genes with no names. A pairwise alignment between the linear sequences was rendered based upon BLAST algorithm with cut-off values from 64% to 100%. Regions with higher nucleotide identity were displayed with a shaded grey. A light-yellow blur square highlighted the variable region among the genomes that interrupted the type III secretion gene cluster. Expanded legend entry acronyms are provided: hrc (hypersensitive response and pathogenicity conserved genes), hrp (hypersensitive response and pathogenicity or hairpin proteins), VgrG (valine-glycine repeat protein G), PAAR (proline-alanine-alanine-arginine repeat protein), dspE (disease-specific effector protein E), dspF (disease-specific chaperone protein F), hecB (hemolysin activation protein HecB), and hecA (hemolysin/hemagglutinin-like protein HecA).
Contrary to the other secretion systems, the type IV secretion system (T4SS) is described as the most versatile delivery nanomachine that can translocate diverse macromolecules, such as DNA, monomeric substrates, and multimeric proteins, to either bacterial or eukaryotic target cells, usually through a direct cell-to-cell contact [74,75]. The virB-T4SS homologs to Agrobacterium tumefaciens were found in P. c. subsp. brasiliense BC1, P. c. subsp. actinidiae KKH3, and P. betavasculorum NCPPB2795 (Table S18 and Figure S7). Intriguingly, we found two virB-T4SS clusters in P. c. subsp. brasiliense BC1, and the genetic features of both the clusters were highly similar (Table S18). We investigated the other two strains of P. brasiliense: SX309 and BZA12, since we found two T4SS clusters in P. c. subsp. brasiliense; however, two copies of this cluster were not detected.
The type V secretion system (T5SS) is defined as the simplest bacterial secretion apparatus and classified into five categories, from Va to Ve [65,76]. Soft rot enterobacteria harbor the type Vb, known as two-partner secretion system or Tps, encoded by the hecA and hecB genes [65]. We found hecAB operon in P. c. subsp. carotovorum, P. c. subsp. brasiliense, P. c. subsp. actinidiae, P. aroidearum, P. polaris, P. peruviense, and C. P. maceratum (Table S19). Additionally, the two-partner secretion (Tps) system cluster in the species mentioned above lies adjacent to T3SS, except for P. c. subsp. actinidiae (Figure 9). This cluster was not found in P. parmentieri, P. wasabiae, and P. betavasculorum, whereas the hecA1 gene was not present in P. c. subsp. carotovorum, P. c. subsp. actinidiae, P. peruviense, and C. P. maceratum (Table S19 and Figure 9). Moreover, the entire system was conformed by pseudogenes in P. c. subsp. odoriferum. In P. c. subsp. brasiliense and P. polaris, the hemagglutinin-coding loci (HecA) were found as pseudogenes. An additional cluster was also found, seems to be homologous to hecAB. However, the genetic content of this cluster was different; it contained a toxin-activating lysine-acyltransferase locus instead of two hecA genes (Table S19 and Figure S8). The species P. wasabiae and P. peruviense lack this operon. Likewise, the locus encoded for toxin-activating lysine-acyltransferase was not present in P. atrosepticum and P. parmentieri, while in P. c. subsp. odoriferum, this gene and gene encoded a filamentous hemagglutinin were present as pseudogenes. The Tps cluster, like hecAB, which was found in some species resembled a CDI system of P. parmentieri SCC3193 within the GI_5 [26] (Table S19).
The type VI secretion system (T6SS), described as an injectosome like weapon structure that resembles to a bacteriophage tail [77], was found in all 12 Pectobacterium species (Table S20 and Figure S9); however, we observed genomic discrepancy; for instance, P. c. susbp. odoriferum harbor an extra set of genes, predicted hypothetical proteins, between hcp and vasL (virulence-associated protein L) genes (Table S20). Furthermore, a part of the T6SS cluster reported in P. atrosepticum (ECA3420–ECA3428) was completely absent in sugar beet pathogen, P. betavasculorum NCPPB2795 (Figure S9). In P. parmentieri and P. wasabiae, the previously mentioned loci, ECA3420–ECA3428, was located to a different place in the chromosomes (Table S20). Besides, ECA3420 locus and Rhs protein were not present in P. c. subsp. carotovorum PCC21. Likewise, the locus ECA3425 was not identified in P. c. susbp. odoriferum, P. c. subsp. actinidiae, P. aroidearum, P. polaris, P. peruviense, and C. P. maceratum. Moreover, the second T6SS cluster identified by Nykyri et al. [26] within a genomic island in P. parmentieri (GI_30) and P. wasabiae was not found in any other species included in this study. The haemolysin-coregulated proteins (Hcp) and valine-glycine repeat proteins (VgrG) have been reported as the major effector proteins injected from this system to the target cells [78,79]. PAAR proteins were later attributed as key components of the T6SS of S. marcescens [80]. Mattinen et al. [81] described the presence of three copies of hcp gene and four copies of the vgrG gene in P. atrosepticum that were shown to be similar to the genes present in main T6SS cluster. In the case of P. parmentieri SCC3193, 26 copies of either hcp or vrgG genes were detected [26]. In the current analysis, we also found several copies of haemolysin-coregulated proteins (Hcp) and valine-glycine repeat proteins (VgrG), the major effector proteins, harbored in genomic islands (GIs), but scattered throughout the genomes (Tables S2–S13). Furthermore, we have described that the T3SS cluster possesses a variable region contained two components of T6SS: VgrG and PAAR proteins, which were shared in some species (Table S17 and Figure 9).
3.8. Pilin Clusters and Attachment Mechanisms
Pili or fimbriae are appendages and flexible filaments that extend through the Gram-negative bacterial surface and linked to main virulence functions [82]. We identified identical type IV pili clusters in P. peruviense IFB5232, while the rest of the species lacked this operon (Table S21 and Figure S10). Furthermore, like P. atrosepticum and P. parmentieri, the pil operon in P. peruviense was harbored in a genomic island (GI-6). However, the function of this cluster to adhesion has not been identified in Pectobacterium. Additionally, we also found a pilW gene, encoded for a “type IV pilus biogenesis and stability protein,” was highly conserved across the Pectobacterium species (Table S21). Aside from the locus pilW, another operon consisted of three genes ppdD, hofB, and hofC was predicted in all species (Table S21). The functionality of both pilW and the gene cluster pilABC is still to be deciphered in Pectobacterium. The Flp/Tad (fimbrial low-molecular-weight protein–tight adherence protein) cluster and the two-component regulators (response regulator–sensor kinase), located upstream of the flp locus, were identified in all 12 species (Table S22 and Figure S11).
3.9. Production of Phytotoxins
The secretion of phytotoxins constitutes another pathogenicity determinant, and they are maintained within the arsenal of pathogenic bacteria to invade, disrupt, and colonize plant tissue [27]. We found that P. peruviense IFB5232 possesses a cfa cluster, a gene that is responsible for the biosynthesis of coronofacic acid—resembling the one reported in P. atroseptium. Moreover, this polyketide phytotoxin cluster was also observed in P. c. subsp. actinidiae and P. betavasculorum (Table S23 and Figure S12). Like P. atrosepticum SCRI1043 [21], the cfa operon in P. c. subsp. actinidiae, P. betavasculorum, and P. peruviense appeared to be acquired through horizontal gene transfer, since this cluster was identified within the predicted genomic islands GI-6, GI-5 and GI-14, respectively. On the other hand, among all species, only P. c. subsp. actinidiae exhibited a similar NRPS system, like syringomycin, which was predicted to be the part of GI-9. In addition, only three genes (KKH3_14020, KKH3_14030, and KKH3_14040) were identified as the main core of NRPS system in P. actinidiae, while other genes, reported in the HAI6 of P. atrosepticum, were absent (Table S24). The locus KKH3_14040 predicted to encode a siderophore biosynthesis NRPS, displayed the highest differences with a nucleotide identity of 78.5% and query cover of 85.8% when compared with NRPS syringomycin of P. atrosepticum (Table S24). So far, NRPS-like syringomycin function and role in virulence have not yet been demonstrated. Therefore, whether this NRPS system in P. actinidiae contributes to the development of canker disease in kiwi fruits, remains uncertain.
3.10. Arsenic Resistance is an Intrinsic Feature of Few Species
Arsenic clusters (ars) confer bacteria the ability to resist against the concentrations of inorganic arsenic present in an environment [83]. We observed that P. c. subsp. brasiliense, P. peruviense, and C. P. maceratum harbored orthologues to arsC, arsB, and arsR genes, which displayed more similarity to P. atrosepticum, by comparing the ars cluster of P. parmentieri and P. atrosepticum with other species (Table S25 and Figure S13). P. c. subsp. carotovorum only contained one gene (arsC, PCC21_022310). Another copy of arsC in P. parmentieri (W5S_3181), was observed to be present in all of the species (Table S25). Additionally, we found another gene (ECA2265) in P. atrosepticum—annotated as arsenic resistance protein (arsH). The mentioned locus showed high similarity among all species (Table S25).
3.11. Polysaccharide Clusters
Most of the bacteria produce cell surface polysaccharides, reported to promote bacterial attachment inside the host, protect bacterial cells from adverse environmental conditions and plant toxins, and enhance colonization and virulence [27]. In our analysis, we observed that the capsular polysaccharide cluster (cps) was highly conserved in eight species (Table S26 and Figure S14). However, the entire cluster was not found in P. c. subsp. brasiliense, P. c. subsp. actinidiae, P. aroidearum, and P. betavasculorum. The enterobacterial common antigen (ECA) that was composed of 11 genes (wec and rff) was present in all species (Table S27 and Figure S15). The EPS and O-antigen cluster (wza and rfb), as documented previously in the HAI5 of P. atrosepticum [21], was only conserved partially across the species (Table S28 and Figure S16). Only the first 4 and last 3 loci were observed to be present in all species. The lipo-oligo/polysaccharide cluster (LOS/LPS) displayed the highest differences among species—with different rearrangements, only five genes found in all species (rfaD, waaF, waaC, waaA, and coaD) (Table S29 and Figure S17).
3.12. Different Iron Piracy Strategies
Iron is a key micronutrient in the bacterial life, as it participates in the cellular signaling, and is a cofactor of many enzymes [27,84]. The siderophore achromobactin, which is composed of the operon cbrABCD, was highly conserved across all species (Table S30). In contrast, the operon entAFBECD, forms the enterobactin siderophore, was not complete in all species. For instance, the locus ECA0476, encodes for a putative iron transporter, and the genes entF and entB were not present in P. wasabiae and P. parmentieri. In addition, both species presented a locus, annotated as a major facilitator superfamily (MFS), was not found in other species (Table S30). The ferric citrate uptake system (fecIRABCDE) was not carried by P. c. subsp. odoriferum, P. wasabiae, P. parmentieri, and P. betavasculorum. Moreover, P. aroidearum PC1 only exhibited a part of this cluster with missing genes of fecA, fecR, and fecI. The cluster hasRADEF, resembled to hemophore of Serratia marcesens [21], was not observed in P. betavasculorum, whereas only hasR and the locus—encodes for an iron siderophore sensor protein—were found in P. parmentieri and P. wasabiae. The operon hmsHFRS, which is involved in the hemin storage similar to Yersinia pestis, was harbored by five species: P. atrosepticum, P. aroidearum, P. parmentieri, P. wasabiae, and P. peruviense. Except for P. betavasculorum, all of the species appeared to harbor the fusABCD operon involved in the plant-ferredoxin uptake system (Table S30). Lastly, the aerobactin siderophore cluster (iuc), as reported by Nikiry et al. [26] in P. parmentieri SCC3193 and previously found in P. c. subsp. carotovorum W3C105 [85], was not present in all species. Besides, as Nikiry et al. [26] described that aerobactin cluster was present in P. parmentieri WPP163, we compared this cluster among other P. parmentieri strains (RNS 08-42-1A, PB20, SS90, CFIA1002)—the cluster was highly conserved across all P. parmentieri strains (Table S31).
