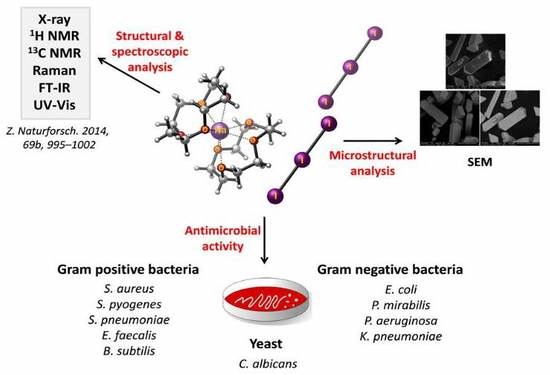
“Smart” Triiodide Compounds: Does Halogen Bonding Influence Antimicrobial Activities?
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Testing by Agar-Well and Disc-Diffusion Studies
2.2. Microstructural Analysis
2.3. Computational Analysis
3. Discussion
3.1. Agar well and Disc Diffusion Assays
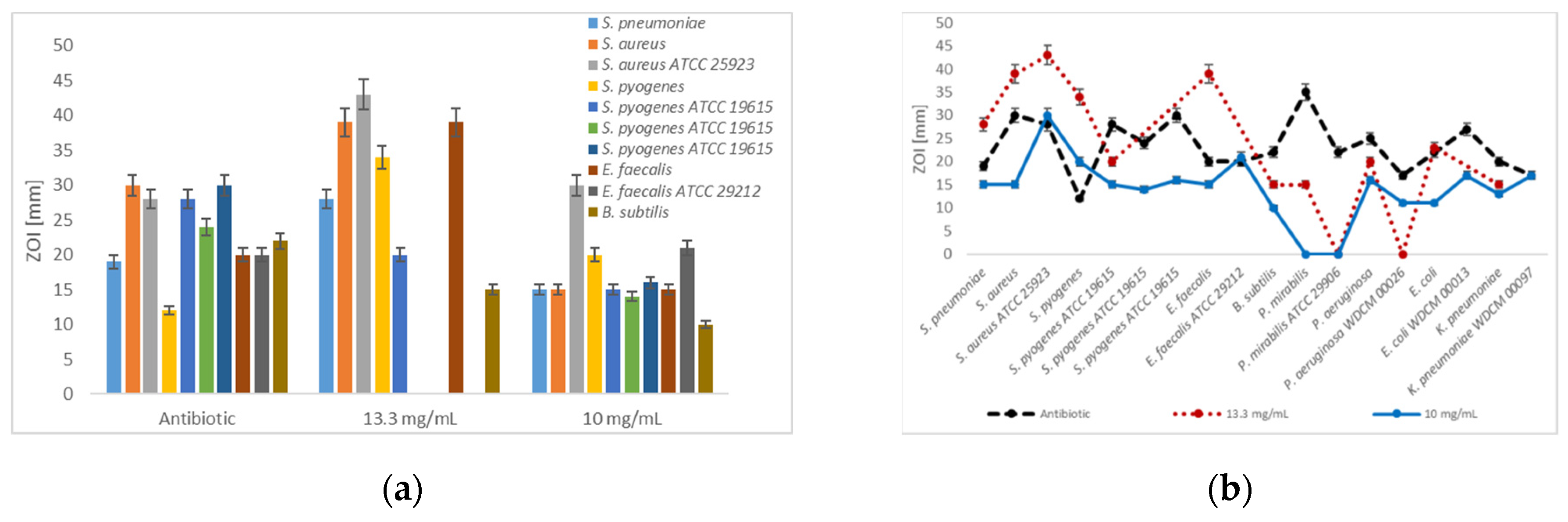
3.2. Antibiotic Screening Tests
4. Materials and Methods
4.1. Chemicals
4.2. Optimized Preparation of [Na(12-crown-4)2]I3
4.3. Bacterial Strains and Culturing
4.4. Investigation/Determination of Antibacterial and Antifungal Properties of [Na(12-crown-4)2]I3
4.4.1. Procedure for Zone of Inhibition Plate Studies
4.4.2. Agar Well Diffusion Method
4.4.3. Disc Diffusion Method
4.5. Scanning Electron Microscope (SEM) Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References and Note
- Vasudevan, P.; Tandon, M. Antimicrobial properties of iodine based products. J. Sci. Ind. Res. 2010, 69, 376–383. [Google Scholar]
- Svensson, P.H.; Kloo, L. Synthesis, structure, and bonding in polyiodide and metal iodide−iodine systems. Chem. Rev. 2003, 103, 1649–1684. [Google Scholar] [CrossRef]
- Blake, A.J.; Li, W.S.; Lippolis, V.; Schröder, M.; Devillanova, F.A.; Gould, R.O.; Parsons, S.; Radek, C. Template self-assembly of polyiodide networks. Chem. Soc. Rev. 1998, 27, 195–206. [Google Scholar] [CrossRef]
- Resnati, G.; Desiraju, G.R.; Ho, P.S.; Kloo, L.; Legon, A.C.; Marquardt, R.; Metrangolo, P.; Politzer, P.; Rissanen, K. Definition of the halogen bond. Pure Appl. Chem. 2013, 85, 1711–1713. [Google Scholar]
- Bartashevich, E.V.; Grigoreva, E.A.; Yushina, I.D.; Bulatova, L.M.; Tsirelson, V.G. Modern level for the prediction of properties of iodine-containing organic compounds: Iodine forming halogen bonds. Russ. Chem. Bull. Int. Ed. 2017, 66, 1–12. [Google Scholar] [CrossRef]
- Kaiho, T. Iodine Chemistry and Applications, 1st ed.; Kaiho, T., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 15–410. ISBN -13: 978-1-118-46629-2. [Google Scholar]
- Giese, M.; Albrecht, M.; Repenko, T.; Sackmann, J.; Valkonen, A.; Rissanen, K. Single-crystal X-ray diffraction and solution studies of anion–π interactions in N-(pentafluorobenzyl)pyridinium salts. Eur. J. Org. Chem. 2014, 12, 2435–2442. [Google Scholar] [CrossRef]
- Van Mengen, M.; Reiss, G.J. I62− Anion composed of two asymmetric triiodide moieties: A competition between halogen and hydrogen bond. Inorganics 2013, 1, 3–13. [Google Scholar] [CrossRef]
- Groenewald, F.; Esterhuysen, C.; Dillen, J. Extensive theoretical investigation: Influence of the electrostatic environment on the I3−···I3− anion–anion interaction. Theor. Chem. Acc. 2012, 131, 1281–1293. [Google Scholar] [CrossRef]
- Watkins, R.R.; Bonomo, R.A. Overview: Global and local impact of antibiotic resistance. Infect. Dis. Clin. N. Am. 2016, 30, 313–322. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2011, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Davydov, A.B.; Belyh, S.I.; Kravets, V.V. Iodine containing coating with prolonged antimicrobial activity based on water insoluble polymer matrix. Biomed. Eng. 2013, 46, 237–240. [Google Scholar] [CrossRef]
