Retrospective Study of Listeria monocytogenes Isolated in the Territory of Inner Eurasia from 1947 to 1999
Abstract
:1. Introduction
2. Results
2.1. Antibiotic Resistance
2.2. L. monocytogenes Virulence
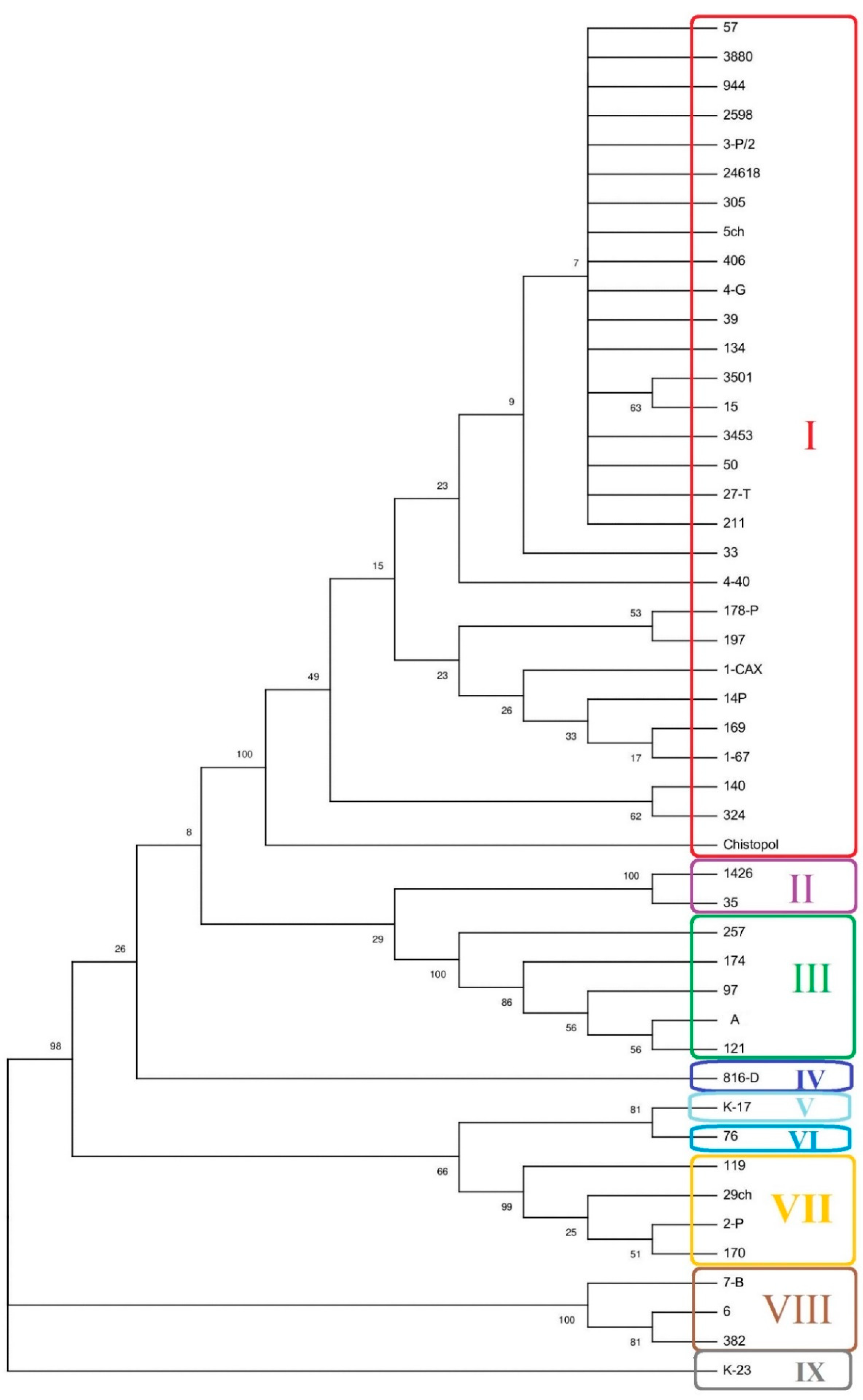
2.3. MLST Typing of L. monocytogenes Strains
2.4. Characterization of CC Distribution among Sources of Isolation
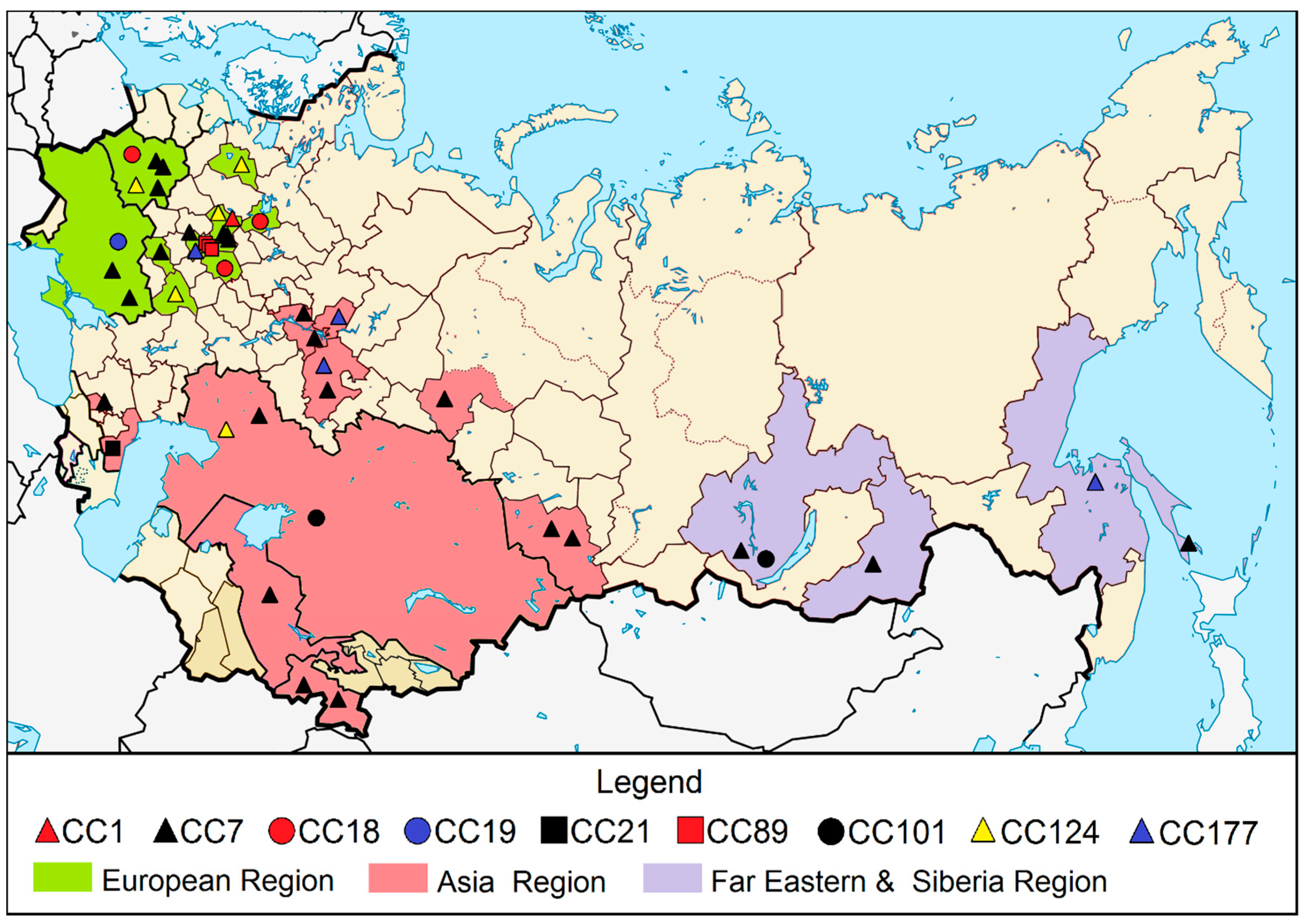
2.5. Distribution of the L. monocytogenes Clonal Complexes on the Territory of Inner Eurasia
2.6. Internalin Gene Diversity in L. monocytogenes Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culturing Conditions
4.2. Antibiotic Resistance Test
4.3. Anton’s Eye Test
4.4. L. monocytogenes Virulence in Mice
4.5. PCR
4.6. PCR Product Sequencing
4.7. Sequence Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walland, J.; Lauper, J.; Frey, J.; Imhof, R.; Stephan, R.; Seuberlich, T.; Oevermann, A. Listeria monocytogenes infection in ruminants: Is there a link to the environment, food and human health? A review. Schweiz. Arch. Tierheilkd. 2015, 157, 319–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Food Safety Authority (EFSA). EU summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, 4634. [Google Scholar]
- Allerberger, F.; Wagner, M. Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef]
- De Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Schukken, Y.H.; Nightingale, C.R.; Fortes, E.D.; Ho, A.J.; Her, Z.; Grohn, Y.T.; Mcdonough, P.L.; Wiedmann, M. Ecology and Transmission of Listeria monocytogenes Infecting Ruminants and in the Farm Environment. Appl. Environ. Microbiol. 2004, 70, 4458–4467. [Google Scholar] [CrossRef]
- Zaytseva, E.; Ermolaeva, S.; Somov, G.P. Low genetic diversity and epidemiological significance of Listeria monocytogenes isolated from wild animals in the far east of Russia. Infect. Genet. Evol. 2007, 7, 736–742. [Google Scholar] [CrossRef]
- Weis, J.; Seeliger, H.P. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 1975, 30, 29–32. [Google Scholar]
- Ahlstrom, C.A.; Manuel, C.S.; Den Bakker, H.C.; Wiedmann, M.; Nightingale, K.K. Molecular ecology of Listeria spp., Salmonella, Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in pristine natural environments in Northern Colorado. J. Appl. Microbiol. 2018, 124, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Pushkareva, V.I.; Ermolaeva, S.A. Listeria monocytogenes virulence factor Listeriolysin O favors bacterial growth in co-culture with the ciliate Tetrahymena pyriformis, causes protozoan encystment and promotes bacterial survival inside cysts. BMC Microbiol. 