3.13. Other Crucial Virulence Determinants
Other single virulence genes were also compared among the Pectobacterium species (Table S32). The svx gene homologous to the avrXca of Xanthomonas campestris and a putative virulence factor in P. atrosepticum SCRI1043 [86] were highly conserved across all 12 species. The necrosis inducing virulence protein (Nip), where virulence was demonstrated in P. parmentieri SCC3193 and P. atrosepticum SCRI1043 [87], was also present in the rest of the 10 species. The citrate transporter encoded by the cit1 gene, which was observed to reduce the citrate levels and enhanced the growth of P. atrosepticum on potato [88], was also identified in all species. The phytase gene appA formerly identified in P. wasabiae [89]—possible role in pathogenicity has not been revealed yet—appeared to be encoded by all species, except P. c. subsp. odoriferum. Welte et al. [90] found SaxA enzyme involved in isothiocyanates breakdown, was already observed in P. c. subsp. carotovorum, P. c. subsp. brasiliense, P. c. subsp. odoriferum, and P. atrosepticum. In our analysis, we found the sax gene in P. polaris, P. peruviense, and C. P. maceratum. Furthermore, all of the species were predicted to maintain a copy of the rply gene, which encodes for the ribosomal protein RplY, a verified determinant for full virulence in P. carotovorum PccS1, the expression of which is induced by plant extract [91]. Among all species, the Erwinia virulence factor (evf), involved in enhancing interactions of certain P. carotovorum strains with Drosohila melanogaster [9], was only present in C. P. maceratum PB69 (PB69LOC_02095). The bud operon, encoded for acetoin or 3-hydroxy-2-butanone pathway (3H2B) [92], was conserved among all species; however, the budC gene was located far away from budRAB operon (Table S33 and Figure S14).
The 50 flagella genes were conserved across all species: P. atrosepticum SCRI1043 (ECA1685–ECA1740), P. parmentieri (W5S_1760–W5S_1810), P. c. subsp. carotovorum PCC21 (PCC21_026650–PCC21_027210), P. c. subsp. brasiliense BC1 (NC16_13535–NC16_13255), P. c. subsp. odoriferum BC S7 (BCS7_13110–BCS7_12835), P. c. subsp. actinidiae KKH3 (KKH3_25780–KKH3_25130), P. aroidearum PC1 (PC1_2615–PC1_2560), P. wasabiae CFBP 3304 (A7983_22275–A7083_22035), P. betavasculorum NCPPB2795 (KP22_07745–KP22_07480), P. polaris NIBIO1392 (BJK05_08375–BJK05_08100), P. peruviense IFB5232 (A0G03_04140–A0G03_03865), and C. P. maceratum PB69 (PB69LOC_00365–PB69LOC_00421). In all bacterial species, the flagellar genes were located within genomic islands (Tables S2–S13), which indicate that the common ancestor of all Pectobacterium species acquired the set of flagellar genes through horizontal gene transfer.
3.14. Virulence Mechanism in Pectobacterium Species Appears to be Regulated by Several Factors
The study of regulatory mechanisms in Pectobacterium has revealed that specific regulators that modulate transcription positively or negatively mediate the function of pathogenicity determinants; at the same time, these regulators are activated or suppressed by the other molecules, thus constituting an extensive complex network of regulators [27,65]. Overall, all of the proteins involved in QS (quorum sensisng: ExpI, ExpR1, VirR/ExpR2, CarR/ExpR3, LuxS), global regulators (RsmA, KdgR, ExpM, Hor, and HexY), two-component regulators (PehR-PehS, PmrA-PmrB, and ExpS-ExpA), phosphorelay system (RcsC, RcsD, and RcsB), and other regulatory compounds (HexA and RexZ) were almost conserved in all 12 Pectobacterium species (Table S34), with a few exceptions, including the presence of CarR (ExpR3) in only those carbapenem-producing strains [93], the lack of VirS in P. parmentieri SCC3193, and the absence of PmrA-PmrB, and rexZ in P. betavasculorum. The response virulence regulator (evr), formerly described in P. atrosepticum [8], was found only in P. c. subsp. carotorum PCC21. Furthermore, the Fur regulator, which controls ferric uptake [27], was conserved across all species. All of the species carried SirB1, which is necessary for virulence in P. parmentieri SCC3193 [26], in their genomes. Finally, the Cytidine repressor (CytR) of P. aroidearum PC1, which controls biofilm formation [94], was predicted in all 12 Pectobacterium species.
3.15. Diverse Antimicrobial Compounds Secreted by Pectobacterium Species
The mining of Pectobacterium species genomes has revealed several genes that are involved in the synthesis of secondary metabolites with antimicrobial function. Based on this information, our objective was to analyze all Pectobacterium species for the presence or absence of the previouly mentioned antimicrobial compounds.
Carotovoricin synthesis—Carotovoricin (Ctv) has been defined as a phage tail bacteriocin of high molecular weight [31,67]. All Pectobacterium species possess the Ctv cluster (Table S35) adjacent to the T1SS (Figure S19). However, the genes fibA and fibB, encoded for a bacteriophage tail fiber, were missing in P. c. subsp. actinidiae, and P. betavasculorum. Likewise, the ferredoxin (2Fe-2S) protein was not identified in P. atrosepticum, P. c. subsp. brasiliense, P. parmentieri and P. wasabiae. Moreover, in P. c. subsp. carotovorum, P. c. subsp. odoriferum and P. polaris, the fibB gene was not located within the ctv operon but present at another region (Table S35). Additionally, the organization of the ctv gene cluster in P. atrosepticum and P. betavasculorum showed divergence as compared with other species. Specifically, 14 genes in P. atrosepticum and 17 genes in P. betavasculorum located far away from first part of the ctv cluster (yellow blur rectangle in Figure S19) and in reverse orientation. The ctv cluster of P. betavasculorum displayed the highest divergence among the species, with three hypothetical genes only found in this bacterium (Figure S19).
Carbapenem synthesis—First time P. c. subsp. carotovorum ATTC 39048 was described as a carbapenem producer among Pectobacterium species [95]. This cluster is composed of an operon of eight genes, denotated as carABCDEFGH, with carA, carB, carC, carD, and CarE, assigned as crucial encoding enzymes genes, whose function is the biosynthesis of carbapenem, while the carF and carG genes were observed to encode intrinsic elements related to resistance against the carbapenem and carH with an unknown function [28,96]. Only P. c. subsp. actinidiae and P. betavasculorum contained the complete car operon (Table S36). We cross checked with other strains of both species, and all of them also maintained identical carbapenem genes, thus showing that carbapenem synthesis seems to be an innate characteristic of kiwi and sugar beet bacterial pathogens (Figure S20). We also included the carbapenem cluster of P. c. subsp. brasiliense strains 1692 and ICMP 19477, displayed high similarity with other carbapenem producing species and with P. c. subsp. carotovorum ATCC 3908. Surprisingly, P. c. subsp. brasiliense BC1, P. parmentieri, and P. wasabiae only possess partial car operon—carF, carG, and carH (Table S36 and Figure S20).
Phenazine synthesis—It has been described that phenazines activity triggers an excessive accumulation of reactive oxygen species, which affect cellular redox and end up inhibiting the cell growth [97]. All the strains of both P. c. subsp. actinidiae and P. betavasculorum carried phenazine biosynthetic cluster, as previously observed in P. atrosepticum strains SCRI1043 and ICMP1526, and P. c. subsp brasiliense ICMP19477 (Table S37 and Figure S21). However, the size of the ehpG varied—1218 bp in P. c. subsp. brasiliense and P. betavasculorum, 1410 bp in P. atrosepticum SCRI1043 and all P. c. subsp. actinidiae strains, and 1383 bp in P. atrosepticum ICMP 1526. This gene encodes one of the key enzymes involved in the biosynthesis of phenazine.
Colicin-like bacteriocins—Colicin-like bacteriocins exhibited powerful toxic effects against closely related bacteria [98]. We found few uncharacterized bacteriocins in few Pectobacterium species (Table S38). For instance, homologous to the Carocin D [69] were observed in P. c. susbp. brasiliense, P. c. subsp. odoriferum, and P. betavasculorum. The previous characterized pectocin M1 [98], in P. aroidearum, was also predicted in P. carotovorum subsp. carotovorum PCC21. A type of colicin/pyocin-like bacteriocins were also identified in three P. carotovorum subspecies (carotovorum, brasiliense, and odoriferum) and P. wasabiae. Likewise, a colicin like-bacteriocin protein, and its corresponding immunity protein, were found in P. atrosepticum SCRI1043. The rest of the species, P. parmentieri, P. c. subsp. actinidiae, P. peruviense, P. polaris, and C. P. maceratum, apparently did not carry carocin-like bacteriocin operons.
3.16. A unique Phosphotransferase System (PTS) Cluster Conserved in P. parmentieri
A 4.5 kb region, which contained five genes, was exclusively present in P. parmentieri SCC3193. We compared this region with other P. parmentieri strains available in the NCBI GenBank database, and observed that all strains (WPP163, RNS 08-42-1A, PB20, SS90, and CFIA1002) maintained this highly conserved cluster (Table S39). The cluster corresponds to a cellobiose-specific phosphotransferase system (PTS), which contained five genes (Figure S22). The cluster found in P. parmentieri, first three genes encoded for three domains EIIA, EIIB, and EIIC, which together conformed substrate specific permease to transport cellobiose from cell membrane to cytoplasm. The gene next to the EIIC, encoded a beta-glucosidase with cellobiose/specific PTS activity, predicted function—conversion of cellobiose to glucose. The LacI family transcriptional regulator adjacent to this gene is most likely responsible for the regulation of the transcription of cellobiose PTS permease subunits EIIA, B, and C, as well as beta-glucosidase.