- Moulay, S. Molecular iodine/polymer complexes. J. Polym. Eng. 2013, 33, 389–443. [Google Scholar] [CrossRef]
- Zhao, G.; He, C.; Zhou, W.; Hooper, J.P.; Imler, G.H.; Parrish, D.A.; Shreeve, J.M. Control of Biohazards: A High Performance Energetic Polycyclized Iodine-Containing Biocide. Inorg. Chem. 2018, 57, 8673–8680. [Google Scholar] [CrossRef]
- Wang, S.; Schoenitz, M.; Grinshpun, S.A.; Yermakov, M.; Dreizin, E.L. Biocidal effectiveness of combustion products of iodine-bearing reactive materials against aerosolized bacterial spores. J. Aerosol Sci. 2018, 116, 106–115. [Google Scholar] [CrossRef]
- Singh, J.; Singh, P. Synthesis, spectroscopic characterization, and in vitro antimicrobial studies of pyridine-2-carboxylic acid N′-(4-chloro-benzoyl)-hydrazide and its Co(II), Ni(II), and Cu(II) complexes. Bioinorg. Chem. Appl. 2012, 2012, 104549. [Google Scholar] [CrossRef]
- Starosta, R.; Stokowa, K.; Florek, M.; Krol, J.; Chwilkowska, A.; Kulbacka, J.; Saczko, J.; Skala, J.; Jezowska-Bojczuk, M. Biological activity and structure dependent properties of cuprous iodide complexes with phenanthrolines and water soluble tris (aminomethyl) phosphanes. J. Inorg. Biochem. 2011, 105, 1102–1108. [Google Scholar] [CrossRef]
- Bor, T.; Gyawali, R.; Ibrahim, S.A. Evaluating the effectiveness of essential oils and combination of copper and lactic acid on the growth of E. coli O157:H7 in Laboratory Medium. Food 2016, 5, 14. [Google Scholar] [CrossRef]
- Bingjun, Q.; Jung, J.; Zhao, Y. Impact of acidity and metal ion on the antibacterial activity and mechanisms of β- and α-chitosan. Appl. Biochem. Biotechnol. 2015, 175, 2972–2985. [Google Scholar] [CrossRef]
- Zhitnitsky, D.; Rose, J.; Lewinson, O. The highly synergistic, broad spectrum, antibacterial activity of organic acids and transition metals. Sci. Rep. 2017, 7, 44554. [Google Scholar] [CrossRef] [PubMed]
- Zain, N.M.; Stapley, A.G.F.; Shama, G. Green synthesis of silver and copper nanoparticles using ascorbic acid and chitosan for antimicrobial applications. Carbohydr. Polym. 2014, 112, 195–202. [Google Scholar] [CrossRef]
- Arora, S.; Yadav, V.; Kumar, P.; Gupta, R.; Kumar, D. Polymer based antimicrobial coatings as potential biomaterial: A review. Int. J. Pharm. Sci. Rev. Res. 2013, 23, 279–290. [Google Scholar]
- Danilovas, P.P.; Rutkaite, R.; Zemaitaitis, A. Thermal degradation and stability of cationic starches and their complexes with iodine. Carbohydr. Polym. 2014, 112, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Klimaviciute, R.; Bendoraitiene, J.; Rutkaite, R.; Siugzdaite, J.; Zemaitaitis, A. Preparation, stability and antimicrobial activity of cationic cross-linked starch-iodine complexes. Int. J. Biol. Macromol. 2012, 51, 800–807. [Google Scholar] [CrossRef]
- Gao, T.; Fan, H.; Wang, X.; Gao, Y.; Liu, W.; Chen, W.; Dong, A.; Wang, Y.J. Povidone-Iodine-Based Polymeric Nanoparticles for Antibacterial applications. ACS Appl. Mater. Interfaces 2017, 9, 25738–25746. [Google Scholar] [CrossRef]
