Abstract
The bacterium Aggregatibacter actinomycetemcomitans is associated with aggressive forms of periodontitis and with systemic diseases, such as endocarditis. By assessing a Ghanaian longitudinal adolescent cohort, we earlier recognized the cagE gene as a possible diagnostic marker for a subgroup of JP2 and non-JP2 genotype serotype b A. actinomycetemcomitans strains, associated with high leukotoxicity as determined in a semi-quantitative cell assay. This group of A. actinomycetemcomitans is associated with the progression of attachment loss. In the present work, we used conventional polymerase chain reaction (PCR) and quantitative PCR to perform the cagE genotyping of our collection of 116 selected serotype b A. actinomycetemcomitans strains, collected over a period of 15 years from periodontitis patients living in Sweden. The A. actinomycetemcomitans strains carrying cagE (referred to as cagE+; n = 49) were compared to the cagE-negative strains (n = 67), present at larger proportions in the subgingival plaque samples, and were also much more prevalent in the young (≤35 years) compared to in the old (>35 years) group of patients. Our present results underline the potential use of cagE genotyping in the risk assessment of the development of periodontal attachment loss in Swedish adolescents.
1. Introduction
Aggregatibacter actinomycetemcomitans is a Gram-negative opportunistic pathogen associated with rapidly progressing periodontitis and with extra-oral diseases, such as endocarditis [1,2,3]. Several longitudinal studies have demonstrated that adolescents colonized with A. actinomycetemcomitans, as compared to those that are not, have a significantly increased risk of the development of periodontal attachment loss (AL) [4,5,6,7]. A. actinomycetemcomitans produces an array of virulence factors that allow this bacterium to evade and suppress the host immune response, including two exotoxins, i.e., leukotoxin and cytolethal distending toxin (CDT) [8,9,10]. A large genetic diversity within the A. actinomycetemcomitans species has been found, and seven different serotypes (a–g) exist, representing genetically divergent lineages [11,12,13]. A. actinomycetemcomitans genotypes can have extensively different pathogenic potentials [5,14,15]. For example, carriers of the JP2 serotype b-specific genotype of A. actinomycetemcomitans are at higher risk of development of AL compared to carriers of a non-JP2 genotype of A. actinomycetemcomitans. Typical for JP2 genotype strains is the deletion of 530 base pairs (bp) in the promoter region of the ltxCABD gene operon, which encodes leukotoxin (LtxA), and an enhanced leukotoxicity [16,17]. LtxA is a virulence factor of A. actinomycetemcomitans with the capacity to cause imbalance in the host inflammatory response [9]. The ltx promoter deletion has been frequently used as genetic marker to identify A. actinomycetemcomitans carriers with an increased risk for periodontal disease onset and progression [17], and this genotype is easily detected using a DNA-based assay (PCR) [18]. In addition to the JP2 genotype, a subgroup of non-JP2 genotype serotype b strains exhibits a similar disease association and high leukotoxicity, as has been shown in a semi-quantitative cell assay [14]. Genetic characterization has revealed that this particular subgroup of non-JP2 genotype of A. actinomycetemcomitans strains of serotype b are genetically closely related to the JP2 genotype by sharing the same arbitrarily-primed (AP) PCR gel electrophoresis banding pattern, referred to as AP-PCR genotype 1 in the present work [14]. Another property shared between the JP2 genotype and highly leukotoxic non-JP2 genotype serotype b strains was recently recognized, i.e., the carriage of the cagE gene sequence [19]. The cagE gene in A. actinomycetemcomitans was initially characterized by Teng and Hu [20], presenting evidence that the encoded CagE protein could induce apoptosis on primary human epithelial cells. However, consistent with leukotoxicity being a major virulence property of cagE-positive A. actinomycetemcomitans serotype b strains, one JP2 genotype bacterial cell was enough to lyse the majority of macrophage cells in vitro [20], whereas, as in comparison, a ratio of 50,000 JP2 genotype bacterial cells per epithelial cell was used to detect the CagE-induced apoptotic effects in vitro [21]. This suggests that CagE may have limited overall contribution to the virulence at biologically-relevant bacterial levels. In the present study, we utilized the cagE gene sequence as a diagnostic risk marker for the PCR detection of highly leukotoxic JP2 and non-JP2 genotypes of A. actinomycetemcomitans serotype b [19].
Furthermore, a type IV secretion system (T4SS) is a large macromolecular complex in Gram-negative bacteria which mediates conjugation, DNA transport and the secretion of virulence factors. Experimental work on model organisms, such as Agrobacterium tumefaciens and Helicobacter pylori, has revealed an archetypal T4SS system composed of 12 proteins, referred to as VirB1–VirB11, and VirD4 [22,23]. A. actinomycetemcomitans T4SS gene clusters are found in approximately 50% of strains and can be encoded both on the chromosome and on plasmids [24,25,26]. Interestingly, CagE exhibits homology to two T4SS proteins. The CagE N-terminus is homologous to VirB1 (lytic transglycosylase; also known as MagB01), and the CagE C-terminus is homologous to VirB4 (ATP:ase; MagB03) [19]. As judged by in silico analysis of the serotype b genomes available in the National Center for Biotechnology (NCBI) database, the cagE gene locus is not present in any of the strains encoding VirB1 and VirB4 on the chromosome. Whether the cagE and virB1/virB4 genes are consistently inversely carried in serotype b strains has not earlier been thoroughly assessed but would support the notion that CagE may represent the result of a recombination event in which parts of the virB1 and virB4 genes were fused together to encode a chimeric VirB1–VirB4 protein in A. actinomycemcomitans [19].