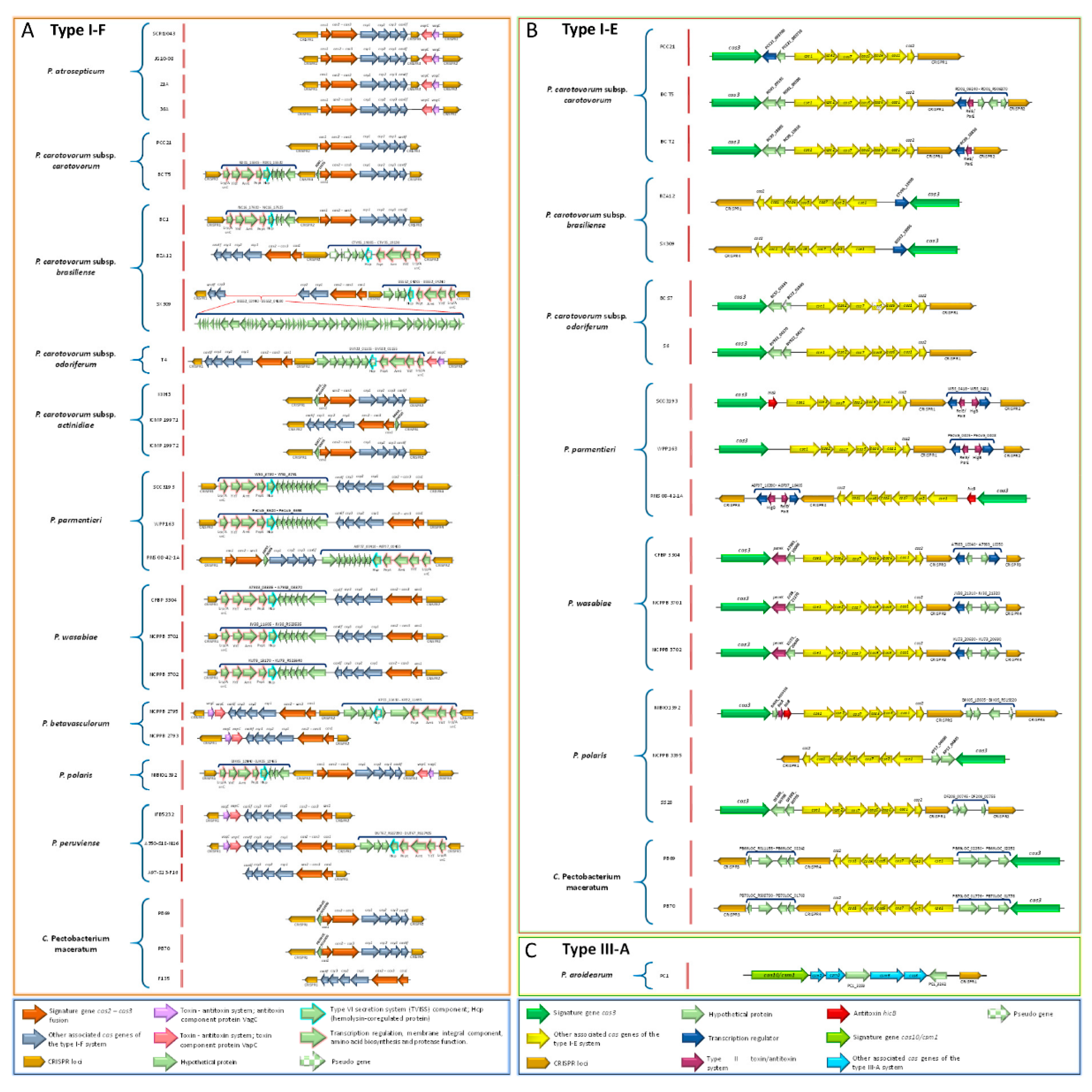
3.17. Pectobacterium Species Harbor Different CRISPR-Cas Systems
The CRISPR-CAS constitutes the adaptive immune system in bacteria [99]. We found three types of CRISPR-Cas systems across the Pectobacterium species: subtype I-F, subtype I-E, and type III-A (Figure 10). All of the strains within species P. atroseoticum, P. c. subsp. actinidiae, P. betavasculorum, and P. peruviense contained the subtype CRISPR-Cas system I-F (Figure 10A). P. aroidearum was the only species harbored type III-A CRISPR-Cas system (Figure 10B). The rest of the species exhibited great heterogeneity, with some strains containing both I-F and type I-E CRISPR-Cas systems, while others, such as P. c. subsp. brasiliense BC1, P. c. susbp. odoriferum, P. polaris strains NCPPB 3395 and SS28, and C. P. maceratum F135 merely presented the subtype I-E CRISPR-Cas (Figure 10C). We also found strains with no CRISPR-Cas systems, namely P. polaris NIBIO1006 and P. peruviense IFB5229. The presence of putative toxin and antitoxin proteins were observed in subtype I-F CRISPR-Cas systems (Figure 10A). For instance, the analyzed strains in P. atrosepticum revealed the presence of VapC toxin and VagC antitoxin between CRISPR3 and CRISPR2. Likewise, same toxin/antitoxin components were observed in P. c. subsp. odoriferum T4, both strains of P. betavasculorum, and in P. peruviense IFB5232 and A350-S18-N16. Other species and certain strains contained a set of sequences downstream of cas6f operon (P. parmentieri and P. wasabiae), or upstream of cas1 gene and after the CRISPR loci (P. c. subsp. carotovorum BC T5, P. c. subsp. brasiliense all three strains, P. c. subsp. odoriferum T4, P. betavasculorum NCPPB2795, P. polaris NIBIO1392, and P. peruviense A350-S18-N16). These genes were identified as coding sequences of the Lrp/AsnC transcriptional regulator protein, YitT integral membrane protein, an M18 peptidase, the type VI secretion component Hcp (hemolysin-coregulated protein), and other hypothetical proteins. Interestingly, we found a region of 49 CDS that interrupted the cas operon between the genes csy3 and csy2 in P. c. subsp. brasiliense SX309. This region harbored proteins that were related to phage structures, suggesting that the subtype I-F CRISPR-Cas system, in this strain, has a horizontally transferred origin. Unlike the subtype I-F CRISPR-Cas system, the subtype I-E possessed different types of toxin-antitoxin systems localized either downstream of the cas2 gene or between the cas3 and cse1 operons (Figure 10B). Specifically, we found P. c. susbp. carotovorum T5 and T2 contained RelE/ParE toxin protein that was located between the CRISPR-1 and CRISPR-2, whereas all of the strains within P. parmentieri presented the same protein with additional toxin protein HigB. In P. wasabiae, a toxin protein, called pemK, was observed downstream of cas3. P. polaris NIBIO1392 harbored a hypothetical protein, HicA toxin, and HicB antitoxin next to Cas3 operon. Strains SCC3193 and RNS 08-42-1A of P. parmentieri contained the same antitoxin HicA adjacent to the cas3 operon. Furthermore, other species, such as P. c. subsp. carotovorum, P. c. subsp. brasiliense, P. parmentieri, and P. wasabiae, contained locus predicted to encode a transcriptional regulator (blue arrows in Figure 10B). The type III-A CRISPR-Cas system was exclusively found in P. arodiearum PC1 (Figure 10C). This system harbored two hypothetical proteins, one between the genes cms3 and cms5 and the other between cas6 and CRISPR locus.

Figure 10.
Schematic sequence organization of the type I-F, type I-E, and type III-A clustered regularly interspaced short palindromic repeats-Cas (CRISPR-Cas) across the genomes of the 12 Pectobacterium species. (A) CRISPR-Cas subtype I-F marked in orange. (B) CRISPR-Cas subtype I-E system marked in green. (C) CRISPR-Cas type III-A marked in yellow. Brown horizontal pentagons represented the CRISPR loci while the arrows depicted specific gene composition such as the cas or csm and other surrounding genes with either known or unknown function. The arrow and pentagon positions represented sequence orientation. Descriptions of gene names are provided inside the arrows or pentagons as well as at the top or bottom of each linear graph; genes without a name or hypothetical proteins labeled with locus-tag. The linear graphs were drawn manually and represent the scale of the respective gene lengths. Key acronyms used in the linear sequences: CRISPR (clustered regularly interspaced short palindromic repeats), cas, csy, and csm (genes encoding CRISPR-associated proteins), Lrp/AsnC (leucine responsive regulatory protein/asparagine synthase C transcription regulator family); YitT (membrane protein YitT), Amt (aspartate/tyrosine/aromatic aminotransferase), Pept (M48 family peptidase), Hcp (hemolysin-coregulated protein). VapC (toxin protein), VagC (antitoxin/virulence-associated protein), HigB (mRNA interferase toxin HigB), RelE/ParE (toxin-antitoxin system, mRNA interferase toxin RelE/toxin ParE), HicA (mRNA interferase toxin HicA), HicB (antitoxin HicB), pemK (mRNA interferase toxin pemK).
We performed a BLASTn analysis since the entire CRISPR-Cas system was found within a genomic island (GI-22). The corresponding results showed a 90% nucleotide identity with Serratia sp. ATCC39066 with over 99% of query cover. A similar homology percentage was observed with D. zeae Ech586, but with lower query cover (97%). These results suggest that P. aroidearum acquired the entire type III-A CRISPR-Cas system from Serratia sp. through a horizontally transferred mechanism. A set of features was observed in predicted CRISPRs loci. Table S40 provides specific information regarding the main features of each CRISPR, such as position, length of direct repeats, number of spacers, and orientation. Generally, the length of direct repeats within all CRISPRs was 28- to 29-bp except directed repeats of CRISPR located within type III-A CRISPR-Cas system of P. aroidearum, whose size was 37 bp. In terms of the total size of the CRISPRs, we observed a plethora of variabilitfrom shortest CRIPSRs of just 208 bp, CRISPR-1 of P. wasabiae strains CFBP 3304 and NCPPB 3701, and the largest CRISPRs with a size of 2250 bp—predicted for CRISPR-1 in P. betavasculorum NCPPB 2793. A size of 2488 bp found in CRISPR-2 of C. P. maceratum F135.
However, the highest number of 41 spacers were detected in CRISPR-2 of C. P. maceratum (F135), and P. c. subsp. carotovorum (PCC21). P. polaris NIBIO1392, and P. betavasculorum NCPPB 2793 reached the highest number of six CRISPRs loci, while P. aroidearum and P. peruviense A97-S13-F16 only showed one CRISPR. Finally, we observed the presence of orphan CRISPRs that were distant from the cas operon regions—P. betavasculorum NCPPB 2793 contained four orphan CRISPRs loci. Other species, such as P. c. subsp. odoriferum strains (BC S7 and S6), and C. P. maceratum (F135) harbored two orphan CRSIPRs, whereas P. c. subsp. carotovorum BC T2 and P. polaris (NCPPB 3395 and SS28) possessed one orphan CRISPR.
In summary, Figure 11 illustrates a circos plot that describes primary genetic features as well as all homolog genes that are associated with pathogenicity determinants and the synthesis of antimicrobial compounds mentioned in this study.
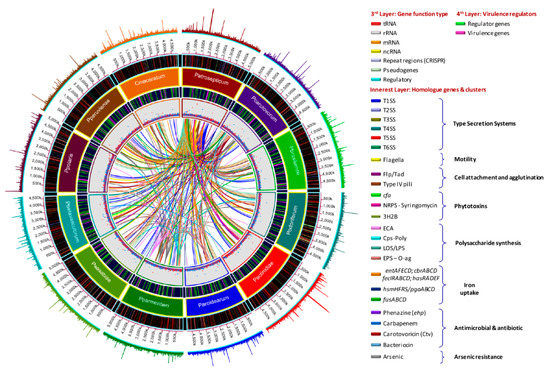
Figure 11.