- He, C.; Parrish, D.A.; Shreeve, J.M. Alkyl ammonium cation stabilized biocidal polyiodides with adaptable high density and low pressure. Chem. Eur. J. 2014, 20, 6699–6706. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S.H. Preparation and structural and spectroscopic characterization of triiodides [M(12-crown-4)2]I3 with M = Na and Rb. Z. Nat. 2014, 69, 995–1002. [Google Scholar]
- Edis, Z.; Bloukh, S.H. Preparation and structural and spectroscopic characterization of a pentaiodide [Rb(12-crown-4)2]I5. Z. Nat. 2013, 68, 1340–1346. [Google Scholar]
- Wang, H.; Jian, G.; Zhou, W.; DeLisio, J.B.; Lee, V.T.; Zachariah, M.R. Metal iodate-based energetic composites and their combustion and biocidal performance. ACS Appl. Mater. Interfaces 2015, 7, 17363–17370. [Google Scholar] [CrossRef]
- Manchanda, G.; Sodhi, R.K.; Jain, U.K.; Chandra, R.; Madan, J. Iodinated curcumin bearing dermal cream augmented drug delivery, antimicrobial and antioxidant activities. J. Microencapsul. 2018, 35, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. Available online: http://pubchem.ncbi.nlm.nih.gov (accessed on 2 December 2018). [CrossRef] [PubMed]
- Hansch, C.; Leo, A.; Hoekman, D. Exploring QSAR—Hydrophobic, Electronic, and Steric Constants; American Chemical Society: Washington, DC, USA, 1995; p. 3. [Google Scholar]
- Tetko, I.V.; Tanchuk, V.Y. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J. Chem. Inf. Comput. Sci. 2002, 42, 1136–1145. Available online: http://www.vcclab.org/web/alogps/ (accessed on 2 December 2018). [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Hehre, W.J.; Radom, L.; Schleyer, P.V.R.; Pople, J. Ab Initio Molecular Orbital Theory; Wiley: New York, NY, USA, 1986. [Google Scholar]
- Adamo, C.; Barone, V.J. Toward reliable density functional methods without adjustable parameters: The PBE0 model. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Pamidighantam, S.; Nakandala, S.; Abeysinghe, E.; Wimalasena, C.; Rathnayakae, S.; Marru, S.; Pierce, M. Community science exemplars in SEAGrid Science Gateway: Apache Airavata based implementation of advanced infrastructure. Procedia Comput. Sci. 2016, 80, 1927–1939. [Google Scholar] [CrossRef]
- Shen, N.; Fan, Y.; Pamidighantam, S. E-Science infrastructures for molecular modeling and parametrization. J. Comput. Sci. 2014, 5, 576–589. [Google Scholar] [CrossRef]
- Dooley, R.; Milfeld, K.; Guiang, C.; Pamidighantam, S.; Allen, G. From proposal to production: Lessons learned developing the computational chemistry grid cyberinfrastructure. J. Grid Comput. 2006, 4, 195–208. [Google Scholar] [CrossRef]
- Milfeld, K.; Guiang, C.; Pamidighantam, S.; Giuliani, J. Cluster computing through an application-oriented computational chemistry grid. In Proceedings of the 2005 Linux Clusters: The HPC Revolution, April 2005; Available online: http://www.linuxclustersinstitute.org/conferences/archive/2005/PDF/presentations/Milfeld_K.pdf (accessed on 15 December 2018).