2010, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Wacheck, S.; Fredriksson-Ahomaa, M.; König, M.; Stolle, A.; Stephan, R. Wild boars as an important reservoir for foodborne pathogens. Foodborne Pathog. Dis. 2010, 7, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Goshima, T.; Mori, T.; Murakami, M.; Haruna, M.; Ito, K.; Yamada, Y. Prevalence and antimicrobial susceptibility of foodborne bacteria in wild boars (Sus scrofa) and wild deer (Cervus nippon) in Japan. Foodborne Pathog. Dis. 2013, 10, 985–991. [Google Scholar] [CrossRef]
- Castro, H.; Jaakkonen, A.; Hakkinen, M.; Korkeala, H.; Lindström, M. Occurrence, Persistence, and Contamination Routes of Listeria monocytogenes Genotypes on Three Finnish Dairy Cattle Farms: A Longitudinal Study. Appl. Environ. Microbiol. 2018, 84, e02000–e02017. [Google Scholar] [CrossRef]
- Voronina, O.L.; Ryzhova, N.N.; Kunda, M.S.; Kurnaeva, M.A.; Semenov, A.N.; Aksenova, E.I.; Egorova, I.Y.; Kolbasov, D.V.; Ermolaeva, S.A.; Gintsburg, A.L. Diversity and Pathogenic Potential of Listeria monocytogenes Isolated from Environmental Sources in the Russian Federation. IJMER 2015, 5, 5–15. [Google Scholar]
- Locatelli, A.; Spor, A.; Jolivet, C.; Piveteau, P.; Hartmann, A. Biotic and Abiotic Soil Properties Influence Survival of Listeria monocytogenes in Soil. PLoS ONE 2013, 8, e75969. [Google Scholar] [CrossRef]
- Dreyer, M.; Aguilar-Bultet, L.; Rupp, S.; Guldimann, C.; Stephan, R.; Schock, A.; Otter, A.; Schüpbach, G.; Brisse, S.; Lecuit, M.; et al. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 2016, 6, 36419. [Google Scholar] [CrossRef] [Green Version]
- Dell’Armelina Rocha, P.R.; Lomonaco, S.; Bottero, M.T.; Dalmasso, A.; Dondo, A.; Grattarola, C.; Zuccon, F.; Iulini, B.; Knabel, S.J.; Capucchio, M.T.; et al. Ruminant rhombencephalitis-associated Listeria monocytogenes strains constitute a genetically homogeneous group related to human outbreak strains. Appl. Environ. Microbiol. 2013, 79, 3059–3066. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef]
- Esteban, J.I.; Oporto, B.; Aduriz, G.; Juste, R.A.; Hurtado, A. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet. Res. 2009, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Schoder, D.; Melzner, D.; Schmalwieser, A.; Zangana, A.; Winter, P.; Wagner, M. Important Vectors for Listeria monocytogenes Transmission at Farm Dairies Manufacturing Fresh Sheep and Goat Cheese from Raw Milk. J. Food Prot. 2011, 74, 919–924. [Google Scholar] [CrossRef]
- Reu, K.; Grijspeerdt, K.; Herman, L. A Belgian Survey of hygiene indicator bacteria and pathogenic bacteria in raw milk and direct marketing of raw milk farm products. J. Food Saf. 2004, 24, 17–36. [Google Scholar] [CrossRef]