Circos plot description of main genetic features, relevant virulence, and antibiotic biosynthesis clusters among all Pectobacterium species. Seven circular layers have been rendered to show unique features based upon the comparative genomic analyses. From the outermost layer to the innermost layer: the area of each genome; sizes of each genome (axis in kb); gene function types across the species, name of each species indicated in 3rd layer—acronyms used in the plot starting with Pectobacterium atrospecticum (Patrosepticum), P. carotovorum subsp. carotovorum (Pcarotovorum), P. c. subsp. brasiliense (Pbrasiliense), P. c. subsp. odoriferum (Podoriferum), P. c. subsp. actinidiae (Pactinidiae), P. aroidearum (Paroidearum), P. parmentieri (Pparmentieri), P. wasabiae (Pwasabiae), P. betavasculorum (Pbetavasculorum), P. polaris (Ppolaris), P. peruviense (Pperuviense), and Candidatus Pectobacterium maceratum (Cmaceratum); virulence genes (pink lane) and key gene regulators (green lane), as indicated in the legend (5th layer); scattered dots of all genes, where the red and blue dots depict negative and positive gene orientation, respectively; and the inner layer showing the connection among homologues genes of all gene clusters covered in this study. The colors of the connector lanes of each cluster are labeled on the right side of the figure. Expanded legend entry acronyms are provided: T1SS (Type I secretion system), T2SS (Type II secretion system), T3SS (Type III secretion system), T4SS (Type IV secretion system), T5SS (Type V secretion system), T6SS (Type VI secretion system), Flp/Tad (Fimbrial low-molecular-weight protein/Tight adherence protein), cfa (coronofacic acid), NRPS (Non-Ribosomal Peptide-Synthetase) like syringomycin, 3H2B (3-hydroxy-2-butanone pathway), ECA (Enterobacteria Common Antigen), Cps-Poly (capsular polysaccharide), LOS/LPS (lipopolysaccharide/lipooligosaccharide), EPS—O-ag (Exopolysaccharide O-antigen), entAFECD (enterobactin), cbrABCD (achromobactin), fecIRABCD (ferric citrate uptake), hasRADEF (heme acquisition), hsmHFRS/pgaABCD (hemin storage), and fusABCD (ferredoxin uptake).
4. Discussion
In this study, we performed genome-wide comparative analyses of 12 Pectobacterium species. The genetic relationships among the Pectobacterium species were evaluated using different approaches: ANI, dDDH, phylogenetics, codon and amino acid usage, and pan- and core-genome. In addition, different virulence determinants were analyzed. A highly conserved novel cluster, composed of five genes that were involved in phosphotransferase system (PTS) of cellobiose, was identified exclusively in P. parmentieri.
Among the 12 Pectobacterium species, P. betavasculorum exhibited the shortest genome size and the lowest amount of coding sequences (CDS) (Table 1). P. c. subsp. odoriferum possessed the highest number of pseudogenes, which were observed within important clusters, including type secretion systems I, III, V, and VI, some PCWDE and polysaccharides clusters—indicating a genomic degeneration in the coding ability of this species. Similar features were observed in the comparative genomics of five Clavibacter michiganensis subspecies, where C. michiganensis subsp. sepedonicus contained the fewest number of coding sequences, which was suggested to be the result of disposable genes to adapt into a new niche [100]. P. c. susbp. odoriferum could possibly invade the plant tissues and cause soft rot in potato as well as chicory [25] with fewer number of genes. Contrary to P. c. subsp. odoriferum, P. parmentieri presented the largest genome and highest number of coding genes. These features may be the consequence of horizontal gene transfer. Nykyri et al. [26] reported a total of 56 genomic islands in this species. By analyzing the genomic islands throughout the genus Pectobacterium (Tables S2–S13), P. parmentieri presented the highest number of GIs among the species.
DNA structural properties, namely intrinsic curvature, stacking energy, and position reference, which were visualized in genome atlases (Figure S1), showed significant differences among the species. Regions with high gene expression are the genomic sequences with very low position preference, and not easy to condense [50]. P. parmentieri showed the lowest high position preference areas; thus, it can be concluded that P. parmentieri would have more highly expressed regions. P. c. subsp. carotovorum, followed by P. aroidearum and P. c. subsp. odoriferum led the content of the highest stacking energy and intrinsic curvature areas. Genomic regions with high levels of intrinsic curvature and stacking energy are thought to display high transcriptional levels mediated by nucleosome [50]. In Pseudomonas putida, high intrinsic curvature and stacking energy were linked with high recombination levels that lacked synteny [101]. Hence, it could be possible that the genome atlases of aforementioned Pectobacterium species indicate relevant hotspots that were involved in recombination and transcriptional events. DNA curvature participates in crucial biological functions, such as replication, transcription, recombination, and nucleosome positioning [102]. The intrinsic DNA curvature might indicate fingerprints that demonstrate how different Pectobacterium species have evolved and adapted in a new environment. A reduction of intrinsic DNA curvature has been proven to increase local mutation rates [103]; thus, these regions may be associated with mutation levels and speciation in Pectobacterium. P. atrosepticum harbored the highest number of either direct or indirect repeats—an indication of carrying more unstable regions within the genome [104].
ANI, dDDH, and proteome comparison (Figure 1 and Figure 3) showed that P. parmentieri and P. wasabiae shared lowest nucleotide identity with other species of Pectobacterium–similar results were obtained with codon and amino acid usage profiles (Figure 4) as well as with pan- and core-genome dendrogram (Figure 5B). This information showed that both bacteria might have evolved differently, and that could be the reason for not harboring T3SS. Phylogenetic analysis distinguished all 12 species clearly (Figure 2); two strains—PCC21 and ATCC 39048, named as P. c. subsp. carotovorum, but grouped within the cluster of P. c. subsp brasiliense, indicating that both strains were incorrectly named. ANI and dDDH values showed the highest homology of C. P. maceratum with P. c. subsp. odoriferum, which agreed with previous findings [20]. Nevertheless, the rest of in-silico analyses, including the phylogenetic tree, BLAST matrix, codon and amino acid usages, and pan and core genome dendrogram revealed that C. P. maceratum clustered close to P. polaris.
The pan-core genome analysis showed a pan and core genome values of 9296 and 2414 genes, respectively (Figure 5A). Glasner et al. [8] compared P. atrosepticum, P. c. subsp. carotovorum and P. c. subsp. brasiliense and reported pan and core genome sizes of 6065 and 3264, respectively. In a recent study conducted in 2018 [24], compared P. c. subsp. carotovorum, P. c. subsp. brasiliense and P. c. subsp. odoriferum and revealed a pan and core genome sizes of 3107 and 3903, respectively—indicating great similarity between these subspecies. The significant increase and decrease in pan and core genomes, respectively, indicate high heterogeneity.
Three types of T1SS have been described in Pectobacterium: the prtDEF operon involved in the secretion of hydrolases, a T1SS that modulates iron acquisition through the secretion of hemophore HasA, and a T1SS that secretes a proteinaceous multi-repeat adhesin protein (MRP) [65]. Only T1SS involved in iron uptake was not present in P. parmentieri, P. wasabiae, and P. betavasculorum, which suggests the inefficiency of these Pectobacterium species over the other species in iron-scavenging process. On the other hand, the length of the gene encoding the adhesin protein (MRP) significantly differed among the species (Figure S5). This MRP has been confirmed to mediate bacterial attachment into the plant tissue [71]. We hypothesized that the differences within this gene could be associated with host specificity since P. atrosepticum, P. betavasculorum, P. c. subsp. actinidiae, and P. wasabiae are restricted to potato, sugar beet, kiwi, and Japanish horseradish, respectively. The other two T1SS and T2SS clusters appeared to be conserved in all species (Tables S14 and S15). Only the outN gene was not present in P. parmentieri and P. wasabiae (Table S15 and Figure S6). However, this gene has not been considered to be an essential element of T2SS [73]. The primary function of T2SS is the translocation of the PCWDE to the extracellular medium [27,72]. Almost all PCWDE contributed to the core genome. However, the HrpK protein with a pectate lyase domain, a pectate lyase located nearby the T2SS cluster (Figure S6), and the polygalacturonase PehK were absent in both P. parmentieri and P. wasabiae. These results are congruent with the outcomes by Nykyri et al. [26]. Contrary to the PCWDE, the proteases are secreted by T1SS, known as prtDEF operon [27,65]. Except some proteases commonly found in all—mainly metalloproteases PrtW and Prt1—others were observed to be unique in P. aoridearum, P. parmentieri, and P. wasabiae (Table S16). These findings suggest that some proteases, unlike the PCWDE, tend to vary and are specific to certain species.
In the case of T3SS, only P. parmentieri and P. wasabiae lacked this cluster (Table S17). Kim et al. [105] demonstrated that P. parmentieri WPP163 was able to cause disease in plant stems and tubers; thus, confirming that T3SS is not essential in Pectobacterium species for disease development. The other 10 species harbor T3SS exhibited variable regions that were composed of unknown proteins and components of T6SS between the loci hrpN and hrpW (Figure 9). The influence of this variable region on the function of the T3SS has yet to be proved. Additionally, a hrpK gene, adjacent to the T3SS cluster and predicted to encode a T3SS dependent effector protein, was found in P. c. subsp. carotovorum, P. c. subsp. brasiliense, P. c. subsp. actinidiae, P. aroidearum, P. polaris, P. peruviense, and C. P. maceratum (Table S17). Glasner et al. [8] also found this gene in P. c. susbp. carotovorum WPP14 and P. c. subsp. brasiliense 1692. However, the role of this gene during plant infection is still unknown; indeed, the HrpK-like effector was found not to be fundamental in the promotion of cell death in tobacco or translocation of the effector DspE [8,106].
Some Pectobacterium species harbor the conjugation system virB-T4SS of Agrobacterium tumefaciens—this was the case of P. atrosepticum [21], P. c. subsp. brasiliense 1692 [8], P. parmentieri SCC3193, and P. wasabiae CFBP 3304 [26]. P. c. subsp. carotovorum WPP14 was described to possess an incomplete virB operon [8], while P. parmentieri WPP163 and P. aroidearum lacked this cluster [26]. In our analyses, we identified a virB-T4SS in P. c. subsp. actinidiae KKH3 and P. betavasculorum NCPPB2795, and surprisingly, P. c. subsp. brasiliense BC1 harbor two virB-T4SS clusters (Figure S7 and Table S18). However, two other strains of P. c. subsp. brasiliense (SX309 and BZA12) did not carry two copies of this cluster. The virB-T4SS has been reportedly harbored within a genomic island in P. atrosepticum [21]. This indicates that the virB-T4SS is distributed sporadically throughout the species and even strains within Pectobacterium genus.
Pectobacterium harbors a T5SS known as the two-partner secretion system (type Vb or Tps) [65]. In P. atrosepticum, the T5SS, located next to the T3SS, consisted a hecB gene that encodes for a small transporter TpsB, and two hecA genes that encode an exoprotein, called hemagglutinin [21]. The same pattern was observed for the remaining species harbored the T3SS, except for P. c. subsp. actinidiae, where the cluster was found elsewhere in the chromosome. Likewise, the sizes and number of copies of hecA gene varied widely among the species. In D. chrysanthemy EC16, the hemagglutinin that was encoded by hecA was observed to provide attachment, aggregation, and promote cell death in the leaves of Nicotiana clevelandii [107]. The fact that the HecA protein is involved in cell host adhesion, like the MRP protein, reinforces our hypothesis that the genetic variability of both proteins lies in the Pectobacterium resulted in host specificity. Besides the HecA, TpsB reportedly secretes other cytotoxins and growing inhibitors [76]. In P. parmentieri strains WPP163 and SCC3193, for instance, a contact-dependent growth inhibition (CDI) was described [26,108]. Poole et al. [108] reported a novel Tps system, called rearrangement hotspot (Rhs), whose function is involved in toxicity and intercellular competition against other bacteria. Additionally, we identified a novel Tps cluster that resembles the hecAB operon that presents a toxin between the hecB and hecA loci instead of harboring two copies of the hecA gene (Table S14 and Figure S8). This cluster was not found in P. wasabiae and P. peruviense. The different types of T5SS reported in Pectobacterium, and the new cluster that was identified in this study, highlighted the possibility that uncharacterized T5SS might play important role in infection and disease development.