In our previous work, delineating the role of cagE as a potential diagnostic marker, we studied a collection of A. actinomycetemcomitans strains collected from a prospective cohort of Ghanaian adolescents [19]. To further evaluate the role of cagE and virB1/virB4 as diagnostic tools, we assessed our collection of A. actinomycetemcomitans strains that were collected during 15 years from periodontitis patients living in Sweden [27]. Data from microbiological analyses of this collection revealed that the young individuals (≤35 years) had a higher prevalence of A. actinomycetemcomitans and larger proportions of it in the samples compared to the older patients (>35 years). Moreover, serotype b was highly prevalent in the samples collected from young patients [27]. The aim of the present work was to determine the prevalence of the cagE genotype among the serotype b strains from this aforementioned collection (n = 116) and also to evaluate the potential use of cagE as a diagnostic marker for the carriage of highly leukotoxic serotype b strains among periodontitis patients living in Sweden.
2. Results
2.1. Validation of PCR Assays to Detect VirB1 and VirB4 Sequences in A. actinomycetemcomitans Serotype B Reference Strains
All A. actinomycetemcomitans serotype b strains assessed in the present study were grouped according to their AP-PCR genotype—1, 2 or “other” (i.e., AP-PCR types 3–11 as defined earlier [27]) (Figure 1A).
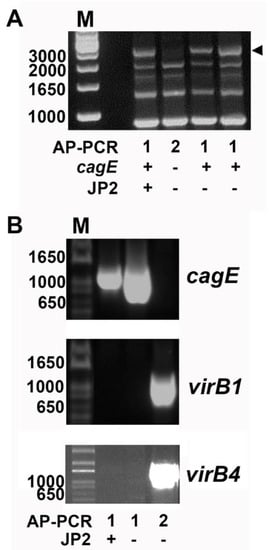
Figure 1.
PCR genotyping of Aggregatibacter actinomycetemcomitans serotype b strains. (A) Distinct arbitrarily-primed (AP)-PCR banding patterns distinguish cagE-positive and cagE-negative serotype b strains of A. actinomycetemcomitans. The approximately 3000-bp DNA-band (arrowed) detected in AP-PCR type 1 is unique for this genotype and was earlier demonstrated to contain the cagE gene sequence [19]. Typically, this DNA band reflects the difference between AP-PCR types 1 and 2. The presence/absence of the cagE gene and ltxA JP2 promoter type in AP-PCR types 1 and 2 is indicated. (B) PCR detection of cagE, virB1, and virB4, respectively. An amplicon specific for cagE was revealed in both JP2 and non-JP2 AP-PCR genotype 1 strains. In AP-PCR genotype 2 strains, amplicons specific for virB1 and virB4 were detected, whereas cagE was not. Sizes (bp) of selected bands in the DNA molecular weight marker (M) are indicated. Figures illustrate representative experiments.
To test the hypothesis that the presence of chromosomal virB1 and virB4 genes can serve as genetic markers that are suitable for the detection of cagE-negative serotype b strains, PCR was employed as described in the Materials and Methods section. To evaluate the PCR approach, we initially assessed 25 A. actinomycetemcomitans strains of serotype b which have previously been subject to whole genome sequencing (Table 1) (Figure 1B). As expected, this revealed presence of both virB1 and virB4 in the cagE-negative strains only (n = 7; 4 type 2 AP-PCR and 3 “other” AP-PCR type), whereas neither virB1 nor virB4 were detected by PCR in the cagE-positive strains (n = 18; all AP-PCR type 1). This finding prompted us to further investigate this apparent inverse relationship between the carriage of cagE and virB1/virB4 in the assessment of our local collection of serotype b A. actinomycetemcomitans strains. As virB1 and virB4 were carried simultaneously in the strains studied, we continued our analyses, mainly screening for the presence of virB4.

Table 1.
Genotyping of A. actinomycetemcomitans serotype b strains (n = 25) that were earlier subjected to whole genome sequencing.
2.2. Screening of CagE and VirB4 in Serotype B A. actinomycetemcomitans Strains Collected from Patients with Periodontitis Living in Sweden
We screened the 116 serotype b A. actinomycetemcomitans strains, collected from periodontitis patients living in Sweden, using qPCR to determine the prevalence of the cagE and virB4 genes (Table 2) (Table S1) (Figure 2).

Table 2.
Inverse relationship in the carriage of cagE and virB4. Presence of chromosomal cagE and virB4 genes in serotype b strains of A. actinomycetemcomitans (n = 116) in different AP-PCR genotypes. The number of strains and percent (%) of all strains are indicated. The cagE-positive strains all (100%) belong to AP-PCR type 1 and lack the virB4 gene.
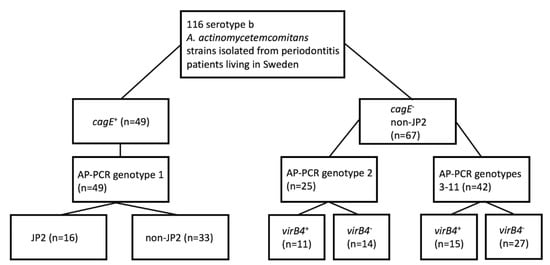
Figure 2.