- Jayarao, B.M.; Donaldson, S.C.; Straley, B.A.; Sawant, A.A.; Hegde, N.V.; Brown, J.L. A Survey of Foodborne Pathogens in Bulk Tank Milk and Raw Milk Consumption among Farm Families in Pennsylvania. J. Dairy Sci. 2006, 89, 2451–2458. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, L.; Lan, R.; Salazar, J.K.; Liu, J.; Xu, J.; Ye, C. Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg. Microbes Infect. 2017, 6, e44. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, L.; Larsen, H.E.; Christiansen, T.; Jensen, M.M.; Eriksen, E. An outbreak of meningo-encephalitis in fallow deer caused by Listeria monocytogenes. Vet. Rec. 1988, 122, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Pewsner, M.; Origgi, F.C.; Frey, J.; Ryser-Degiorgis, M.P. Assessing fifty years of general health surveillance of roe deer in Switzerland: A retrospective analysis of necropsy reports. PLoS ONE 2017, 12, e0170338. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. World wide distribution of major clones of listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, K.K.; Windham, K.; Wiedmann, M. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 2005, 187, 5537–5551. [Google Scholar] [CrossRef] [PubMed]
- Cantinelli, T.; Chenal-Francisque, V.; Diancourt, L.; Frezal, L.; Leclercq, A.; Wirth, T.; Lecuit, M.; Brisse, S. “Epidemic clones” of listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 2013, 51, 3770–3779. [Google Scholar] [CrossRef] [PubMed]
- Piffaretti, J.C.; Kressebuch, H.; Aeschbacher, M.; Bille, J.; Bannerman, E.; Musser, J.M.; Selander, R.K.; Rocourt, J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 1989, 86, 3818–3822. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; van Cauteren, D.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef]
- Li, Z.; Pérez-Osorio, A.; Wang, Y.; Eckmann, K.; Glover, W.A.; Allard, M.W.; Brown, E.W.; Chen, Y. Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol. 2017, 17, 134. [Google Scholar] [CrossRef]
- Belén, A.; Pavón, I.; Maiden, M.C.J. Multilocus Sequence Typing. Mol. Epidemiol. Microorg. 2009, 551, 129–140. [Google Scholar] [Green Version]
- Aanensen, D.M.; Spratt, B.G. The multilocus sequence typing network: Mlst.net. Nucleic Acids Res. 2005, 33, 728–733. [Google Scholar] [CrossRef]
- Jeong, C.; Balanovsky, O.; Lukianova, E.; Kahbatkyzy, N.; Flegontov, P.; Zaporozhchenko, V.; Immel, A.; Wang, C.C.; Ixan, O.; Khussainova, E.; et al. The genetic history of admixture across inner Eurasia. Nat. Ecol. Evol. 2019, 3, 966–976. [Google Scholar] [CrossRef]