Genomic analysis revealed a T6SS in all Pectobacterium species (Table S20 and Figure S9). However, a part of this cluster was not present in P. betavascolorum, while the T6SS of P. c. subsp. odoriferum harbor an extra set of genes that encoded for unknown proteins. Nykyri et al. [26] reported that P. parmentieri strains SCC3193, WPP163, and P. wasabiae CFBP3304 contain two T6SS whose function seems to be redundant. A handful of functions have been associated with T6SS, including interaction with host eukaryotic cells, pathogenicity, antibacterial activity, symbiosis, metal ion acquisition, and biofilm formation [77,80,109]. In P. atrosepticum, this secretion system allegedly alters the plant defense mechanisms [110]. However, Mattinen et al. [81] stated that the T6SS function is to secret Hcp proteins, rather than virulence. Haemolysin-coregulated protein (Hcp) and valine-glycine repeat protein (VgrG), the major effector proteins of the T6SS [78,79], have been described to be present in several copies and scattered in the chromosomes of P. atrosepticum, P. parmentieri, and P. wasabiae [26,81]. The same pattern was observed for the other species in the current study. Nykyri et al. [26] indicated that the number of hcp and vgrG genes differs among species and at the strain level. The distinct genetic organization of this cluster indicates the need for further studies to clarify the functions of T6SS and decipher how the T6SS-effectors act during disease development.
P. atrosepticum and P. parmentieri have a type IV pili or fimbriae cluster, predicted to confer adhesion into the cell hosts [111]. We identify a similar pil operon in P. peruviense within a genomic island (Figure S10). Furthermore, we found a gene termed pilW and another operon consisted of three genes—ppdD, hofB, and hofC—that are homologues in all species (Table S21). In Nisseria meningitis, pilW stabilized the pilus fiber [112] and, in Pseudomonas aeruginosa, involved in the insertion and multimerization of PilQ [113]. The pilABC operon, homologous to the ppdD, hofB, and hofC, has been associated with bacterial adherence in P. aeruginosa [114,115]. In Pectobacterium, the functionality of pilW and pilABC is still to be elucidated. The Flp/Tad pilus [116], another class of fimbriae, was harbored by the 12 species (Table S22 and Figure S11). Nykyri et al. [116] proved the association of flp/tad operon with maceration of potato tubers, highlighted the importance of this cluster during disease development, which would explain why this was highly conserved in Pectobacterium.
The biosynthesis of phytotoxins, such as coronofacic acid (cfa) and syringomycin, are described as important pathogenicity determinants in Pectobacterium [27]. The cfa cluster was first reported in P. atrosepticum within a genome island, and its capacity to boost blackleg disease development has already been proven [21]. Panda et al. [117] showed a similar cfa operon in the genome islands of P. atrosepticum IMCP 1526, P. c. subsp. brasiliense IMCP19477, and P. peruviense IFB5232 (formerly P. c. subsp. carotovorum UGC32). We identified the same cfa cluster in P. peruviense IFB5229 (Figure 7) as well as in the kiwi and sugar beet pathogens P. c. subsp. actinidiae and P. betavasculorum (Figure S12), and, like the other species, this phytotoxin operon was contained in GIs, which indicates that this cluster was acquired by other Pectobacterium species and strains via horizontal genetic transfer. Consistent with this inference, Panda et al. [117] suggested that cfa could be acquired prior speciation of P. atrosepticum. The NRPS, like syringomycin, was also predicted into a GI in P. atrosepticum [21] and P. c subsp. brasiliense ICMP 19477. In our analysis, only P. c. subsp. actinidiae had a similar NRPS-syringomycin cluster (Table S14), which was harbored into a GI. Although the syringomycin toxin was reported to induce electrolyte leakage, leading to necrosis symptoms in P. syringae [118], the strict virulence role of this toxin has not been clarified in Pectobacterium. Our finding of syringomycin cluster in P. c. susbp. actinidiae brings new research to decipher whether this phytotoxin contributes to the development of canker disease in kiwi fruits.
The acquisition of arsenic clusters (ars) confers the ability to resist arsenic concentration in the environment [83]. These ars genes can be part of the chromosome or plasmid or have acquired through horizontal gene transfer [83]. P. parmentieri SCC3193 possess an arsenic resistance cluster of 13,686 bp harbored within a GIs [26]. P. atrosepticum presents four genes of this cluster, also contained into the HAI7 [21]. We observed that P. c. subsp. brasiliense, P. peruviense, and C. P. maceratum have orthologues to the arsC, arsB, and arsR genes of P. atrosepticum. However, the complete ars operon of P. parmentieri SCC3193 was not identified in any of the Pectobacterium species (Figure S13) or other P. parmentieri strains. Nykyri et al. [26] reported that even P. parmentieri WPP163 and its close relative P. wasabiae lacked this arsenic cluster. Since P. parmentieri SCC3193 was isolated from southern Finland, whose cultivated soils exhibits the highest arsenic concentrations [26], it is logical to assume that this phytobacteria have evolved and acquired a complete ars cluster to adapt into this niche. The arsC gene of P. parmentieri was described to present two additional copies that were located elsewhere in the genome [26]. One locus is shared across all species (Table S25). Likewise, we identified an arsenic resistance protein (arsH) that displayed a high identity in all species. The presence of these two genes suggests that they are vital for living and bacterial growth inside ecological niches surrounded by arsenic components.
Members of the Pectobacterium genus have been reported to produce cell surface polysaccharides [27]. P. atrosepticum, for instance, harbors a capsular polysaccharide cluster (cps) [21] that—according to our data—is highly conserved, but only present in few species (Table S26 and Figure S14). Except ECA cluster—shared by all species (Table S27 and Figure S15), the EPS (extracellular polysaccharides), and O-antigen cluster of P. atrosepticum [21] and lipo-oligo/polysaccharide cluster (LOS/LPS) of P. parmentieri SCC3193 [26] were partially conserved and displayed distinct genetic rearrangements (Tables S28 and S29 and Figures S16 and S17). Evans et al. [119] observed that mutants in the LPS biosynthesis cluster of P. atrosepticum SCRI1043 led to a reduction in potato soft rot. We assumed that divergences that were observed in EPS and LPS of Pectobacterium species could be the result of specialized adaptive mechanisms that each species has developed to survive.
Iron sequestration constitutes a crucial step for bacterial pathogens during the infection process [84]. Bell et al. [21] reported five different iron uptake systems in P. atrosepticum SCRI1043, including achromobactin, enterobactin, ferric citrate, hemophore, and hemin storage. The achromobactin (cbrABCD) was present in all species (Table S30), which suggests that this operon is crucial for iron assimilation, especially in niches with poor iron content. Contrary to cbrABCD, the enterobactin (entAFBECD), heme operon (hasRADEF), and ferric citrate uptake system (fecIRABCDE) were either partially present or absent in P. parmentieri and P. wasabiae. The hemin storage cluster (hmsHFRS) was identified in few species: P. atrosepticum, P. aroidearum, P. parmentieri, P. wasabiae, and P. peruviense—this operon was involved in biofilm matrix formation [120]. The aerobactin cluster (iuc), found first in P. c. subsp. carotovorum W3C105 [85] and later in P. parmentieri SCC3193 [26], also found in other P. parmentieri strains (Table S31). This finding suggests that aerobactin is a distinctive characteristic of P. parmentieri, and might subtitute the absence of ferric citrate and heme acquisition clusters. A recently plant-ferredoxin uptake system (fusABCD), characterized in P. c. subsp. carotovorum PCC21 [121,122], was harbored in almost all species except for P. betavasculorum, which also lacked fecIRABCDE, hasRADEF, and hmsHFRS clusters.
Medium alkalization was described to be crucial for the accurate activity of pectate lyases during potato tubers infection in P. c. subsp. carotovorum WPP14 [92]. This function was described to be governed by acetoin or the 3-hydroxy-2-butanone pathway (3H2B) that was encoded by the bud operon [92]. In our analysis, this operon was highly conserved in all 12 Pectobacterium species (Table S33 and Figure S18), which suggests that the 3H2B pathway is a vital component at the early stages of infection not only in potato, but also in other Pectobacterium hosts, such as kiwi, sugar beet, cucumber, chicory, and Chinese cabbage.
Motility is a fundamental virulence factor in motile bacteria [27]. The flagella cluster was highly conserved in the entire Pectobacteirum genus. In accordance with Nykyri et al. [26], we also observed that, in all species, the flagella genes were located within predicted GIs (Tables S2–S13). Therefore, it seems that the common ancestor of all Pectobacterium species acquired the flagellar apparatus via horizontal DNA transfer in order to move inside the plant cells, rapidly colonize the plant host tissues, or scape from harmful environmental conditions. Previous reports demonstrated that flagellar genes were involved in chemotaxis, and motility is fundamental for pathogenicity in D. dadantii, whereas, in Pectobacterium, FliC elicited the plant cell defenses [65]. The non-motile mutants of P. parmentieri SCC3193 displayed a diminished virulence in tobacco [123]. Furthermore, it observed that the flagella were involved in colicin delivery, a toxin that kills closely related species, suggesting that the flagella operon in Pectobacterium can also be used as a weapon against other bacteria [65].
The single gene encoded virulence factors carried by all Pectobacterium species (Table S32). These loci include: the necrosis inducing virulence protein (Nip)—observed in P. parmentieri SCC3193, P. atrosepticum SCRI1043, and P. c. subsp. carotovorum ATTn10 essential for virulence in tuber [87,124]; the svx gene, whose inactivation in P. atrosepticum SCRI1043 decreased virulence in potato [86]; the citrate transporter encoded by cit1 gene, which is involved in citrate uptake and tuber maceration [88]; the rplY gene, proved to be crucial in pathogenesis in P. carotovorum PccS1 [91]. We speculate that these genes constitute fundamental elements for the virulence lifestyle of entire genus. In contrast to these loci, the sax operon [90], the phytase gene appA [89], and the evf [9] were absent in few Pectobacterium species (Table S32). The Erwinia virulence factor (evf), for instance, was proven to allow the interactions of some P. carotovorum strains and enhanced bacterial persistence into the gut of D. melanogaster, thus contributing the spreading of these bacteria [9]. In our analysis, only C. P. maceratum harbored this gene. Similarly, the evf gene was also found in P. carotovorum SCC1 [34], as proposed by Shirshikov et al. [20], as the type strain of C. P. maceratum.
Signal molecules regulate most of the pathogenicity factors, which act as a complex network system [27,65]. Mutagenesis conducted in some of these regulators significantly affected the virulence and growth rate of Pectobacterium sp. [27,65,110]. In our data, nearly all of the virulence regulators were harbored by all Pectobacterium species (Table S34), indicating their crucial role in bacterial survival, growth, and disease development.