Genotype patterns of A. actinomycetemcomitans serotype b strains. Schematic overview of AP-PCR type, as well as the JP2- and virB4-genotype patterns of the cagE-positive and cagE-negative strains, respectively. The collection of 116 serotype b strains was earlier sampled from periodontitis patients living in Sweden [27].
According to our results, cagE was present in 49 (42.2%) strains, including all 16 JP2 genotype strains, and hence absent in 67 (57.8%) strains. Of the cagE-positive strains, all (100%) belonged to AP-PCR genotype 1. Interestingly, three cagE-positive strains (all non-JP2 genotypes) were found to carry the virB4 gene. However, PCR analysis, using the primers magB01-F and ssb-R, supported that all three strains most likely carried virB4 on a plasmid rather than on the chromosome (data not shown). Thus, we concluded that a property common among the cagE-positive strains is an apparent lack of a chromosomal virB4 gene. Of the cagE-negative A. actinomycetemcomitans strains, 25 (37.3%) belonged to AP-PCR genotype 2, and 42 (62.7%) belonged to AP-PCR genotypes 3–11. The prevalence of virB4 was somewhat higher among the AP-PCR genotype 2 A. actinomycetemcomitans strains (44%) compared to the strains belonging to AP-PCR genotypes 3–11 (35.7%), suggesting that virB4 might be usable as a genetic marker for a subgroup of cagE-negative strains. Thus, taken together, as none of the 116 strains studied encoded both cagE and virB4 on the chromosome, we concluded that there is an apparent inverse relationship in the carriage of these genes in the A. actinomycetemcomitans strains of serotype b.
2.3. Higher Proportions of CagE-Positive A. actinomycetemcomitans Serotype B in Subgingival Plaque Samples
Furthermore, we assessed whether the cagE genotype may correlate with the proportion of A. actinomycetemcomitans found in the subgingival plaque samples. For this, the serotype b A. actinomycetemcomitans strains (n = 116) were divided into two groups, i.e., cagE-positive (n = 49) and cagE-negative (n = 67), and then they were matched with the determined total viable counts (%) of A. actinomycetemcomitans in the respective samples [27]. This clearly revealed that cagE-positive strains were carried in patients at significantly higher (p < 0.001) proportions than cagE-negative A. actinomycetemcomitans (Figure 3A). However, among the cagE-positive, the proportion of A. actinomycetemcomitans in samples with a JP2 genotype strain (n = 16) was not significantly different from that with a non-JP2 genotype strain (n = 33) (Figure 3B).
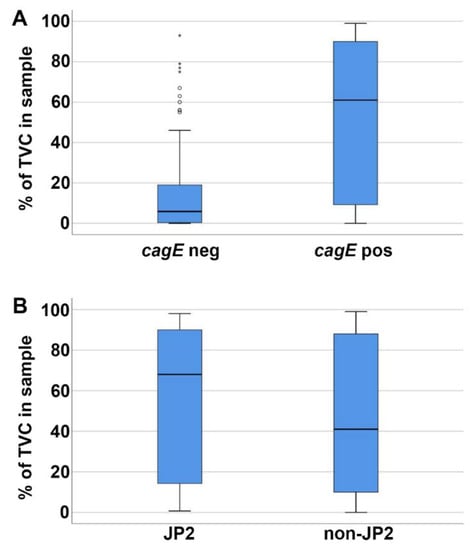
Figure 3.
Higher proportions of A. actinomycetemcomitans in subgingival plaque samples containing a cagE-positive serotype b. The proportion of A. actinomycetemcomitans (total viable count—TVC; %) in the subgingival plaque samples was determined earlier for each of the serotype b strains (n = 116) [27]. (A) The cagE-positive strains (n = 49) were present in significantly higher (p < 0.001) proportions than the cagE-negative strains (n = 67). (B) The JP2 (n = 16) and non-JP2 (n = 33) strains were present at similar proportions. Median and quartiles from the samples are shown in each panel.
2.4. Higher Prevalence of CagE-Positive A. actinomycetemcomitans Serotype B in Young Patients
To further evaluate the virulence of the cagE-positive serotype b A. actinomycetemcomitans strains (n = 49) among periodontitis patients living in Sweden, we also assessed the age-associated prevalence of these strains. For this purpose, the patients (n = 116) were grouped into young (≤35 years; n = 62) and old (>35 years; n = 54) groups (Table 3). This revealed that among the young patients, cagE+ A. actinomycetemcomitans strains (n = 40; 64.5%) were much more common than among the older patients (n = 9; 16.7%), i.e., these strains had a significantly higher (p < 0.001, odds ratio (OR) = 9.1, 95% CI: 3.8–22.0) prevalence among the young patients.

Table 3.
Age-associated distribution of the cagE genotype of serotype b. The prevalence of cagE-positive and cagE-negative strains among the A. actinomycetemcomitans serotype b strains (n = 116) sampled from young (≤35 yr; n = 62) and from old patients (>35 yr; n = 54). The numbers and percentages (%) of young, old, and all patients are indicated. The prevalence of cagE-positive strains was significantly higher (p < 0.001; odds ratio (OR) = 10.5, 95% CI: 4.2–26.1) in the young compared to old patients.