- Adgamov, R.; Zaytseva, E.; Thiberge, J.-M.; Brisse, S.; Ermolaev, S. Genetically Related Listeria Monocytogenes Strains Isolated from Lethal Human Cases and Wild Animals. Genet. Divers. Microorg. 2012. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, K.; Smith, M.A.; Doyle, M.P. Pathogenicity of Food and Clinical Listeria monocytogenes Isolates in a Mouse Bioassay. J. Food Prot. 2016, 66, 2362–2366. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.K.; Singh, D.V.; Dubey, S.K. Pregnancy—Associated human listeriosis: Virulence and genotypic analysis of Listeria monocytogenes from clinical samples. J. Microbiol. 2015, 53, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Jacquet, C.; Gerner-Smidt, P.; Graves, L.M.; Loncarevic, S.; Mathisen, T.; Morvan, A.; Salcedo, C.; Torpdahl, M.; Vazquez, J.A.; et al. Multicenter Validation of a Multiplex PCR Assay for Differentiating the Major Listeria monocytogenes Serovars 1/2a, 1/2b, 1/2c, and 4b: Toward an International Standard. J. Food Prot. 2016, 68, 2648–2650. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.; Singh, M.; Singh, D.; Dubey, S. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014, 14, 241. [Google Scholar] [CrossRef]
- Lyautey, E.; Topp, E.; Hartmann, A.; Piveteau, P.; Rieu, A.; Pagotto, F.; Tyler, K.; Lapen, D.R.; Wilkes, G.; Robertson, W.J.; et al. Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can. J. Microbiol. 2007, 53, 1158–1167. [Google Scholar] [CrossRef]
- Jamali, H.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015, 15, 144. [Google Scholar] [CrossRef]
- Tamburro, M.; Ripabelli, G.; Fanelli, I.; Grasso, G.M.; Sammarco, M.L. Typing of Listeria monocytogenes strains isolated in Italy by inlA gene characterization and evaluation of a new cost-effective approach to antisera selection for serotyping. J. Appl. Microbiol. 2010, 108, 1602–1611. [Google Scholar] [CrossRef]
- Weindl, L.; Frank, E.; Ullrich, U.; Heurich, M.; Kleta, S.; Ellerbroek, L.; Gareis, M. Listeria monocytogenes in Different Specimens from Healthy Red Deer and Wild Boars. Foodborne Pathog. Dis. 2016, 13, 391–397. [Google Scholar] [CrossRef]
- Parihar, V.S.; Lopez-Valladares, G.; Danielsson-Tham, M.-L.; Peiris, I.; Helmersson, S.; Unemo, M.; Andersson, B.; Arneborn, M.; Bannerman, E.; Barbuddhe, S.; et al. Characterization of Human Invasive Isolates of Listeria monocytogenes in Sweden 1986–2007. Foodborne Pathog. Dis. 2008, 5, 755–761. [Google Scholar] [CrossRef]
- Lopez-Valladares, G.; Danielsson-Tham, M.-L.; Goering, R.V.; Tham, W. Lineage II (Serovar 1/2a and 1/2c) Human Listeria monocytogenes Pulsed-Field Gel Electrophoresis Types Divided into PFGE Groups Using the Band Patterns Below 145.5 kb. Foodborne Pathog. Dis. 2016, 14, 8–16. [Google Scholar] [CrossRef]