The secretion of antimicrobial compounds boosts ecological fitness and allows for bacteria to outcompete endophytes or secondary invaders [8]. In Pectobacterium, four components have been described: carotovoricin (Ctv) [31,32], carbapenem [28], phenazine [21], and bacteriocins (carocins) [29,30,68]. Only the carotovoricin (ctv), which was composed of 21 genes encoding phage structures [67], was found in all Pectobacterium species (Table S35) and adjacent to the T1SS (the prtDEF operon) (Figure S19). The location of the ctv operon close to T1SS shows that this bactericide may have evolved together with prtDEF operon. Significant genomic conservation of T1SS+Ctv block was found in 96.7% of Pectobacterium genomes, which suggests that T1SS might export Ctv elements through the periplasm [39]. Contrary to the carotovoricin, the carbapenem cluster was observed in few species (Table S36). P. c. subsp. carotovorum ATTC 39048 was the first species reported as carbapenem producer bacterium [95]. Later, Glasner et al. [8] reported in P. c. subsp. brasiliense 1692 the same car cluster, which inhibited the growth of P. c. subsp. carotovorum WPP14 and P. atrosepticum SCRI1043 [92]. We identified the car operon in all P. c. subsp. actinidiae and P. betavasculorum strains (Table S36 and Figure S20). Surprisingly, P. c. subsp. brasiliense BC1, P. parmentieri, and P. wasabiae only presented carF, carG, and carH genes (Figure S20). As the carF and carG genes confer protection against the carbapenem [96], it seems that these three species may be immune against the antibiosis effect of carbapenem. The phenazine (ehp), on the other hand, has been found within genome islands of P. c. subsp. brasiliense ICMP19477 and P. atrosepticum strains SCRI1043 and ICMP1526 [21]. In the present study, a similar synteny of phenazine cluster was observed in all P. c. subsp. actinidiae and P. betavasculorum strains (Table S37 and Figure S21).
Several bacteriocins have already been identified within Pectobacterium genus, including Carocin S1 [29], Carocin D produced by P. c. subsp. carotovorum PCC21 [69], Carocin S2 of P. c. subsp. carotovorum F-rif-18 [68], two colicin M-like bacteriocins with a ferredoxin domain termed Pectocin M1 and M2 characterized in P. aroidearum PC1 and P. c. subsp. brasiliense 1692, respectively [98]. We found that P. c. susbp. brasiliense BC1, P. c. subsp. odoriferum BC S7, and P. betavasculorum harbor homologous Carocin D. Further, a type of unreported colicin/pyocin-like bacteriocins were observed in P. carotovorum subspecies (carotovorum, brasiliense, and odoriferum) and in P. wasabiae (Table S38). P. c. subsp. actinidiae and P. betavasculorum contained the phenazine and carbapenem clusters, which have a broad antibiosis spectrum [125,126], might explain why other Pectobacterium species have not been isolated yet from kiwi and sugar beet plants.
Multiple genome alignment led us to discern a ~4.54 kb novel cellobiose-specific phosphotransferase system (PTS) specific for all P. parmentieri strains (Figure S22). In Klebsiella pneumoniae, the celB gene, encoding component EIIC within the cellobiose-specific phosphotransferase system, was reported to be involved in biofilm formation as well as in cellobiose transport; showed a dual role of this cellobiose-specific PTS and its contribution in virulence [127]. The fructose-specific PTS permease IIA was proven to facilitate the biofilm formation and mediate the stress response in Listeria monocytogenes [128]. When considering this previous data, it can be speculated that the uncharacterized cellobiose-PTS found in our study might have involved in virulence functions and, therefore, could be a pathogenic determinant in P. parmentieri.
The CRISPR-Cas is defined as the innate and adaptive immune system of bacteria [99]. We articulated that the presence of two CRISPR-Cas systems in some Pectobacterium species (Figure 10) might confer a better immune system against invasive elements. P. aroidearum PC1 was the only species carried type III-A CRISPR-Cas (Figure 10C); this CRISPR type was found with less frequency among the bacterial species [129]. Interestingly, the entire CRISPR-Cas in PC1 was located within a genomic island (GI-22; Table S7) and exhibited 90% homology with Serratia sp. ATCC 39066; thus, indicating a DNA transfer event of this system. Consistent with this outcome, the CRISPR-Cas systems have been suggested to be mobile genetic elements acquired via horizontal gene transfer [130], where plasmids, and even phages, seem to have facilitated the spreading of this immunity system across the bacterial taxa [131,132,133]. We observed a peculiar region of 49 CDS between the csy3 and csy2 genes in P. c. subsp. brasiliense SX309 (Figure 10A). This region encoded proteins related to structures of phages, suggesting that the subtype I-F CRISPR-Cas might have a phage origin in SX309. Moreover, the lack of CRISPR-Cas in P. polaris NIBIO1006 and P. peruviense IFB5229 supports the theory that CRISPR-Cas can be acquired by certain species, but not by all. Different toxin-antitoxin systems were distinguished between the CRISPR loci and the cas genes (Figure 10A,B). Kooni and Makarova [134] observed that prokaryotic toxins are tightly associated with CRISPR-Cas and they trigger cell death or dormancy in the case of an immune system failure. Numerous toxin and antitoxin genes were solely found surrounding the type I CRISPR-Cas system [135]—these findings are in concordant to our outcomes, and reinforce the hypothesis that CRISPR-Cas and suicide response mediated by toxin-antitoxin systems. Another aspect of being highlighted was the presence of an Hcp protein, a T6SS component (Figure 10A), within the subtype I-F CRISPR-Cas. Congruently with our information, a ~28 kb GI containing a CRISPR-Cas system and proteins of the T6SS (VgrG, Hcp, and PAAR) were reported in Vibrio cholerae S12; showed the importance of GIs as propagators of CRISPR-Cas and T6SS [136]. Different CRISPR arrays, distinct sizes, amounts, and locations were observed within genus Pectobacterium (Table S40). We identified strains with just three spacers, while others with 41. Usually, CRISPR loci are next to the cas genes [137]. However, CRISPR arrays have also been reported to be located distantly from the cas operon in Listeria monocytogenes, Aggregatibacter actinomycetemcomitans, and Enterococcus faecalis [138]. The same pattern was observed with some Pectobacterium species (Figure 11). In our analyses, 13 orphans CRISPRs were found throughout the 34 analyzed Pectobacterium genomes (Table S40). Zhang et al. [138] reported that some CRISPRs seem to exert their function far away from the cas locus, although they might not be functional. Molecular assays are necessary for proving whether the orphan CRISPRs found in some Pectobacterium species are functional or just remnants of previous CRISPR-Cas systems.
5. Conclusions
To conclude, the general genomic features showed that P. c. subsp. odoriferum possessed the highest number of pseudogenes, indicating a coding ability degeneration, while P. parmentieri harbored the largest number of CDS. Zones of high expression levels that seem to be controlled by the nucleosome were found in the genome atlases of P. c. subsp. carotovorum, P. aroidearum, and P. c. subsp. odoriferum, whereas P. atrosepticum appeared to encompass more unstable regions. Our comparative analysis unveiled that the T2SS, T6SS, T1SS, some PCWDE and proteases, bud operon, the ECA cluster, achromobactin, the flagella genes, single virulence locus, the pilW and pilABC genes, Flp/Tad, carotovoricin, and the majority of virulence regulators appear to be crucial components for the survival and virulence lifestyle of the entire Pectobacterium genus. In contrast, T3SS, T4SS, T5SS, phytotoxins, type IV pilus, capsular polysaccharide, lipopolysaccharides, exopolysaccharides, iron uptake systems, phenazine, carbapenem, and colicin-like bacteriocins were exclusively observed in some species; highlighting that species harboring these elements might have an advantage over the others. Our study found the novel cellobiose phosphotransferase system (PTS) exclusively present in P. parmentieri, and an unreported T5SS conserved in almost all Pectobacterium species. Three CRISPR-Cas systems: I-E, I-F, and III-A, along with toxin-antitoxin systems and Hcp proteins, were found. Several pathogenicity determinants, antimicrobial clusters, and the type III-A CRISPR-Cas system were located within genomic islands, indicating the influence of horizontal gene transfer in this genus. We believe that our findings reported in this manuscript constitute significant insights regarding how different Pectobacterium species are equipped, and have evolved to outcompete with their relatives, and adapted host specificity. Further, the findings of novel clusters and general description of CRISPR-Cas systems provide ample opportunities for future investigations on Pectobacterium.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/247/s1.
Author Contributions
Conceptualization, M.A.; methodology, D.A. and M.A.; software, D.A. and M.A.; validation, D.A. and M.A.; formal analysis, D.A.; investigation, D.A. and M.A.; resources, M.A.; data curation, D.A.; writing—original draft preparation, D.A.; writing—review and editing, M.A.; visualization, D.A. and M.A.; supervision, M.A.; project administration, M.A.; funding acquisition, M.A.
Funding
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 9038H, managed by the College of Tropical Agriculture and Human Resources.
Conflicts of Interest
Authors declare no competing interest.