- This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number OCI-1053575
- Punyani, S.; Narayana, P.; Singh, H.; Vasudevan, P. Iodine based water disinfection: A review. J. Sci. Ind. Res. 2006, 65, 116–120. [Google Scholar]
- Lachapelle, J.M.; Castel, O.; Casado, A.F.; Leroy, B.; Micali, G.; Tennstedt, D.; Lambert, J. Antiseptics in the era of bacterial resistance: A focus on povidone iodine. Clin. Pract. 2013, 10, 579–592. [Google Scholar] [CrossRef]
- Aoki, S.; Yamakawa, K.; Kubo, K.; Takeshita, J.; Takeuchi, M.; Nobuoka, Y.; Wada, R.; Kikuchi, M.; Sawai, J. Antibacterial properties of silicone membranes after a simple two-step immersion process in iodine and silver nitrate solutions. Biocontrol Sci. 2018, 23, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.C.; Morse, A.M.; Mietzner, T.; Miller, S. Jawetz, Melnick and Adelberg’s Medical Microbiology, 27th ed.; Mc Graw-Hill Education: New York, NY, USA, 2016; ISBN 978-1-25-925534-2. [Google Scholar]
- Lamberts, K.; Handels, P.; Englert, U.; Aubert, E.; Espinos, E. Stabilization of polyiodide chains via anion⋯anion interactions: Experiment and theory. CrystEngComm 2016, 18, 3832–3841. [Google Scholar] [CrossRef]
- Wiley, J.M.; Sherwood, L.M.; Woolverton, C.J. Prescott’s Microbiology, 10th ed.; Mc Graw-Hill Education: New York, NY, USA, 2016; ISBN 978-1259281594. [Google Scholar]
- Pianalto, K.M.; Alspaugh, J.A. New horizons in antifungal therapy. J. Fungi 2016, 2, 26. [Google Scholar] [CrossRef]