- Yoshida, T.; Sugimoto, T.; Sato, M.; Hirai, K. Incidence of Listeria monocytogenes in Wild Animals in Japan. J. Vet. Med. Sci. 2000, 62, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Lomonaco, S.; Verghese, B.; Gerner-Smidt, P.; Tarr, C.; Gladney, L.; Joseph, L.; Katz, L.; Turnsek, M.; Frace, M.; Chen, Y.; et al. Novel Epidemic Clones of Listeria United States, 2011. Emerg. Infect. Dis. 2013, 19, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The Epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Xie, J.; Li, Y.; Wang, H. Listeriosis in mainland China: A systematic review. Int. J. Infect. Dis. 2019, 81, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maury, M.M.; Tsai, Y.; Charlier, C.; Touchon, M.; Europe PMC funders group. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, M.M.; Engelen-Lee, J.Y.; Brouwer, M.C.; Jaspers, V.; Man, W.K.; Vall Seron, M.; van de Beek, D. Characterization of a Listeria monocytogenes meningitis mouse model. J. Neuroinflamm. 2018, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory Activities and Experiences Associated with Whole-Genome Sequencing Analysis of Isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef]
- Sobyanin, K.A.; Sysolyatina, E.V.; Chalenko, Y.M.; Kalinin, E.V.; Ermolaeva, S.A. Route of Injection Affects the Impact of InlB Internalin Domain Variants on Severity of Listeria monocytogenes Infection in Mice. BioMed Res. Int. 2017, 2017, 2101575. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals, 4th ed.; CLSI supplement VET08; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Bakulov, I.A.; Vasylyev, D.A.; Kolbasov, D.B.; Kovaleva, E.N.; Egorova, I.Y.; Selyaniniov, Y.O. Listeria and Listeriosis. Monograph, 2nd ed.; Revised and Enlarged; RDICMB: Ulyanovsk, Russia, 2016; p. 334. [Google Scholar]
- Kumar, S.; Dudley, J.; Nei, M.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]

| Antibiotic | Number of Resistant Strains |
|---|---|
| Penicillin G | 14 |
| Enrofloxacin | 4 |
| Ampicillin | 7 |
| Tetracycline | 1 |
| Chloramphenicol | 1 |
| Kanamycin | 0 |
| Tylosin | 2 |
| Streptomycin | 0 |
| Polymyxin B | 44 |
| Neomycin | 0 |
| LD50 | Number of Strains in a Particular Source | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Humans | Cattle | Goat | Pig | Mouse | Rat | Sheep | Rabbit | Tick | |
| 104–107 | 1 | 3 | 1 | 2 | 2 | 1 | 2 | 2 | 0 |
| 107–109 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ≥109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Phylogenetic Lineage | Clonal Complex (CC) | Sequence Type (ST) | Number of Strains in a Particular Source | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Humans | Cattle | Small Ruminants | Pigs | Rodents | Horse | Arthropods | |||