3. Discussion
In the present work, we used conventional PCR and qPCR to genotypically analyze our collection of 116 A. actinomycetemcomitans serotype b strains, collected during 15 years from periodontitis patients living in Sweden, and our present results underline the potential use of cagE genotyping in the risk assessment of the development of periodontal attachment loss in adolescents living in Sweden.
Each of the 116 serotype b strains were matched both with its load (% of total viable count) in the respective subgingival plaque sample and with its age-associated prevalence category [27]. As cagE-positive, in contrast to cagE-negative serotype b A. actinomycetemcomitans strains, were found at larger proportions in the plaque samples and exhibited a much higher prevalence in the young compared to in the old patients of this population, our present results are consistent with our findings assessing the longitudinal Ghanaian adolescent cohort [19]. Whereas the proportions of A. actinomycetemcomitans genotypes in the total viable counts of subgingival plaque samples had not earlier been assessed in patient cohorts, the ratio of cagE-positive among serotype b strains carried by young patients in the Swedish population (64.5%) was similar to that of the adolescents in the Ghanaian cohort, exhibiting an association between the progression of attachment loss and exposure to this particular cagE-positive genotype [14,19]. As the cagE-positive serotype b strains sampled in both Ghana and Sweden were found at larger proportions in the plaque samples and exhibited a much higher prevalence in the young group of patients [19,27], the results from our present study are consistent with the notion that cagE-positive strains (including both the JP2 and non-JP2 genotypes) represent a subgroup of highly virulent A. actinomycetemcomitans serotype b.
The genetic similarity of cagE+ serotype b strains is supported by the observation that they share the same AP-PCR genotype, as well as the fact that they have all a complete cdtABC gene operon [30]. We speculated earlier that they may belong to a clonal lineage that is closely related to the JP2 genotype ancestor [19]. As cagE-positive strains include both the JP2 and non-JP2 genotypes but no identified JP2-genotype strain has thus far been found to be cagE-negative, it is hypothesized that the JP2 genotype-associated deletion in the ltxCABD promoter once originated in a cagE-positive serotype b strain (Figure 4). It is tempting to speculate that high leukotoxicity may have been a characteristic of this ancestral A. actinomycetemcomitans strain, as that is a property common among cagE-positive strains, regardless of whether they are of the JP2 genotype or not.
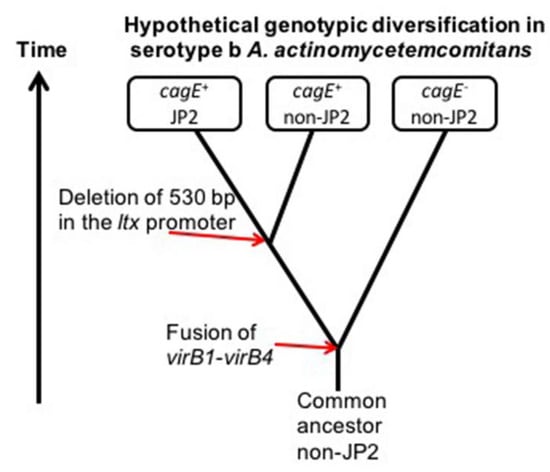
Figure 4.
Hypothetical origin of the cagE and JP2 genotypes in serotype b A. actinomycetemcomitans. The genetic similarity between cagE-positive strains and the apparent absence of the JP2 genotype among cagE-negative strains suggests the possibility that the JP2-associated 530-bp deletion in the ltx promoter might have originated in a cagE+ strain. The JP2 genotype of A. actinomycetemcomitans initially appeared as a distinct genotype in the Mediterranean part of Africa approximately 2400 years ago [29]. As cagE+ strains consistently lack chromosomal copies of virB1 and virB4, an earlier recombination event causing fusion of a virB1- and a virB4-like gene sequence resulting in the cagE determinant might have taken place in a common serotype b A. actinomycetemcomitans ancestral strain.
Consistent with our earlier in silico analysis of the genome-sequenced serotype b strains in the NCBI database [19], another property shared between the cagE-positive strains assessed in the present work was a lack of chromosomal genes encoding the T4SS-associated proteins VirB1 and VirB4. Based on the homology between VirB1 and VirB4 with the CagE N-, and C-terminus, respectively, we suggested earlier that CagE may represent a fusion product of a VirB1- and a VirB4-like amino acid sequence [19]. A scenario where the origin of the cagE+ serotype b A. actinomycetemcomitans strains is a recombination event on the chromosome, generating a fusion of parts of the genes encoding virB1 and virB4 (as illustrated in Figure 4), is plausible considering that chimeric proteins do exist in a number of bacterial T4SS gene clusters. For example, it was reported that H. pylori VirB3 and VirB4 is a fusion product, i.e., the first 150 amino acids of VirB4 have weak similarity with VirB3 although the motifs are conserved [31]. Similarly, a Western blot assay indicated a CagE-like protein pattern when prototypical virB3 and virB4 genes of A. tumefaciens were fused together and expressed [32]. Chimeric proteins are also encoded in a number of T4SS gene clusters of other species, including VirB3–VirB4 in Campylobacter jejuni [33,34], VirB1–VirB8 in Bordetella pertussis [35], and VirB11–VirD4 (MagB11–MagB12) in at least one strain of A. actinomycetemcomitans [25]. Moreover, observations with H. pylori are consistent with the notion that T4SS gene clusters can include regions that are prone to genetic rearrangements, resulting in the disruption or activation of the secretion system [36]. Results from our present work show that CagE and VirB1/VirB4 can be encoded in the same A. actinomycetemcomitans strain, albeit, as supported by PCR, with the T4SS genes most likely encoded on plasmids. The carriage of plasmids encoding T4SS genes, such as virB1 and virB4, has been demonstrated in some A. actinomycetemcomitans strains [25,26]. In contrast, cagE appears not to be encoded on plasmids. According to the sequences available in the NCBI database, no hitherto sequenced A. actinomycetemcomitans plasmid carries a cagE gene locus. We were unable to detect by PCR the presence of a T4SS-encoding plasmid in the cagE-positive serotype b strain HK1651, which was earlier reported [25]. The reason for this discrepancy is not known but may reflect the possibility that this plasmid was lost in the strain preserved in our stocks upon repeated in vitro cultivation. The loss of plasmids of A. actinomycetemcomitans strains during in vitro cultivation is a phenomenon that has been reported earlier, albeit then related to growth in an antibiotic free medium [37].