- Spitzer, M.; Robbins, N.; Wright, G.D. Combinatorial strategies for combating invasive fungal infections. Virulence 2017, 8, 169–185. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef]
- Kong, M.; Chen, X.G.; Xing, K.; Park, H.J. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int. J. Food Microbiol. 2010, 144, 51–63. [Google Scholar] [CrossRef]
- Park, S.C.; Nah, J.W.; Park, Y. pH-dependent mode of antibacterial actions of low molecular weight water-soluble chitosan (LMWSC) against various pathogens. Macromol. Res. 2011, 19, 853–860. [Google Scholar] [CrossRef]
- Park, S.C.; Nam, J.P.; Kim, J.H.; Kim, Y.M.; Nah, J.W.; Jang, M.K. Antimicrobial action of water-soluble β-chitosan against clinical multi-drug resistant bacteria. Int. J. Mol. Sci. 2015, 16, 7995–8007. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, B.; Parandhaman, T.; Das, S.K. Antibacterial effects of biosynthesized silver nanoparticles on surface ultrastructure and nanomechanical properties of Gram-negative bacteria viz. Escherichia coli and Pseudomonas aeruginosa. ACS Appl. Mater. Interfaces 2016, 8, 4963–4976. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.S. The contribution of capsule polysaccharide genes to virulence of Klebsiella Pneumoniae. Virulence 2017, 8, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Amikacin: Uses, resistance, and prospects for inhibition. Molecules 2017, 22, 2267. [Google Scholar] [CrossRef]
- Pan, X.; Wang, Y.; Chen, Z.; Pan, D.; Cheng, Y.; Liu, Z. Investigation of antibacterial activity and related mechanism on a series of nano-Mg(OH)2. ACS Appl. Mater. Interfaces 2013, 5, 1137–1142. [Google Scholar] [CrossRef]