| II | CC7 | ST7 | 1 | 0 | 3 | 3 | 5 | 0 | 0 |
| ST12 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ST23 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| ST85 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| ST98 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| ST106 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ||
| ST519 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| ST1534 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| ST1535 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| ST1536 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ST1537 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| ST1538 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ST1539 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| ST1540 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| ST1541 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| II | CC124 | ST124 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ST1550 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ||
| ST1551 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ST1552 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| II | CC177 | ST177 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ST1542 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ST1543 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| II | CC18 | ST481 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ST1544 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| ST1545 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| II | CC89 | ST1547 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| II | CC101 | ST101 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ST1548 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| II | CC19 | ST1546 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| II | CC21 | ST21 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| II | CC307 | ST1549 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| I | CC1 | ST252 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Clonal Complexes (Number of Strains) | Gene Allele | ||||
|---|---|---|---|---|---|
| inlA | inlB | inlC | inlE | ||
| I | CC7 (29) | 4 | 14 | 6 | 8 |
| II | CC101 (2) | 14 | 20 | 15 | 8 |
| III | CC124 (5) | 6 | 12 | 6 | 6 |
| IV | CC21 (1) | 12 | 14 | 6 | 8 |
| V | CC19 (1) | 9 | 14 | 6 | 6 |
| VI | CC89 (1) | 15 | 15 | 17 | 6 |
| VII | CC177 (4) | 8 | 14 | 6 | 8 |
| VIII | CC18 (3) | 12 | 13 | 6 | 6 |
| IX | CC1 (1) | 1 | 9 | 1 | 3 |
