Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors
Abstract
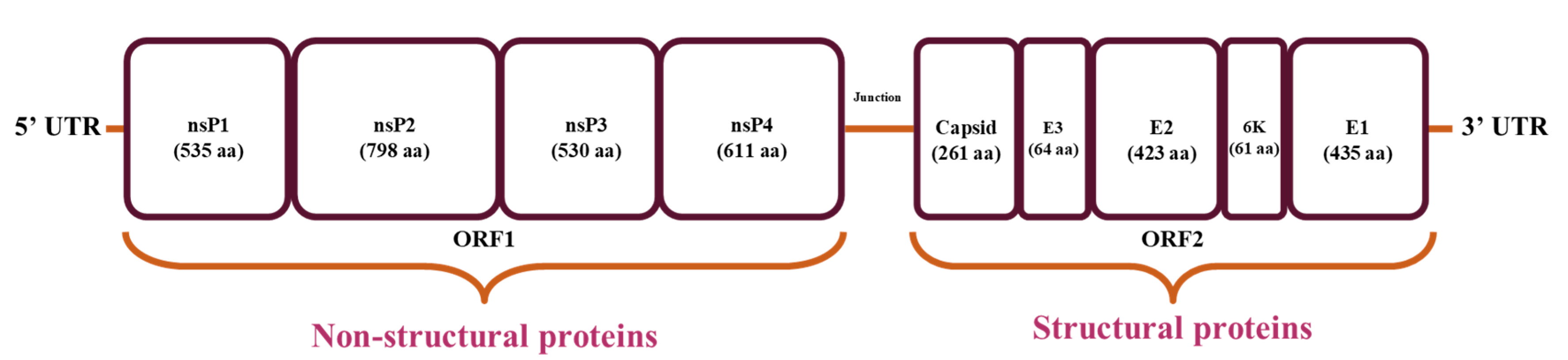
1. Introduction
2. Result and Discussion
2.1. Pharmacokinetic Properties
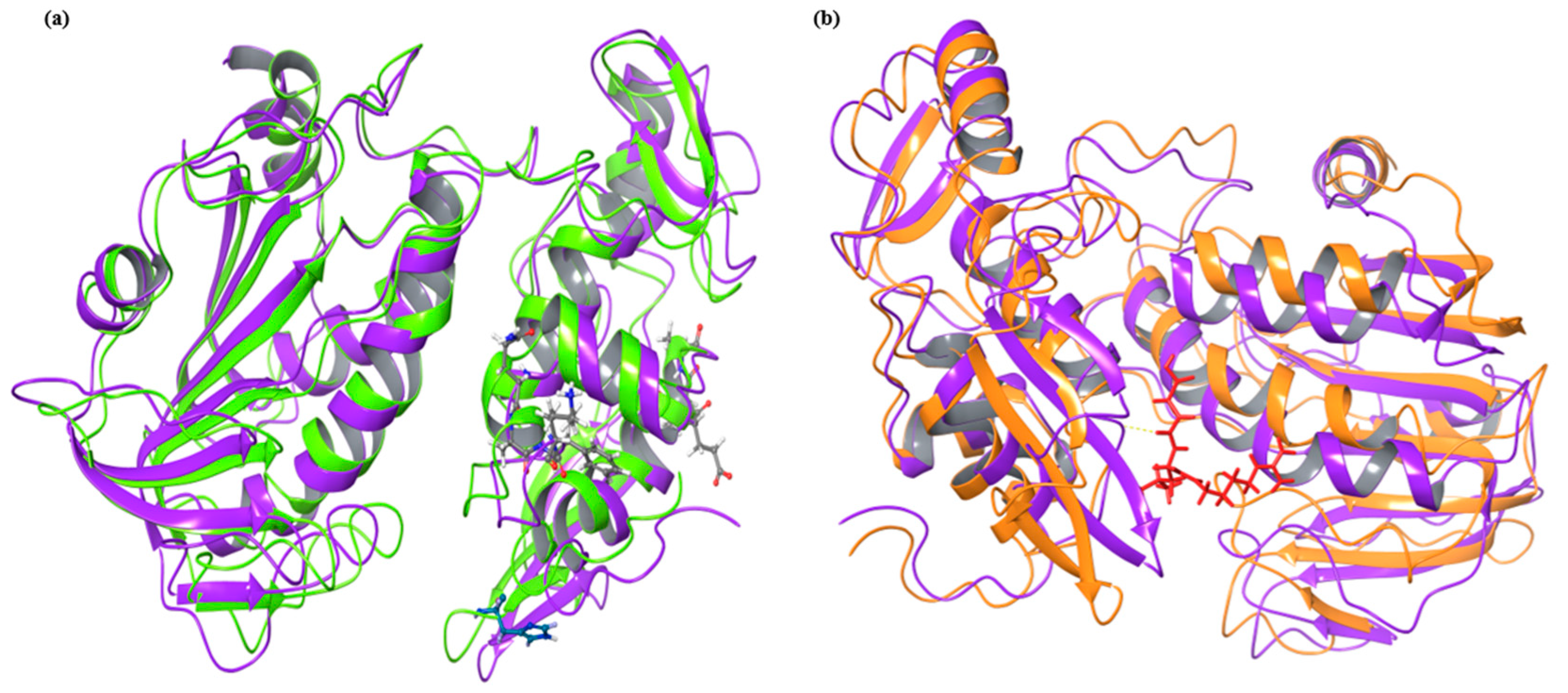
2.2. MD Simulation Analysis
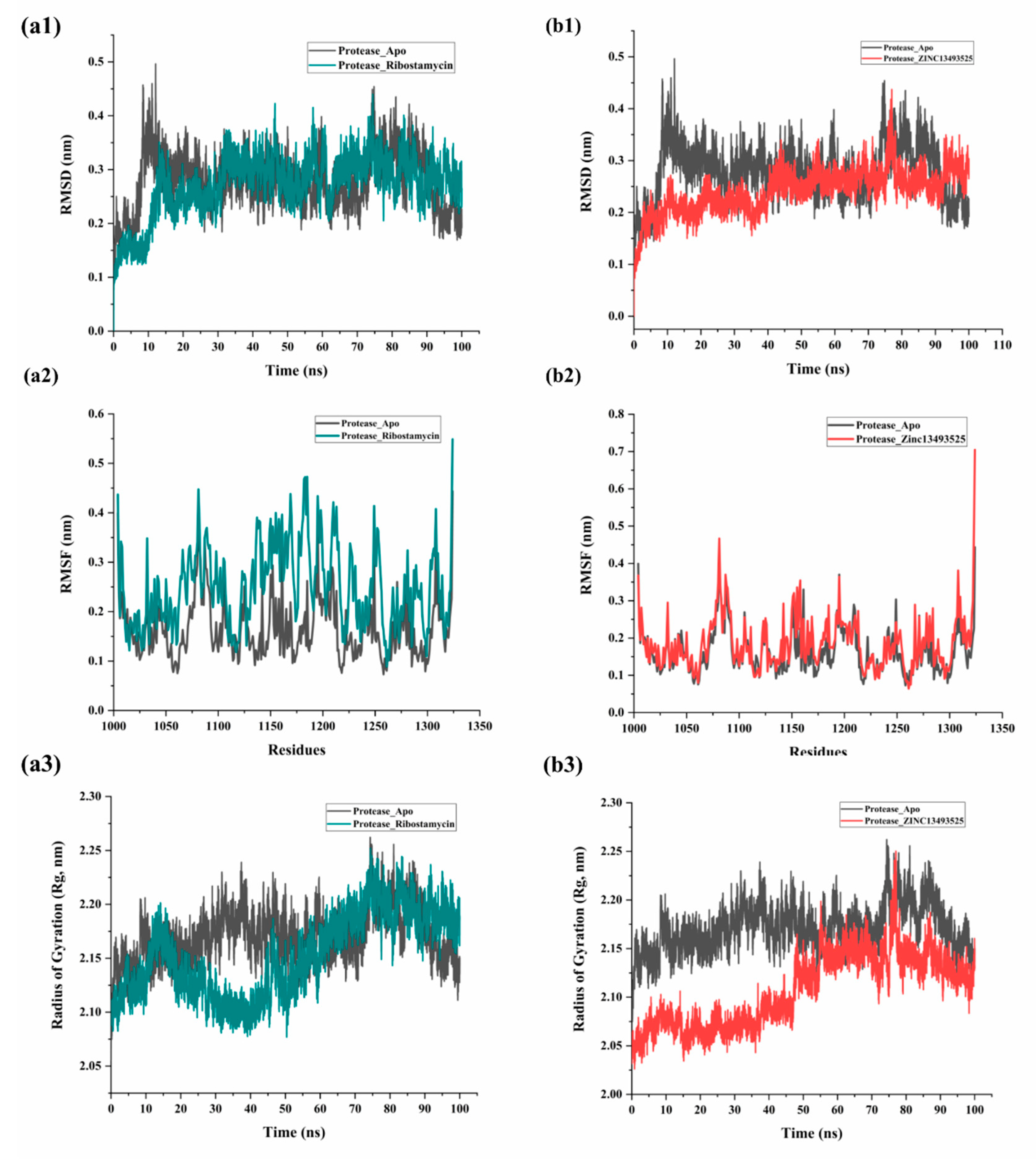
2.2.1. RMSD, RMSF and Radius of Gyration (Rg) Evaluation