Taken together, our present results further support the usefulness of the cagE gene as a potential diagnostic marker in the risk assessment of the development of attachment loss among young individuals. We conclude that cagE positive A. actinomycetemcomitans strains of serotype b among periodontitis patients living in Sweden consist of the JP2 and non-JP2 genotypes with phenotypic characteristics similar to the ones seen for the JP2 genotype strains but with a leukotoxin promoter region lacking the 530-bp deletion. Their origin, evolution, and extent of genetic similarity will be further explored by whole genome sequencing.
4. Materials and Methods
4.1. Collection of A. actinomycetemcomitans Strains and Clinical Data Used in the Present Study
For the present work, we used data from our microbiological analyses of 3459 subgingival plaque samples, collected from 1445 patients during 15 years (2000–2014) that included 337 ‘younger’ patients (≤35 years of age) and 1108 ‘older’ patients (>35 years of age) [27]. At the specialist clinics, it is recommended that microbial analysis is performed to study the microbial biofilm profiles of individuals ≤35 years affected by periodontal attachment loss, and of patients >35 years with rapidly progressive periodontitis, not responding to conventional periodontal therapy. The samples were sent from the Specialist Clinic of Periodontology at the Dental School in Umeå, Sweden, and from external specialist dental clinics throughout Sweden to be analyzed at the laboratory for microbiological diagnostics, Dental School, Umeå. The samples were collected from individuals between 9 and 92 years of age that were all diagnosed with periodontitis and referred to specialist clinics for periodontal treatment. However, due to the many clinics involved and the retrospective nature of the present study, clinical and other parameters were not systematically reported in the patient information attached to the referral to the laboratory for microbiological diagnostics. Therefore, the classification of the patients was dichotomized only and was based on the old definition of early onset periodontitis, which distinguished patients ≤35 years versus those >35 years of age [38]. An A. actinomycetemcomitans strain was collected and isolated from 347 patients [27]. PCR characterization revealed that 118 (34.0%) of the A. actinomycetemcomitans strains were serotype b, and 17 (14.4% of the serotype b strains) were characterized by 530-bp deletion in the promoter region of the leukotoxin gene operon (JP2 genotype). Among these 118 serotype b strains, we were able to cultivate and characterize 116 for use in the present study: 100 non-JP2 genotype and 16 JP2 genotype. For the present work, each of these 116 unique A. actinomycetemcomitans strains was combined with recorded clinical data, i.e., the age group of the patient (>35 or ≤35 years), and proportion of A. actinomycetemcomitans of the total cultivable microflora (TVC) in the sample.
4.2. Bacterial Strains and Growth Conditions
In the present work, we used a collection of 116 unique A. actinomycetemcomitans serotype b strains that were collected from periodontitis patients living in Sweden [27]. A list of these strains is presented in Table S1. The sampling of this collection and the subsequent characterization of the serotype, the AP-PCR genotype, and the leukotoxin promoter type (JP2/non-JP2 genotype) has been described earlier [27]. In the present study, 25 serotype b A. actinomycetemcomitans strains were used as reference, as they were subject to prior whole genome sequencing [15,19,39,40,41] (Table 1). Among these, five belong to a collection of oral A. actinomycetemcomitans strains previously reported on by Prof. Sirkka Asikainen: ANH9381, I23C, S23A, SCC1398 and SCC4092. Nine strains belong to the collection of serotype b A. actinomycetemcomitans strains, sampled from periodontitis patients living in Sweden: 133A1-08U, 196A1-10U, 115A-11U, 245-12U, 338A1-13U, 304A1-14U, 299A1-15U, 456A1-13U, and 520A-01U [28]. A. actinomycetemcomitans strains 443G, 486G, 575G, 605G, and 638G were sampled from a Ghanaian cohort of adolescents [6,42]. Finally, six type strains were included in the study: HK908 [29], HK909 [43], HK912 [29], HK921 [43], HK1651 [39], and Y4 [44,45]. All strains were cultured on blood agar plates (5% defibrinated horse blood, 5 mg of hemin/l, 10 mg of vitamin K/l, Columbia agar base) and incubated in air supplemented with 5% CO2 at 37 °C.