References
- Czajkowski, R.; Pérombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Davidsson, P.R.; Kariola, T.; Niemi, O.; Palva, T. Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; Bell, K.S.; Holeva, M.C.; Birch, P.R.J. Soft rot erwiniae: From genes to genomes. Mol. Plant Pathol. 2003, 4, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Hibbing, M.E.; Kim, H.-S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, F.; Wu, Y.; Hu, H.; Dai, X. Progress of potato staple food research and industry development in China. J. Integr. Agric. 2017, 16, 2924–2932. [Google Scholar] [CrossRef]
- Nabhan, S.; Boer, S.H.; Maiss, E.; Wydra, K. Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J. Appl. Microbiol. 2012, 113, 904–913. [Google Scholar] [CrossRef]
- Glasner, J.D.; Marquez-Villavicencio, M.; Kim, H.-S.; Jahn, C.E.; Ma, B.; Biehl, B.S.; Rissman, A.I.; Mole, B.; Yi, X.; Yang, C.-H.; et al. Niche-specificity and the variable fraction of the Pectobacterium pan-genome. Mol. Plant Microbe Interact. 2008, 21, 1549–1560. [Google Scholar] [CrossRef]
- Basset, A.; Tzou, P.; Lemaitre, B.; Boccard, F. A single gene that promotes interaction of a phytopathogenic bacterium with its insect vector, Drosophila melanogaster. EMBO Rep. 2003, 4, 205–209. [Google Scholar] [CrossRef]
- Hauben, L.; Moore, E.R.B.; Vauterin, L.; Steenackers, M.; Mergaert, J.; Verdonck, L.; Swings, J. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 1998, 21, 384–397. [Google Scholar] [CrossRef]
- Gardan, L.; Gouy, C.; Christen, R.; Samson, R. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Duarte, V.; De Boer, S.H.; Ward, L.J.; Oliveira, A.M.R. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 2004, 96, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.; Legendre, J.B.; Christen, R.; Saux, M.F.-L.; Achouak, W.; Gardan, L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1415–1427. [Google Scholar] [PubMed]
- Brady, C.L.; Cleenwerck, I.; Venter, S.N.; Engelbeen, K.; De Vos, P.; Coutinho, T.A. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2430–2440. [Google Scholar]
- Koh, Y.J.; Kim, G.H.; Lee, Y.S.; Sohn, S.H.; Koh, H.S.; Kwon, S.; Heu, S.; Jung, J.S. Pectobacterium carotovorum subsp. actinidiae subsp. nov., a new bacterial pathogen causing canker-like symptoms in yellow kiwifruit, Actinidia chinensis. N. Z. J. Crop Hortic. Sci. 2012, 40, 269–279. [Google Scholar]
- Nabhan, S.; De Boer, S.H.; Maiss, E.; Wydra, K. Pectobacterium aroidearum sp. nov., a soft rot pathogen with preference for monocotyledonous plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2520–2525. [Google Scholar] [CrossRef]
- Khayi, S.; Cigna, J.; Chong, T.M.; Quêtu-Laurent, A.; Chan, K.-G.; Hélias, V.; Faure, D. Transfer of the potato plant isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5379–5383. [Google Scholar]
- Dees, M.W.; Lysøe, E.; Rossmann, S.; Perminow, J.; Brurberg, M.B. Pectobacterium polaris sp. nov., isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2017, 67, 5222–5229. [Google Scholar] [CrossRef]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Wielgomas, B.; Waleron, K. Transfer of Pectobacterium carotovorum subsp. carotovorum strains isolated from potatoes grown at high altitudes to Pectobacterium peruviense sp. nov. Syst. Appl. Microbiol. 2018, 41, 85–93. [Google Scholar] [CrossRef]
- Shirshikov, F.V.; Korzhenkov, A.A.; Miroshnikov, K.K.; Kabanova, A.P.; Barannik, A.P.; Ignatov, A.N.; Miroshnikov, K.A. Draft genome sequences of new genomospecies “Candidatus Pectobacterium maceratum” Strains, Which Cause Soft Rot in Plants. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.G.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.C.; Mungall, K.; et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 2004, 101, 11105–11110. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.O.; Lind, J.; Rubio-Salazar, I. Genomics of plant-associated bacteria: The soft rot Enterobacteriaceae. In Genomics of Plant-Associated Bacteria; Gross, D.C., Lichens-Park, A., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 37–58. [Google Scholar]
- Lee, D.H.; Lim, J.-A.; Lee, J.; Roh, E.; Jung, K.; Choi, M.; Oh, C.; Ryu, S.; Yun, J.; Heu, S. Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology 2013, 159, 1487–1496. [Google Scholar] [PubMed]
- Huang, Y.; Liu, C.; Wang, H.; Guan, T.; Liu, L.; Yu, S. Bioinformatic analysis of the complete genome sequence of Pectobacterium carotovorum subsp. brasiliense BZA12 and candidate effector screening. J. Plant Pathol. 2019, 101, 39–49. [Google Scholar] [CrossRef]
- Waleron, M.; Waleron, K.; Lojkowska, E. Characterization of Pectobacterium carotovorum subsp. odoriferum causing soft rot of stored vegetables. Eur. J. Plant Pathol. 2014, 139, 457–469. [Google Scholar] [CrossRef]
- Nykyri, J.; Niemi, O.; Koskinen, P.; Nokso-Koivisto, J.; Pasanen, M.; Broberg, M.; Plyusnin, I.; Törönen, P.; Holm, L.; Pirhonen, M.; et al. Revised phylogeny and novel horizontally acquired virulence determinants of the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Toth, I.K.; Pritchard, L.; Birch, P.R.J. Comparative genomics reveals what makes an enterobacterial plant pathogen. Annu. Rev. Phytopathol. 2006, 44, 305–336. [Google Scholar] [CrossRef]
- McGowan, S.J.; Sebaihia, M.; Porter, L.E.; Stewart, G.S.; Williams, P.; Bycroft, B.W.; Salmond, G.P.C. Analysis of bacterial carbapenem antibiotic production genes reveals a novel β-lactam biosynthesis pathway. Mol. Microbiol. 1996, 22, 415–426. [Google Scholar] [CrossRef]
- Chuang, D.; Chien, Y.; Wu, H.-P. Cloning and expression of the Erwinia carotovora subsp. carotovora gene encoding the low-molecular-weight bacteriocin Carocin S1. J. Bacteriol. 2007, 189, 620–626. [Google Scholar] [CrossRef]
- Chan, Y.; Wu, H.-P.; Chuang, D. Extracellular secretion of Carocin S1 in Pectobacterium carotovorum subsp. carotovorum occurs via the type III secretion system integral to the bacterial flagellum. BMC Microbiol. 2009, 9. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Tomita, T.; Hirota, M.; Sato, T.; Kamio, Y. A simple purification method and morphology and component analyses for carotovoricin Er, a phage-tail-like bacteriocin from the plant pathogen Erwinia carotovora Er. Biosci. Biotechnol. Biochem. 1999, 63, 1360–1369. [Google Scholar] [CrossRef]
- Itoh, Y.; Izaki, K.; Takahashi, H. Purification and characterization of a bacteriocin from Erwinia carotovora. J. Gen. Appl. Microbiol. 1978, 24, 27–39. [Google Scholar] [CrossRef]
- Park, T.-H.; Choi, B.-S.; Choi, A.-Y.; Choi, I.-Y.; Heu, S.; Park, B.-S. Genome sequence of pectobacterium carotovorum subsp. carotovorum strain pcc21, a pathogen causing soft rot in Chinese cabbage. J. Bacteriol. 2012, 194, 6345–6346. [Google Scholar] [CrossRef] [PubMed]
- Niemi, O.; Laine, P.; Koskinen, P.; Pasanen, M.; Pennanen, V.; Harjunpää, H.; Nykyri, J.; Holm, L.; Paulin, L.; Auvinen, P.; et al. Genome sequence of the model plant pathogen Pectobacterium carotovorum SCC1. Stand. Genom. Sci. 2017, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Bai, Y.; Guo, C. Comparative genomics of 151 plant-associated bacteria reveal putative mechanisms underlying specific interactions between bacteria and plant hosts. Genes Genom. 2018, 40, 857–864. [Google Scholar] [CrossRef]
- McAdam, P.R.; Richardson, E.J.; Fitzgerald, J.R. High-throughput sequencing for the study of bacterial pathogen biology. Curr. Opin. Microbiol. 2014, 19, 106–113. [Google Scholar] [CrossRef]
- Xu, J.; Wang, N. Where are we going with genomics in plant pathogenic bacteria? Genomics 2019, 111, 729–736. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Rollins, J.R.; Sudarshana, M.R.; Vinatzer, B.A. Disease management in the genomics era—summaries of focus issue papers. Phytopathology 2016, 106, 1068–1070. [Google Scholar] [CrossRef]
- Bellieny-Rabelo, D.; Tanui, C.K.; Miguel, N.; Kwenda, S.; Shyntum, D.Y.; Moleleki, L.N. Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in Pectobacterium and Dickeya spp. Appl. Environ. Microbiol. 2018, 85. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, 581–591. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, 535–542. [Google Scholar] [CrossRef]
- Vesth, T.; Lagesen, K.; Acar, Ö.; Ussery, D. CMG-Biotools, a free workbench for basic comparative microbial genomics. PLoS ONE 2013, 8, e60120. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ussery, D.W.; Wassenaar, T.M.; Borini, S. Computing for Comparative Microbial Genomics: Bioinformatics for Microbiologists; Springer: London, UK, 2009. [Google Scholar]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Brinkman, F.S.L. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Hsiao, W.W.; Brinkman, F.S. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinform. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Hudson, C.M.; Lau, B.Y.; Williams, K.P. Islander: A database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res. 2015, 43, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. AntiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, 278–281. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, 246–251. [Google Scholar] [CrossRef]
- Biswas, A.; Staals, R.H.J.; Morales, S.E.; Fineran, P.C.; Brown, C.M. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef]
- Uddin, A. Codon Usage Bias: A tool for understanding molecular evolution. J. Proteom. Bioinform. 2017, 10. [Google Scholar] [CrossRef]
- Faye, P.; Bertrand, C.; Pédron, J.; Barny, M.-A. Draft genomes of “Pectobacterium peruviense” strains isolated from fresh water in France. Stand. Genom. Sci. 2018, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; Solanilla, E.L.; Low, D.; Moleleki, L.; et al. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Izaki, K.; Takahashi, H. Simultaneous synthesis of pectin lyase and carotovoricin induced by mitomycin c, nalidixic acid or ultraviolet light irradiation in Erwinia carotovora. Agric. Biol. Chem. 1980, 44, 1135–1140. [Google Scholar] [CrossRef]
- Yamada, K.; Hirota, M.; Niimi, Y.; Nguyen, H.A.; Takahara, Y.; Kamio, Y.; Kaneko, J. Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 2006, 70, 2236–2247. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.-C.; Wu, J.-L.; Wu, H.-P.; Tzeng, K.-C.; Chuang, D.-Y. Cloning, purification, and functional characterization of Carocin S2, a ribonuclease bacteriocin produced by Pectobacterium carotovorum. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef]
- Roh, E.; Park, T.-H.; Kim, M.-I.; Lee, S.; Ryu, S.; Oh, C.-S.; Rhee, S.; Kim, D.-H.; Park, B.-S.; Heu, S. Characterization of a new bacteriocin, Carocin D, from Pectobacterium carotovorum subsp. carotovorum Pcc21. Appl. Environ. Microbiol. 2010, 76, 7541–7549. [Google Scholar] [CrossRef]
- Delepelaire, P. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1694, 149–161. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; Coulthurst, S.J.; Humphris, S.; Campbell, E.; Welch, M.; Toth, I.K.; Salmond, G.P.C. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a Type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase: Complex regulation of a Pectobacterium type I-secreted adhesin. Mol. Microbiol. 2011, 82, 719–733. [Google Scholar] [CrossRef]
- Jha, G.; Rajeshwari, R.; Sonti, R.V. Bacterial type two secretion system secreted proteins: Double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 2005, 18, 891–898. [Google Scholar] [CrossRef]
- Douzi, B.; Filloux, A.; Voulhoux, R. On the path to uncover the bacterial type II secretion system. Phil. Trans. R. Soc. B 2012, 367, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Whitaker, N.; González-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Trokter, M.; Waksman, G. Translocation through the conjugative type IV secretion system requires unfolding of its protein substrate. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Van Ulsen, P.; ur Rahman, S.; Jong, W.S.P.; Daleke-Schermerhorn, M.H.; Luirink, J. Type V secretion: From biogenesis to biotechnology. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1592–1611. [Google Scholar] [CrossRef]
- Bernard, C.S.; Brunet, Y.R.; Gavioli, M.; Lloubes, R.; Cascales, E. Regulation of type VI secretion gene clusters by 54 and cognate enhancer binding proteins. J. Bacteriol. 2011, 193, 2158–2167. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef]