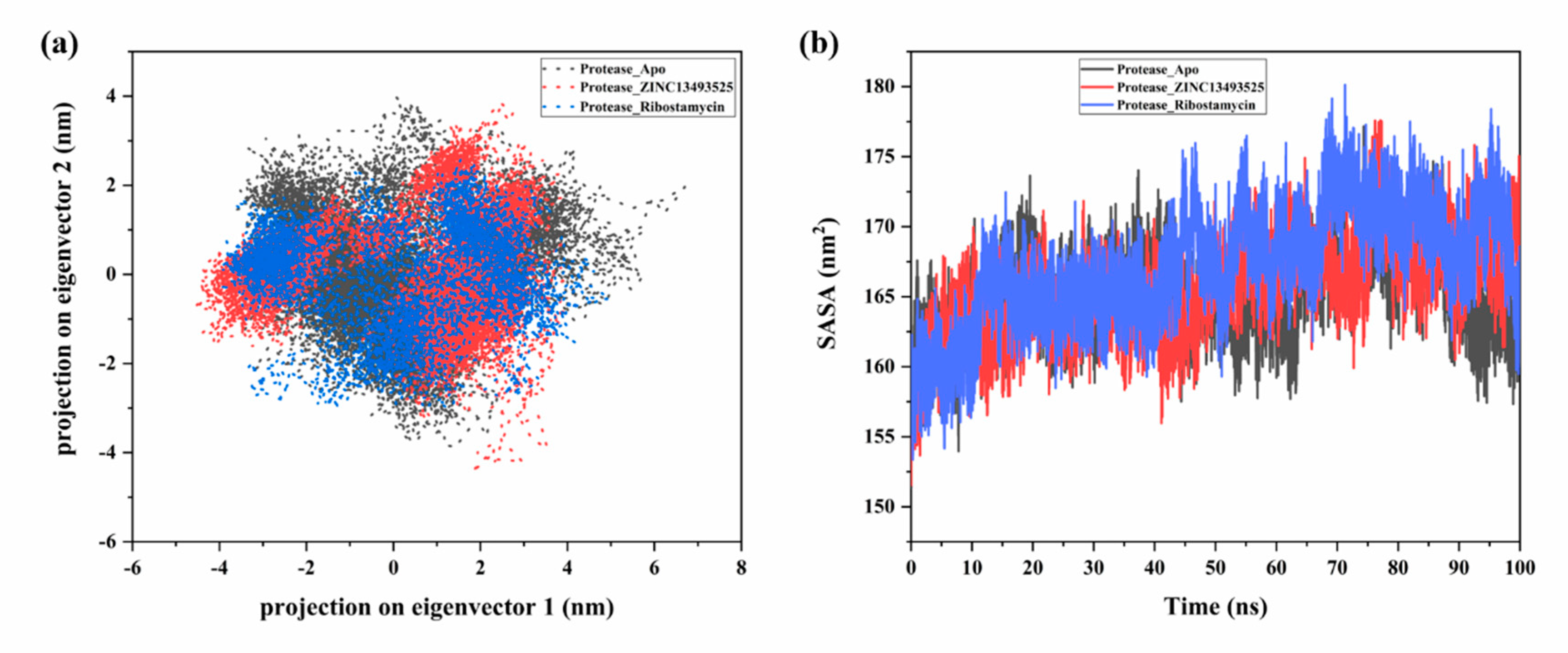
2.2.2. Principal Component Analysis and Solvent Accessible Surface Area (SASA) Prediction
3. Material and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Subudhi, B.; Chattopadhyay, S.; Mishra, P.; Kumar, A. Current Strategies for Inhibition of Chikungunya Infection. Viruses 2018, 10, 235. [Google Scholar] [CrossRef] [PubMed]
- Abdelnabi, R.; Neyts, J.; Delang, L. Towards antivirals against chikungunya virus. Antivir. Res. 2015, 121, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Home: National Vector Borne Disease Control Programme (NVBDCP). Available online: https://www.nvbdcp.gov.in/ (accessed on 20 July 2019).