4.3. DNA Isolation and Polymerase Chain Reaction Analysis
DNA templates for PCR and qPCR analysis were obtained by boiling a loopful of fresh A. actinomycetemcomitans colonies in 100 μl of water. A. actinomycetemcomitans genomic DNA to be used in AP-PCR was isolated using the GenElute™ Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA), following the manufacturer’s instructions. For the isolation of plasmids from A. actinomycetemcomitans strains, a QIAprep® Spin miniprep kit was used (QiaGen, Venlo, The Netherlands). Reaction mixtures for PCR were prepared using illustra™ PuReTaq™ Ready-To-Go™ PCR beads (GF Healthcare, Buckinghamshire, UK), whereas we used a KAPA SYBR® FAST qPCR Kit (KAPA Biosystems, Wilmington, MA, USA) for qPCR. The AP-PCR type was analyzed as earlier described [19,27], using the random sequence oligonucleotide OPB-3 (5′-AGTCAGCCAC-3′) (Invitrogen, Carlsbad, CA, USA) at 0.4 μmol/l and cycling conditions according to Dogan and coworkers [46]. The cagE gene was amplified by PCR as a 1020-bp DNA fragment, using a cagE forward primer (5’-GGATCCGTCCCTGAAATTTTATTAGCTTG-3’) and a cagE reverse primer (5-CTGCAGTTAAACGACCTTTAAACATTTTTTTA-3’) [20]. In qPCR analysis, cagE was detected as earlier described [19] using the cagE_F2 (5’-TGGATTGGGACAAGTGAACA-3’) and cagE_R2 (5’-CAATAATGGCTCGTGCAATATC-3’) primers to amplify a 623-bp internal fragment of the cagE gene. A ≈630-bp fragment of the lytic transglycosylase, virB1 gene was amplified using a cagE forward primer and a virB1 reverse primer (5’-GTTTTTAATCAATCTTCCTGATTG-3’). The amplification of the ATP:ase-encoding virb4 gene, as a ≈900-bp DNA fragment, was carried out by PCR or qPCR using the virB4 forward primer (5’-GTGCAGAAGCCTGTATTCGTGC-3’), and the virB4 reverse primer (5’-CCAGTCATTAGTGGCTTCGCC-3’). The magB01-F (5’-GCCATCTACTACGCCTATCGC-3’) and ssb-R (5’-TTATCGCCGTCAAGCGGAAG-3’) primers [25] were used in PCR to assess the presence of plasmids encoding T4SS genes. PCR cycling conditions were 94 °C for 1 min, followed by 35 cycles of 94 °C for 30 sec, 54 °C for 30 sec, and 72 °C for 1 min, and then finally 72 °C for 7 min. The cycling conditions for qPCR were 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 sec, 54 °C for 5 sec, and 72 °C for 22 sec. The complete genome sequences of serotype b strains SCC1398 (VirB1; GenBank accession KND83482), and I23C (VirB4; KOE53154) [40] were used as reference in oligonucleotide synthesis.
4.4. Statistical Analysis and Image Processing
The rank test was used to calculate the strength of the association between the A. actinomycetemcomitans cagE and JP2 genotypes and proportion of TVC in subgingival plaque samples (IBM SPSS Statistics for Windows, Version 25.0, Armonk, New York). An odds ratio (OR) was used to quantify the strength of the association between the A. actinomycetemcomitans cagE genotype and age group (MedCalc for Windows, MedCalc Software, Ostend, Belgium). No normalization of the data or test unit was used in the present work.
4.5. Ethical Considerations
All procedures were conducted according to the guidelines of the local ethics committee at the Medical Faculty of Umeå University, which are in compliance with the Declaration of Helsinki (64th WMA General Assembly, Fortaleza, October 2013). The characterization of the A. actinomycetemcomitans strains was made utilizing clinical samples from patients visiting the Specialist Clinic of Periodontology at the Dental School in Umeå. Data from specific strains were grouped in relation to age (>35 or ≤ 35 years) and could not be traced to a specific individual.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/3/153/s1. Table S1. The 116 A. actinomycetemcomitans serotype b strains, collected from periodontitis patients living in Sweden, and which were used in the present work.
Author Contributions
Conceptualization, A.J., R.C., and J.O.; methodology, R.C., and J.O.; validation, R.C., M.L., S.J., and J.O; formal Analysis, A.J., R.C., S.J, and J.O.; investigation, R.C., M.L., S.J., and J.O.; resources A.J., R.C., C.H.Å., D.H., and J.O; data curation, A.J., R.C., and J.O.; writing—original draft preparation, A.J., R.C., and J.O.; writing—review and editing, A.J., R.C., C.H.Å., D.H., M.L., S.J., and J.O.; visualization, A.J., R.C., and J.O.; supervision, A.J., R.C., C.H.Å., and J.O.; project administration, A.J. and J.O; funding acquisition, A.J., M.L. and J.O.
Acknowledgments
We are grateful to Elisabeth Granström for valuable technical assistance. This work was supported by TUA grants from the County Council of Västerbotten, Sweden (to J.O. and A.J.), by funds from Insamlingsstiftelsen, Medical Faculty, Umeå University (to J.O. and A.J.), and from Svenska Tandläkare-sällskapet, Kempe Foundation, and Thuréus Foundation (to M.L.).