- Mattinen, L.; Nissinen, R.; Riipi, T.; Kalkkinen, N.; Pirhonen, M. Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics 2007, 7, 3527–3537. [Google Scholar] [CrossRef]
- Cianfanelli, F.R.; Alcoforado Diniz, J.; Guo, M.; De Cesare, V.; Trost, M.; Coulthurst, S.J. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef]
- Mattinen, L.; Somervuo, P.; Nykyri, J.; Nissinen, R.; Kouvonen, P.; Corthals, G.; Auvinen, P.; Aittamaa, M.; Valkonen, J.P.T.; Pirhonen, M. Microarray profiling of host-extract-induced genes and characterization of the type VI secretion cluster in the potato pathogen Pectobacterium atrosepticum. Microbiology 2008, 154, 2387–2396. [Google Scholar] [CrossRef]
- Maier, B.; Wong, G.C.L. How bacteria use type IV pili machinery on surfaces. Trends Microbiol. 2015, 23, 775–788. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef] [PubMed]
- Mosbahi, K.; Wojnowska, M.; Albalat, A.; Walker, D. Bacterial iron acquisition mediated by outer membrane translocation and cleavage of a host protein. Proc. Natl. Acad. Sci. USA 2018, 115, 6840–6845. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, C.A.; Loper, J.E. High-affinity iron uptake systems present in Erwinia carotovora subsp. carotovora include the hydroxamate siderophore aerobactin. J. Bacteriol. 1992, 174, 2993–3003. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Corbett, M.; Virtue, S.; Bell, K.; Birch, P.; Burr, T.; Hyman, L.; Lilley, K.; Poock, S.; Toth, I.; Salmond, G. Identification of a new quorum-sensing-controlled virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Mol. Plant Microbe Interact. 2005, 18, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Mattinen, L.; Tshuikina, M.; Mäe, A.; Pirhonen, M. Identification and characterization of Nip, Necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 2004, 17, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Urbany, C.; Neuhaus, H.E. Citrate uptake into Pectobacterium atrosepticum is critical for bacterial virulence. Mol. Plant Microbe Interact. 2008, 21, 547–554. [Google Scholar] [CrossRef]
- Shao, N.; Huang, H.; Meng, K.; Luo, H.; Wang, Y.; Yang, P.; Yao, B. Cloning, expression, and characterization of a new phytase from the phytopathogenic bacterium Pectobacterium wasabiae DSMZ 18074. J. Microbiol. Biotechnol. 2008, 18, 1221–1226. [Google Scholar]
- Welte, C.U.; Rosengarten, J.F.; de Graaf, R.M.; Jetten, M.S.M. SaxA-Mediated isothiocyanate metabolism in phytopathogenic pectobacteria. Appl. Environ. Microbiol. 2016, 82, 2372–2379. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, M.; Yang, L.; Yao, P.; Ma, L.; Wang, C.; Wang, H.; Qian, G.; Hu, B.; Fan, J. The ribosomal protein RplY is required for Pectobacterium carotovorum virulence and is induced by Zantedeschia elliotiana extract. Phytopathology 2017, 107, 1322–1330. [Google Scholar] [CrossRef]
- Del Pilar Marquez-Villavicencio, M.; Weber, B.; Witherell, R.A.; Willis, D.K.; Charkowski, A.O. The 3-hydroxy-2-butanone pathway is required for Pectobacterium carotovorum pathogenesis. PLoS ONE 2011, 6, e22974. [Google Scholar] [CrossRef]
- McGowan, S.; Sebaihia, M.; Jones, S.; Yu, B.; Bainton, N.; Chan, P.F.; Bycroft, B.; Stewart, G.S.A.B.; Williams, P.; Salmond, G.P.C. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology 1995, 141, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.M.; Oliver, M.M.H.; Nahar, K.; Alam, M.Z.; Hirata, H.; Tsuyumu, S. CytR Homolog of Pectobacterium carotovorum subsp. carotovorum controls air-liquid biofilm formation by regulating multiple genes involved in cellulose production, c-di-GMP signaling, motility, and type III secretion system in response to nutritional and environmental signals. Front. Microbiol. 2017, 8, 972. [Google Scholar] [PubMed]
- Parker, W.L.; Rathnum, M.L.; Wells, J.J.S.; Trejo, W.H.; Principe, P.A.; Sykes, R.B. SQ 27, 860, a simple carbapenem produced by species of Serratia and Erwinia. J. Antibiot. 1982, 35, 653–660. [Google Scholar] [CrossRef] [PubMed]
- McGowan, S.J.; Sebaihia, M.; O’Leary, S.; Williams, P.; Salmond, G.P.C. Analysis of the carbapenem gene cluster of Erwinia carotovora: Definition of the antibiotic biosynthetic genes and evidence for a novel β-lactam resistance mechanism. Mol. Microbiol. 1997, 26. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Peever, T.L.; Mavrodi, O.V.; Parejko, J.A.; Raaijmakers, J.M.; Lemanceau, P.; Mazurier, S.; Heide, L.; Blankenfeldt, W.; Weller, D.M.; et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 2010, 76, 866–879. [Google Scholar] [CrossRef]
- Grinter, R.; Milner, J.; Walker, D. Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS ONE 2012, 7, e33033. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Tambong, J.T. Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS ONE 2017, 12, e0172295. [Google Scholar] [CrossRef]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; van der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef]
- Kozobay-Avraham, L. Involvement of DNA curvature in intergenic regions of prokaryotes. Nucleic Acids Res. 2006, 34, 2316–2327. [Google Scholar] [CrossRef][Green Version]
- Duan, C.; Huan, Q.; Chen, X.; Wu, S.; Carey, L.B.; He, X.; Qian, W. Reduced intrinsic DNA curvature leads to increased mutation rate. Genome Biol. 2018, 19, 132. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, E.A.; Brzostek, A.; Bacolla, A.; Mackiewicz, P.; Vasquez, K.M.; Korycka-Machala, M.; Jaworski, A.; Dziadek, J. Direct and inverted repeats elicit genetic instability by both exploiting and eluding DNA double-strand break repair systems in Mycobacteria. PLoS ONE 2012, 7, e51064. [Google Scholar] [CrossRef]
- Kim, H.-S.; Ma, B.; Perna, N.T.; Charkowski, A.O. Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains. Appl. Environ. Microbiol. 2009, 75, 4539–4549. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Thammarat, P.; Lommel, S.A.; Hogan, C.S.; Charkowski, A.O. Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant–microbe interactions. Mol. Plant Microbe Interact. 2011, 24, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Ham, J.H.; Deng, W.-L.; Doyle, J.J.; Collmer, A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 2002, 99, 13142–13147. [Google Scholar] [CrossRef] [PubMed]
- Poole, S.J.; Diner, E.J.; Aoki, S.K.; Braaten, B.A.; t’Kint de Roodenbeke, C.; Low, D.A.; Hayes, C.S. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef]
- Gallique, M.; Bouteiller, M.; Merieau, A. The type VI secretion system: A dynamic system for bacterial communication? Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Liu, H.; Coulthurst, S.J.; Pritchard, L.; Hedley, P.E.; Ravensdale, M.; Humphris, S.; Burr, T.; Takle, G.; Brurberg, M.-B.; Birch, P.R.J.; et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008, 4. [Google Scholar] [CrossRef]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 2012, 76, 740–772. [Google Scholar] [CrossRef]
- Carbonnelle, E.; Hélaine, S.; Prouvensier, L.; Nassif, X.; Pelicic, V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 2004, 55, 54–64. [Google Scholar] [CrossRef]
- Koo, J.; Tammam, S.; Ku, S.-Y.; Sampaleanu, L.M.; Burrows, L.L.; Howell, P.L. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 2008, 190, 6961–6969. [Google Scholar] [CrossRef] [PubMed]
- Sauvonnet, N.; Gounon, P.; Pugsley, A.P. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; van Schaik, E.J.; Audette, G.F.; Kao, D.; Hodges, R.S.; Hassett, D.J.; Irvin, R.T. The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces. Mol. Microbiol. 2006, 59, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Nykyri, J.; Mattinen, L.; Niemi, O.; Adhikari, S.; Kõiv, V.; Somervuo, P.; Fang, X.; Auvinen, P.; Mäe, A.; Palva, E.T.; et al. Role and regulation of the Flp/Tad pilus in the virulence of Pectobacterium atrosepticum SCRI1043 and Pectobacterium wasabiae SCC3193. PLoS ONE 2013, 8, e73718. [Google Scholar] [CrossRef]
- Panda, P.; Vanga, B.R.; Lu, A.; Fiers, M.; Fineran, P.C.; Butler, R.; Armstrong, K.; Ronson, C.W.; Pitman, A.R. Pectobacterium atrosepticum and Pectobacterium carotovorum harbor distinct, independently acquired integrative and conjugative elements encoding coronafacic acid that enhance virulence on potato stems. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Bender, C.L.; N-Chaidez, F.A.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar]
- Evans, T.J.; Ind, A.; Komitopoulou, E.; Salmond, G.P.C. Phage-selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J. Appl. Microbiol. 2010, 109, 505–514. [Google Scholar]
- Ragunath, C.; Shanmugam, M.; Bendaoud, M.; Kaplan, J.B.; Ramasubbu, N. Effect of a biofilm-degrading enzyme from an oral pathogen in transgenic tobacco on the pathogenicity of Pectobacterium carotovorum subsp. carotovorum. Plant Pathol. 2012, 61, 346–354. [Google Scholar] [CrossRef]
- Grinter, R.; Josts, I.; Mosbahi, K.; Roszak, A.W.; Cogdell, R.J.; Bonvin, A.M.J.J.; Milner, J.J.; Kelly, S.M.; Byron, O.; Smith, B.O.; et al. Structure of the bacterial plant-ferredoxin receptor FusA. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Grinter, R.; Hay, I.D.; Song, J.; Wang, J.; Teng, D.; Dhanesakaran, V.; Wilksch, J.J.; Davies, M.R.; Littler, D.; Beckham, S.A.; et al. FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants. PLoS Biol. 2018, 16. [Google Scholar] [CrossRef]
- Pirhonen, M.; Saarilahti, H.; Karlsson, M.-B.; Palva, E.T. Identification of pathogenicity determinants of Erwinia carotovora subsp. carotovora by transposon mutagenesis. Mol. Plant-Microbe Interact. 1991, 4, 276–283. [Google Scholar] [CrossRef]
- Pemberton, C.L.; Whitehead, N.A.; Sebaihia, M.; Bell, K.S.; Hyman, L.J.; Harris, S.J.; Matlin, A.J.; Robson, N.D.; Birch, P.R.J.; Carr, J.P.; et al. Novel quorum-sensing-controlled genes in Erwinia carotovora subsp. carotovora: Identification of a fungal elicitor homologue in a soft-rotting bacterium. Mol. Plant Microbe Interact. 2005, 18, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.A. Lines of evidence for horizontal gene transfer of a phenazine producing operon into multiple bacterial species. J. Mol. Evol. 2009, 68, 171–185. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhanel, G.G.; Wiebe, R.; Dilay, L.; Thomson, K.; Rubinstein, E.; Hoban, D.J.; Noreddin, A.M.; Karlowsky, J.A. Comparative review of the carbapenems. Drugs 2007, 67, 1027–1052. [Google Scholar] [CrossRef]
- Wu, M.-C.; Chen, Y.-C.; Lin, T.-L.; Hsieh, P.-F.; Wang, J.-T. Cellobiose-specific phosphotransferase system of Klebsiella pneumoniae and its importance in biofilm formation and virulence. Infect. Immun. 2012, 80, 2464–2472. [Google Scholar] [CrossRef]
- Liu, Y.; Yoo, B.B.; Hwang, C.-A.; Suo, Y.; Sheen, S.; Khosravi, P.; Huang, L. LMOf2365_0442 encoding for a fructose specific PTS permease IIA may be required for virulence in L. monocytogenes Strain F2365. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef]
- Godde, J.S.; Bickerton, A. The repetitive DNA elements called CRISPRs and their associated genes: Evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006, 62, 718–729. [Google Scholar] [CrossRef]
- Seed, K.D.; Lazinski, D.W.; Calderwood, S.B.; Camilli, A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 2013, 494, 489–491. [Google Scholar] [CrossRef]
- Watson, B.N.J.; Staals, R.H.J.; Fineran, P.C. CRISPR-Cas-Mediated phage resistance enhances horizontal gene transfer by transduction. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. CRISPR-Cas: Evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013, 10, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Doak, T.G.; Ye, Y. Expanding the catalog of cas genes with metagenomes. Nucleic Acids Res. 2014, 42, 2448–2459. [Google Scholar] [CrossRef] [PubMed]
- Labbate, M.; Orata, F.D.; Petty, N.K.; Jayatilleke, N.D.; King, W.L.; Kirchberger, P.C.; Allen, C.; Mann, G.; Mutreja, A.; Thomson, N.R.; et al. A genomic island in Vibrio cholerae with VPI-1 site-specific recombination characteristics contains CRISPR-Cas and type VI secretion modules. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comp. Biol. 2005, 1. [Google Scholar] [CrossRef]
- Zhang, Q.; Ye, Y. Not all predicted CRISPR–Cas systems are equal: Isolated cas genes and classes of CRISPR like elements. BMC Bioinform. 2017, 18. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).