- Bauer, A.W.; Perry, D.M.; Kirby, W.M.M. Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results. AMA Arch. Intern. Med. 1959, 104, 208–216. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk Susceptibility Testing, 28th ed.; M100S; CLSI: Wayne, PA, USA, 2018; Volume 38. [Google Scholar]
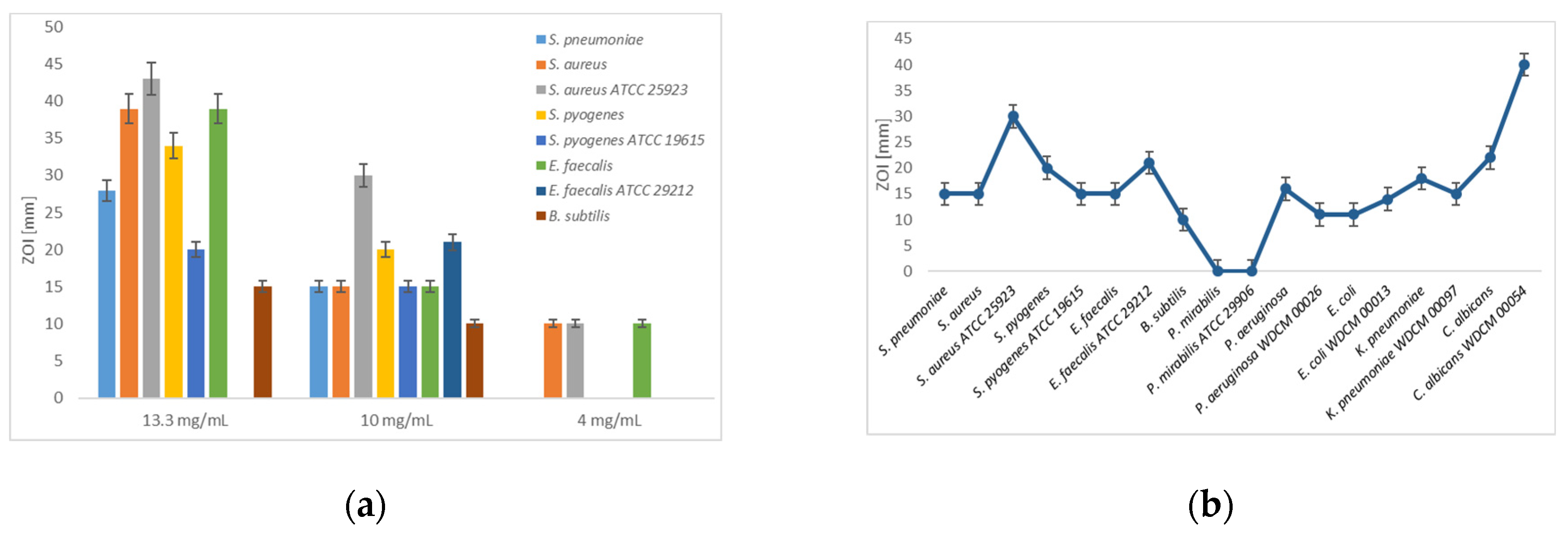
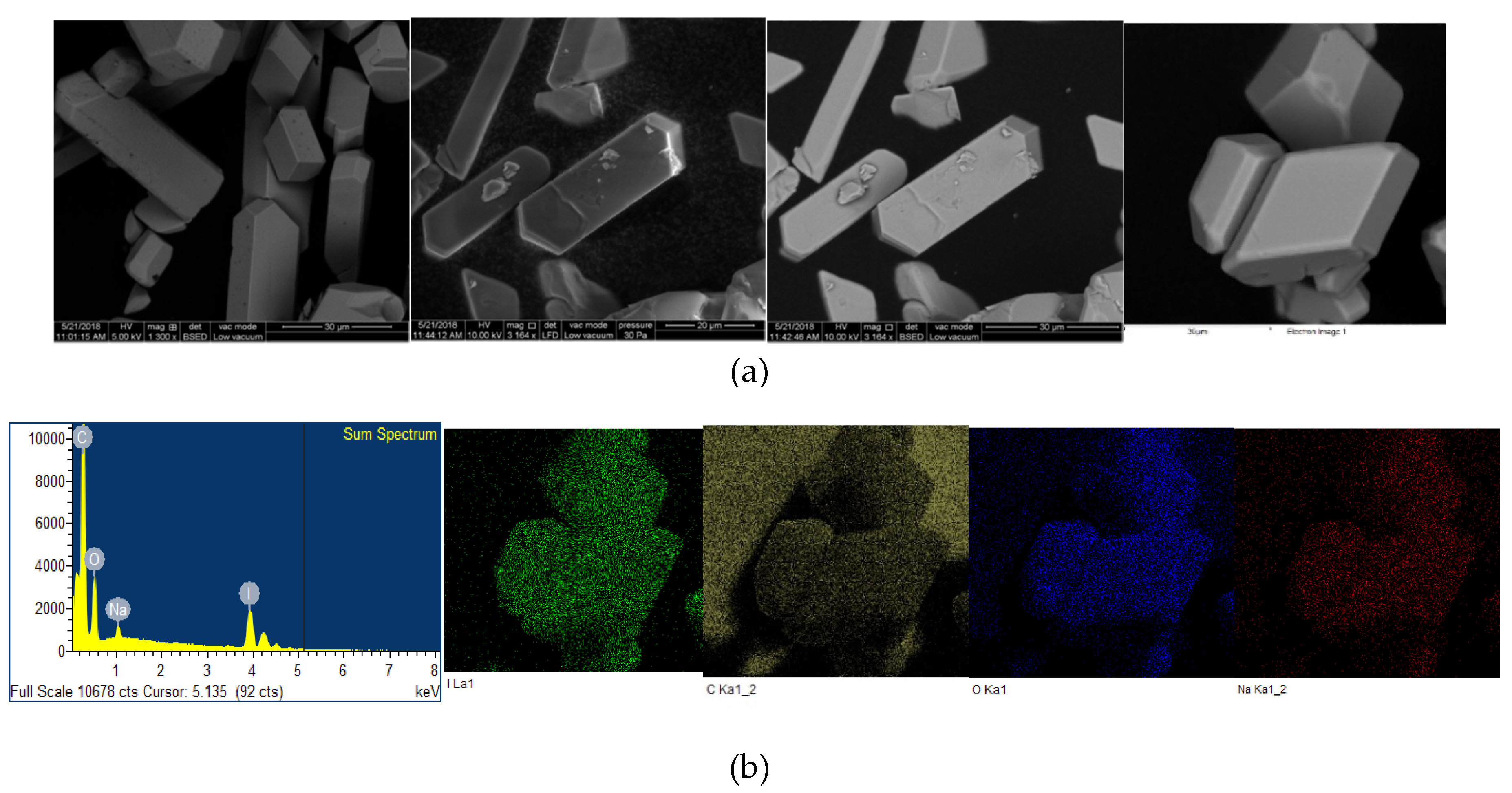
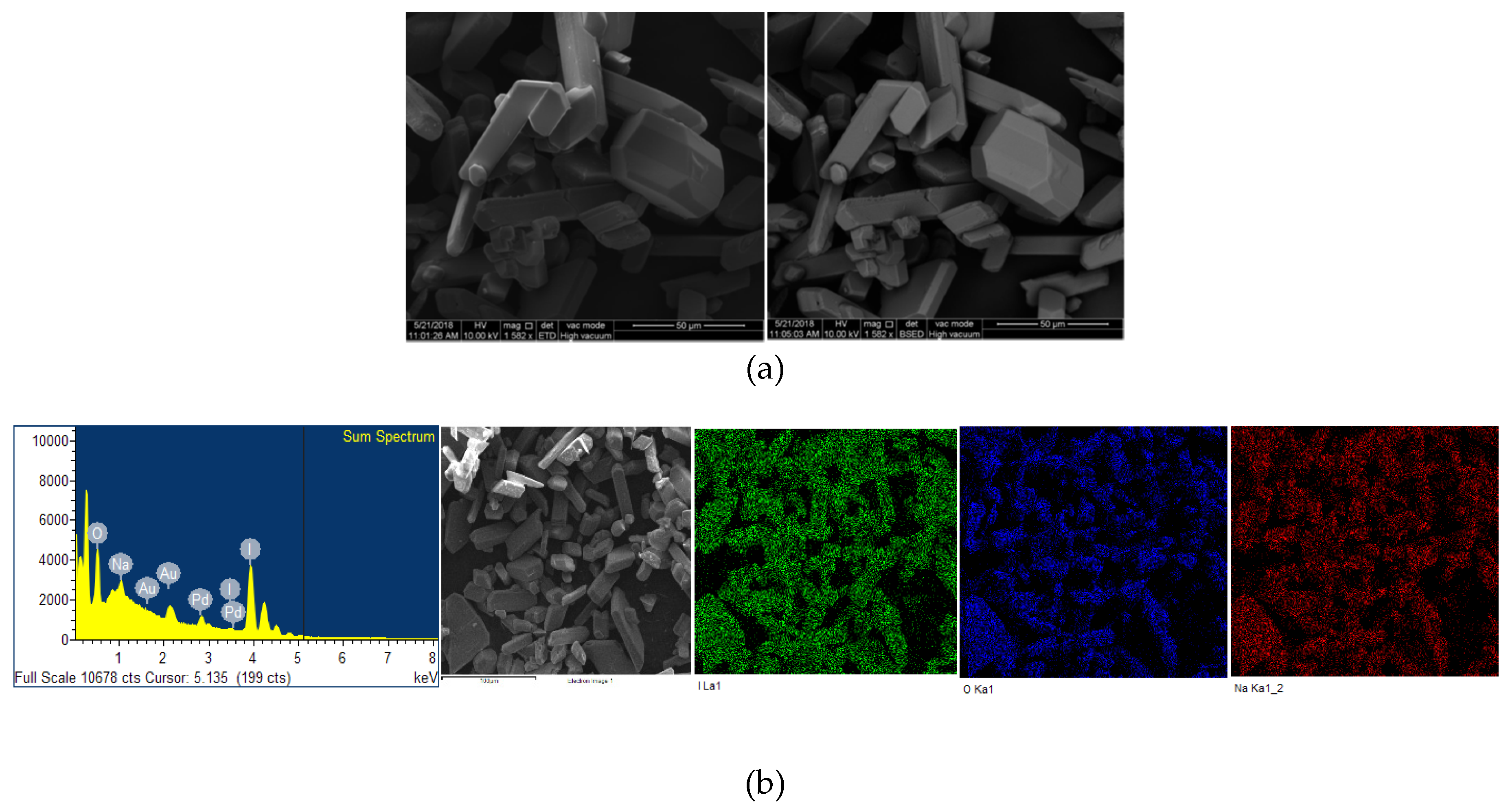
| Strain | I2[a] | NaI[a] | 12C4 [b] | A[a] | A[b] * | A[b] ** | A[b] *** |
|---|---|---|---|---|---|---|---|
| S. pneumonia | 70 | 0 | 0 | 11 | 28 | 15 | 0 |
| S. aureus | 70 | 0 | 10 | 22 | 39 | 15 | 10 |
| S. aureus ATCC 25923 | 70 | 0 | 0 | - | 43 | 30 | 10 |
| S. pyogenes | 60 | 0 | 9 | 15 | 34 | 20 | 0 |
| S. pyogenes ATCC 19615 | 70 | 0 | 0 | - | 20 | 15 | 0 |
| E. faecalis | 80 | 0 | 0 | 16 | 39 | 15 | 10 |
| E. faecalis ATCC 29212 | 52 | 25 | 0 | - | - | 21 | - |
| B. subtilis | 80 | 16 | 10 | 14 | 15 | 10 | 0 |
| P. mirabilis | 64 | 22 | 0 | 0 | 15 | 0 | 0 |
| P. mirabilis ATCC 29906 | 47 | 20 | 0 | - | - | 0 | - |
| P. aeruginosa | 58 | 30 | 15 | 14 | 20 | 16 | 0 |
| P. aeruginosa WDCM 00026 | 63 | 0 | 0 | - | - | 11 | - |
| E. coli | 58 | 27 | 12 | 16 | 23 | 11 | 0 |
| E. coli WDCM 00013 | 60 | 19 | 14 | - | - | 14 | - |
| K. pneumoniae | 65 | 30 | 18 | 9 | 15 | 18 | 0 |
| K. pneumoniae WDCM 00097 | 70 | 35 | 15 | - | - | 15 | - |
| C. albicans | 30 | 0 | 0 | 39 | 50 | 22 | 10 |