Conflicts of Interest
The authors declare no competing interests.
References
- Fine, D.H.; Patil, A.G.; Velusamy, S.K. Aggregatibacter actinomycetemcomitans (Aa) under the radar: Myths and misunderstandings of Aa and its role in aggressive periodontitis. Front. Immunol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Ramich, T.; Asendorf, A.; Nickles, K.; Oremek, G.M.; Schubert, R.; Nibali, L.; Wohlfeil, M.; Eickholz, P. Inflammatory serum markers up to 5 years after comprehensive periodontal therapy of aggressive and chronic periodontitis. Clin. Oral Investig. 2018, 22, 3079–3089. [Google Scholar] [CrossRef] [PubMed]
- van Winkelhoff, A.J.; Slots, J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontol. 2000 1999, 20, 122–135. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Fairlie, K.; Ferrandiz, J.; Nasri, C.; McKiernan, M.; Gunsolley, J. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 2007, 45, 3859–3869. [Google Scholar] [CrossRef] [PubMed]
- Haubek, D.; Ennibi, O.K.; Poulsen, K.; Vaeth, M.; Poulsen, S.; Kilian, M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef]
- Höglund Åberg, C.; Kwamin, F.; Claesson, R.; Dahlen, G.; Johansson, A.; Haubek, D. Progression of attachment loss is strongly associated with presence of the JP2 genotype of Aggregatibacter actinomycetemcomitans: A prospective cohort study of a young adolescent population. J. Clin. Periodontol. 2014, 41, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, U.; Abbas, F.; Armand, S.; Loos, B.G.; Timmerman, M.F.; Van der Weijden, G.A.; Van Winkelhoff, A.J.; Winkel, E.G. Java project on periodontal diseases. The natural development of periodontitis: Risk factors, risk predictors and risk determinants. J. Clin. Periodontol. 2006, 33, 540–548. [Google Scholar] [CrossRef]
- DiRienzo, J.M. Breaking the gingival epithelial barrier: Role of the Aggregatibacter actinomycetemcomitans cytolethal distending toxin in oral infectious disease. Cells 2014, 3, 476–499. [Google Scholar] [CrossRef]
- Johansson, A. Aggregatibacter actinomycetemcomitans leukotoxin: A powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef]
- Oscarsson, J.; Claesson, R.; Lindholm, M.; Höglund Åberg, C.; Johansson, A. Tools of Aggregatibacter actinomycetemcomitans to evade the host response. J. Clin. Med. 2019, 8, 1079. [Google Scholar] [CrossRef]
- Henderson, B.; Ward, J.M.; Ready, D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: A triple A* periodontopathogen? Periodontol. 2000 2010, 54, 78–105. [Google Scholar] [CrossRef]
- Poulsen, K.; Theilade, E.; Lally, E.T.; Demuth, D.R.; Kilian, M. Population structure of Actinobacillus actinomycetemcomitans: A framework for studies of disease-associated properties. Microbiology 1994, 140 Pt 8, 2049–2060. [Google Scholar] [CrossRef][Green Version]
- Tsuzukibashi, O.; Saito, M.; Kobayashi, T.; Umezawa, K.; Nagahama, F.; Hiroi, T.; Hirasawa, M.; Takada, K. A gene cluster for the synthesis of serotype g-specific polysaccharide antigen in Aggregatibacter actinomycetemcomitans. Arch. Microbiol. 2014, 196, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Höglund Åberg, C.; Haubek, D.; Kwamin, F.; Johansson, A.; Claesson, R. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. PLoS ONE 2014, 9, e104095. [Google Scholar] [CrossRef] [PubMed]
- Kittichotirat, W.; Bumgarner, R.E.; Chen, C. Evolutionary divergence of Aggregatibacter actinomycetemcomitans. J. Dent. Res. 2016, 95, 94–101. [Google Scholar] [CrossRef]
- Brogan, J.M.; Lally, E.T.; Poulsen, K.; Kilian, M.; Demuth, D.R. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 1994, 62, 501–508. [Google Scholar] [PubMed]
- Haubek, D.; Johansson, A. Pathogenicity of the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J. Oral Microbiol. 2014, 6, 23980. [Google Scholar] [CrossRef]
- Poulsen, K.; Ennibi, O.K.; Haubek, D. Improved PCR for detection of the highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans in subgingival plaque samples. J. Clin. Microbiol. 2003, 41, 4829–4832. [Google Scholar] [CrossRef]
- Johansson, A.; Claesson, R.; Höglund Åberg, C.; Haubek, D.; Oscarsson, J. The cagE gene sequence as a diagnostic marker to identify JP2 and non-JP2 highly leukotoxic Aggregatibacter actinomycetemcomitans serotype b strains. J. Periodontal Res. 2017, 52, 903–912. [Google Scholar] [CrossRef]
- Teng, Y.T.; Hu, W. Expression cloning of a periodontitis-associated apoptotic effector, cagE homologue, in Actinobacillus actinomycetemcomitans. Biochem. Biophys. Res. Commun. 2003, 303, 1086–1094. [Google Scholar] [CrossRef]