| Gene | Length | N Alleles | Mutations | π a | InDel b | Rm c | |
|---|---|---|---|---|---|---|---|
| Synon | Nonsynon | ||||||
| MLST | |||||||
| abcZ | 537 | 4 | 8 | 1 | 0.00838 | 0 | 0 |
| bglA | 399 | 7 | 11 | 0 | 0.00979 | 0 | 1 |
| cat | 486 | 8 | 8 | 3 | 0.00845 | 0 | 1 |
| dapE | 462 | 9 | 27 | 6 | 0.01852 | 0 | 3 |
| dat | 471 | 4 | 4 | 0 | 0.00425 | 0 | 0 |
| lhkA | 480 | 3 | 1 | 1 | 0.00278 | 0 | 0 |
| ldh | 459 | 18 | 5 | 6 | 0.00768 | 6 | 4 |
| IP d | |||||||
| inlA | 648 | 7 | 13 | 5 | 0.01190 | 0 | 3 |
| inlB | 618 | 5 | 5 | 6 | 0.00906 | 0 | 2 |
| inlC | 587 | 5 | 10 | 5 | 0.01126 | 0 | 0 |
| inlE | 558 | 2 | 1 | 1 | 0.00358 | 0 | 0 |
| No. | Strain | Year | Source | Location | Serovar | ST | CC |
|---|---|---|---|---|---|---|---|
| 1 | 4-40 | 1947 | horse | Moscow | 1/2a | ST106 | CC7 |
| 2 | 134 | 1952 | vole mouse | Moscow region | 1/2a | ST7 | CC7 |
| 3 | 15 | 1952 | pig | Belarus | 1/2a | ST519 | CC7 |
| 4 | 6 | 1952 | pig | Ryazan oblast | 1/2a | ST1544 | CC18 |
| 5 | A | 1952 | ticks | Kazakhstan | 1/2a | ST124 | CC124 |
| 6 | 382 | 1954 | cow | Yaroslav region | 1/2a | ST1545 | CC18 |
| 7 | 197 | 1955 | ticks | Ukraine | 1/2a | ST98 | CC7 |
| 8 | 39 | 1956 | guinea pig | Irkutsk region | 1/2a | ST7 | CC7 |
| 9 | К-17 | 1956 | rabbit | Ukraine | 1/2a | ST1546 | CC19 |
| 10 | 169 | 1957 | louse | Ukraine | 1/2a | ST1534 | CC7 |
| 11 | 944 | 1958 | house mouse | Moscow region | 1/2a | ST7 | CC7 |
| 12 | 2598 | 1960 | rabbit | North Caucasus region | 1/2a | ST7 | CC7 |
| 13 | 97 | 1960 | rabbit | Voronezh region | 1/2a | ST1550 | CC124 |
| 14 | 35 | 1962 | sheep | Kazakhstan | 1/2a | ST101 | CC101 |
| 15 | 406 | 1964 | pig | Kazan | 1/2a | ST7 | CC7 |
| 16 | Chistopol | 1964 | pig | Volga region | 1/2a | ST1541 | CC7 |
| 17 | 121 | 1964 | bovine | Moscow region | 1/2a | ST124 | CC124 |
| 18 | 3501 | 1965 | goat | Moscow | 1/2a | ST85 | CC7 |
| 19 | 3453 | 1965 | pig | Moscow | 1/2a | ST7 | CC7 |
| 20 | 324 | 1965 | pig | Moscow | 1/2a | ST23 | CC7 |
| 21 | 27-T | 1966 | rat | Tajikistan | 1/2a | ST7 | CC7 |
| 22 | 3-P/2 | 1966 | sheep | South Ural region | 1/2a | ST7 | CC7 |
| 23 | 2-P | 1967 | bovine | South Ural region | 1/2a | ST177 | CC177 |
| 24 | 119 | 1967 | cow | Ural region | 1/2a | ST1542 | CC177 |
| 25 | 178-P | 1967 | pig | Uzbekistan | 1/2a | ST1535 | CC7 |
| 26 | 14P | 1969 | sheep | Altai region | 1/2a | ST1534 | CC7 |
| 27 | 3880 | 1970 | pig | Ural region | 1/2a | ST7 | CC7 |
| 28 | 4-G | 1970 | sheep | Kazakhstan | 1/2a | ST7 | CC7 |
| 29 | 1426 | 1970 | cow | Irkutsk region | 1/2a | ST1548 | CC101 |
| 30 | 1-CAX | 1971 | cow | Sakhalin region | 1/2a | ST1536 | CC7 |