| C. albicans WDCM 00054 | 80 | 0 | 0 | - | - | 40 | - |
| Strain | Antibiotic | [mcg/disc] | B | A[b] * | A[b] ** |
|---|---|---|---|---|---|
| S. pneumoniae | E | 15 | 19 | 28 | 15 |
| S. aureus | G | 30 | 30 | 39 | 15 |
| S. aureus ATCC 25923 | G | 30 | 28 | 43 | 30 |
| S. pyogenes | E | 15 | 12 | 34 | 20 |
| S. pyogenes ATCC 19615 | E | 15 15 | 28 29+ | 20 | 15 14+ |
| G | 30 30 | 24 27+ | - | 14 | |
| C | 10 10 | 30 27 | - | 16 14 | |
| E. faecalis | E | 15 | 20 | 39 | 15 |
| E. faecalis ATCC 29212 | E | 15 15 | 20 18+ | - | 21 |
| B. subtilis | S | 10 | 22 | 15 | 10 |
| P. mirabilis | G | 30 | 35 | 15 | 0 |
| P. mirabilis ATCC 29906 | G | 30 | 22 | - | 0 |
| P. aeruginosa | E | 15 | 25 | 20 | 16 |
| P. aeruginosa WDCM 00026 | E | 15 | 17 | - | 11 |
| E. coli | A | 30 | 22 | 23 | 11 |
| E. coli WDCM 00013 | A | 30 | 27 | - | 17 |
| K. pneumoniae | E | 15 | 20 | 15 | 13 |
| K. pneumoniae WDCM 00097 | E | 15 | 17 | - | 17 |
| Element | C (K) | O (K) | Na (K) | I (L) | Total |
|---|---|---|---|---|---|
| Weight% | 50.61 | 20.51 | 1.97 | 26.91 | 100 |
| Atomic% | 72.77 | 22.14 | 1.44 | 3.66 |
| Element | C (K) | O (K) | Na (K) | I (L) | Au (M) | Pd (L) | Total |
|---|---|---|---|---|---|---|---|
| Weight % | 50.61 | 20.51 | 1.97 | 26.91 | 10.13 | 6.41 | 100 |
| Atomic% | 72.77 | 22.14 | 1.44 | 3.66 | 3.10 | 3.63 |
| Species | Consensus log Po/w [a] | log Po/w (XLOGP3) | log Po/w | log Po/w (ab initio) |
|---|---|---|---|---|
| I2 | − | 1.7 [b] | 2.49 [c] | 0.14 [e] |
| I− | − | 0.9 [b] | − | 4.04 [e] |