- Kelk, P.; Claesson, R.; Chen, C.; Sjöstedt, A.; Johansson, A. IL-1β secretion induced by Aggregatibacter (Actinobacillus) actinomycetemcomitans is mainly caused by the leukotoxin. Int. J. Med. Microbiol. 2008, 298, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Whitaker, N.; Gonzalez-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G.; Orlova, E.V. Structural organisation of the type IV secretion systems. Curr. Opin. Microbiol. 2014, 17, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Galli, D.M.; Chen, J.; Novak, K.F.; Leblanc, D.J. Nucleotide sequence and analysis of conjugative plasmid pVT745. J. Bacteriol. 2001, 183, 1585–1594. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, C.C.; Chen, C.H.; Tang, C.Y.; Chen, K.H.; Chen, Z.F.; Chang, S.H.; Tsai, C.Y.; Liou, M.L. Prevalence and comparative analysis of the type IV secretion system in Aggregatibacter actinomycetemcomitan. J. Microbiol. Immunol. Infect. 2018, 51, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Novak, K.F.; Dougherty, B.; Pelaez, M. Actinobacillus actinomycetemcomitans harbours type IV secretion system genes on a plasmid and in the chromosome. Microbiology 2001, 147, 3027–3035. [Google Scholar] [CrossRef][Green Version]
- Claesson, R.; Höglund-Åberg, C.; Haubek, D.; Johansson, A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J. Oral Microbiol. 2017, 9, 1334504. [Google Scholar] [CrossRef]
- Claesson, R.; Gudmundson, J.; Höglund Åberg, C.; Haubek, D.; Johansson, A. Detection of a 640-bp deletion in the Aggregatibacter actinomycetemcomitans leukotoxin promoter region in isolates from an adolescent of Ethiopian origin. J. Oral Microbiol. 2015, 7, 26974. [Google Scholar] [CrossRef]
- Haubek, D.; Poulsen, K.; Kilian, M. Microevolution and patterns of dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 2007, 75, 3080–3088. [Google Scholar] [CrossRef] [PubMed]
- Höglund Åberg, C.; Antonoglou, G.; Haubek, D.; Kwamin, F.; Claesson, R.; Johansson, A. Cytolethal distending toxin in isolates of Aggregatibacter actinomycetemcomitans from Ghanaian adolescents and association with serotype and disease progression. PLoS ONE 2013, 8, e65781. [Google Scholar]
- Kutter, S.; Buhrdorf, R.; Haas, J.; Schneider-Brachert, W.; Haas, R.; Fischer, W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J. Bacteriol. 2008, 190, 2161–2171. [Google Scholar] [CrossRef]
- Mossey, P.; Hudacek, A.; Das, A. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J. Bacteriol. 2010, 192, 2830–2838. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, R.A.; Pearson, B.M.; Friis, L.M.; Guerry, P.; Wells, J.M. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 2004, 150, 3507–3517. [Google Scholar] [CrossRef]
- Fronzes, R.; Christie, P.J.; Waksman, G. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009, 7, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Rambow-Larsen, A.A.; Weiss, A.A. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 2002, 184, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J. The Mosaic Type IV Secretion Systems. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Sreenivasan, P.K.; Fives-Taylor, P. Isolation and characterization of deletion derivatives of pDL282, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle plasmid. Plasmid 1994, 31, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Grossman, N.S. Periodontal disease as a specific, albeit chronic, infection: Diagnosis and treatment. Clin. Microbiol. Rev. 2001, 14, 727–752. [Google Scholar] [CrossRef]
- Haubek, D.; Havemose-Poulsen, A.; Westergaard, J. Aggressive periodontitis in a 16-year-old Ghanaian adolescent, the original source of Actinobacillus actinomycetemcomitans strain HK1651—A 10-year follow up. Int. J. Paediatr. Dent. 2006, 16, 370–375. [Google Scholar] [CrossRef]
- Kittichotirat, W.; Bumgarner, R.E.; Asikainen, S.; Chen, C. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS ONE 2011, 6, e22420. [Google Scholar] [CrossRef]
- Sun, R.; Kittichotirat, W.; Wang, J.; Jan, M.; Chen, W.; Asikainen, S.; Bumgarner, R.; Chen, C. Genomic stability of Aggregatibacter actinomycetemcomitans during persistent oral infection in human. PLoS ONE 2013, 8, e66472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Höglund Åberg, C.; Kwamin, F.; Claesson, R.; Johansson, A.; Haubek, D. Presence of JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans and attachment loss in adolescents in Ghana. J. Periodontol. 2012, 83, 1520–1528. [Google Scholar] [CrossRef]
- Haubek, D.; Dirienzo, J.M.; Tinoco, E.M.; Westergaard, J.; Lopez, N.J.; Chung, C.P.; Poulsen, K.; Kilian, M. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 1997, 35, 3037–3042. [Google Scholar] [PubMed]
- Kiley, P.; Holt, S.C. Characterization of the lipopolysaccharide from Actinobacillus actinomycetemcomitans Y4 and N27. Infect. Immun. 1980, 30, 862–873. [Google Scholar] [PubMed]
- Newman, M.G.; Socransky, S.S. Predominant cultivable microbiota in periodontosis. J. Periodontal Res. 1977, 12, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Dogan, B.; Saarela, M.H.; Jousimies-Somer, H.; Alaluusua, S.; Asikainen, S. Actinobacillus actinomycetemcomitans serotype e-biotypes, genetic diversity and distribution in relation to periodontal status. Oral Microbiol. Immunol. 1999, 14, 98–103. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).