| 31 | 45 | 1971 | sheep | Kazakhstan | 1/2a | ST1459 | CC307 |
| 32 | 140 | 1971 | cow | Belarus | 1/2a | ST23 | CC7 |
| 33 | 257 | 1971 | cow | Novgorod region | 1/2a | ST1551 | CC124 |
| 34 | 24618 | 1971 | human | Moscow | 1/2a | ST7 | CC7 |
| 35 | 57 | 1971 | human | Moscow | 1/2a | ST1539 | CC7 |
| 36 | 174 | 1971 | sheep | Belarus | 1/2a | ST1552 | CC124 |
| 37 | 50 | 1971 | sheep | Belarus | 1/2a | ST7 | CC7 |
| 38 | 1-67 | 1972 | sheep | Altai region | 1/2a | ST1537 | CC7 |
| 39 | 7-B | 1972 | sheep | Belarus | 1/2a | ST481 | CC18 |
| 40 | 170 | 1974 | cow | Khabarovsk region | 1/2a | ST177 | CC177 |
| 41 | 33 | 1975 | sheep | Chita region | 1/2a | ST1540 | CC7 |
| 42 | 305 | 1975 | cow | Kazakhstan | 1/2a | ST12 | CC7 |
| 43 | 816-D | 1975 | goat | Dagestan region | 1/2a | ST21 | CC21 |
| 44 | 5 ch | 1975 | human | Tula region | 1/2a | ST12 | CC7 |
| 45 | К-23 | 1988 | human | Moscow | 4b | ST252 | CC1 |
| 46 | 211 | 1992 | cow | Kursk region | 1/2a | ST1538 | CC7 |
| 47 | 76 | 1997 | human | Tula | 1/2a | ST1547 | CC89 |
| 48 | 29 ch | 1999 | human | Tula region | 1/2a | ST1543 | CC177 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psareva, E.K.; Egorova, I.Y.; Liskova, E.A.; Razheva, I.V.; Gladkova, N.A.; Sokolova, E.V.; Potemkin, E.A.; Zhurilov, P.A.; Mikhaleva, T.V.; Blokhin, A.A.; et al. Retrospective Study of Listeria monocytogenes Isolated in the Territory of Inner Eurasia from 1947 to 1999. Pathogens 2019, 8, 184. https://doi.org/10.3390/pathogens8040184
Psareva EK, Egorova IY, Liskova EA, Razheva IV, Gladkova NA, Sokolova EV, Potemkin EA, Zhurilov PA, Mikhaleva TV, Blokhin AA, et al. Retrospective Study of Listeria monocytogenes Isolated in the Territory of Inner Eurasia from 1947 to 1999. Pathogens. 2019; 8(4):184. https://doi.org/10.3390/pathogens8040184
Chicago/Turabian StylePsareva, Ekaterina K., Irina Yu. Egorova, Elena A. Liskova, Irina V. Razheva, Nadezda A. Gladkova, Elena V. Sokolova, Eugene A. Potemkin, Pavel A. Zhurilov, Tatyana V. Mikhaleva, Andrei A. Blokhin, and et al. 2019. "Retrospective Study of Listeria monocytogenes Isolated in the Territory of Inner Eurasia from 1947 to 1999" Pathogens 8, no. 4: 184. https://doi.org/10.3390/pathogens8040184
APA StylePsareva, E. K., Egorova, I. Y., Liskova, E. A., Razheva, I. V., Gladkova, N. A., Sokolova, E. V., Potemkin, E. A., Zhurilov, P. A., Mikhaleva, T. V., Blokhin, A. A., Chalenko, Y. M., Kolbasov, D. V., & Ermolaeva, S. A. (2019). Retrospective Study of Listeria monocytogenes Isolated in the Territory of Inner Eurasia from 1947 to 1999. Pathogens, 8(4), 184. https://doi.org/10.3390/pathogens8040184