| I3− | − | 2.6 [b] | − | 2.69 [e] |
| 12-crown-4 | 0.59 | −0.04 [a] | −0.34 [d] | 1.66 [e] |
| [Na(12-crown-4)2]+ | −2.80 | −1.55 [a] | − | − |
| [Na(12-crown-4)2]I3 | −1.87 | 1.03 [a] | − | 3.13 [f], 3.60 [g] |
| [Na(12-crown-4)2][(I3)2]− | −1.30 | 3.61 [a] | − | 5.32 [e], 6.74 [f] |
| Chitosan | −15.80 | −21.40 [a], [b] | −2.62 [d] | − |
| Process | Bond Dissociation Energy (kJ/mol) [a] | |
|---|---|---|
| [I–I–I]− → [I–I] + [I]− | (1) | 135.0 |
| [I–I•••I]− → [I–I] + [I]− | (2) | 124.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edis, Z.; Haj Bloukh, S.; Abu Sara, H.; Bhakhoa, H.; Rhyman, L.; Ramasami, P. “Smart” Triiodide Compounds: Does Halogen Bonding Influence Antimicrobial Activities? Pathogens 2019, 8, 182. https://doi.org/10.3390/pathogens8040182
Edis Z, Haj Bloukh S, Abu Sara H, Bhakhoa H, Rhyman L, Ramasami P. “Smart” Triiodide Compounds: Does Halogen Bonding Influence Antimicrobial Activities? Pathogens. 2019; 8(4):182. https://doi.org/10.3390/pathogens8040182
Chicago/Turabian StyleEdis, Zehra, Samir Haj Bloukh, Hamed Abu Sara, Hanusha Bhakhoa, Lydia Rhyman, and Ponnadurai Ramasami. 2019. "“Smart” Triiodide Compounds: Does Halogen Bonding Influence Antimicrobial Activities?" Pathogens 8, no. 4: 182. https://doi.org/10.3390/pathogens8040182
APA StyleEdis, Z., Haj Bloukh, S., Abu Sara, H., Bhakhoa, H., Rhyman, L., & Ramasami, P. (2019). “Smart” Triiodide Compounds: Does Halogen Bonding Influence Antimicrobial Activities? Pathogens, 8(4), 182. https://doi.org/10.3390/pathogens8040182