- Thiberville, S.D.; Moyen, N.; Dupuis-Maguiraga, L.; Nougairede, A.; Gould, E.A.; Roques, P.; De Lamballerie, X. Chikungunya fever: Epidemiology, clinical syndrome, pathogenesis and therapy. Antivir. Res. 2013, 99, 345–370. [Google Scholar] [CrossRef] [PubMed]
- Rezza, G. Dengue and chikungunya: Long-distance spread and outbreaks in naïve areas. Pathog. Glob. Health 2014, 108, 349–355. [Google Scholar] [CrossRef]
- Powers, A.M. Vaccine and therapeutic options to control chikungunya virus. Clin. Microbiol. Rev. 2018, 31, 1–29. [Google Scholar] [CrossRef]
- Chahar, H.S.; Bharaj, P.; Dar, L.; Guleria, R.; Kabra, S.K.; Broor, S. Co-infections with chikungunya virus and dengue virus in Delhi, India. Emerg. Infect. Dis. 2009, 15, 1077–1080. [Google Scholar] [CrossRef]
- Furuya-Kanamori, L.; Liang, S.; Milinovich, G.; Magalhaes, R.J.S.; Clements, A.C.A.; Hu, W.; Brasil, P.; Frentiu, F.D.; Dunning, R.; Yakob, L. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect. Dis. 2016, 16, 84. [Google Scholar] [CrossRef]
- Khan, A.H.; Morita, K.; Parquet, M.D.C.; Hasebe, F.; Mathenge, E.G.; Igarashi, A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J. Gen. Virol. 2015, 83, 3075–3084. [Google Scholar] [CrossRef]
- Singh, S.K.; Unni, S.K. Chikungunya virus: Host pathogen interaction. Rev. Med. Virol. 2011, 21, 78–88. [Google Scholar] [CrossRef]
- Narwal, M.; Singh, H.; Pratap, S.; Malik, A.; Kuhn, R.J.; Kumar, P.; Tomar, S. Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site. Int. J. Biol. Macromol. 2018, 116, 451–462. [Google Scholar] [CrossRef]
- Das, I.; Basantray, I.; Mamidi, P.; Nayak, T.K.; Pratheek, B.M.; Chattopadhyay, S.; Chattopadhyay, S. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS ONE 2014, 9, e100531. [Google Scholar] [CrossRef] [PubMed]
- Hardy, W.R.; Strauss, J.H. Processing the nonstructural polyproteins of sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 1989, 63, 4653–4664. [Google Scholar] [PubMed]
- Pastorino, B.A.M.; Peyrefitte, C.N.; Almeras, L.; Grandadam, M.; Rolland, D.; Tolou, H.J.; Bessaud, M. Expression and biochemical characterization of nsP2 cysteine protease of Chikungunya virus. Virus Res. 2008, 131, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Saisawang, C.; Saitornuang, S.; Sillapee, P.; Ubol, S.; Smith, D.R.; Ketterman, A.J. Chikungunya nsP2 protease is not a papain-like cysteine protease and the catalytic dyad cysteine is interchangeable with a proximal serine. Sci. Rep. 2015, 5, 17125. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, A.; Yadav, R.; Uversky, V.N.; Giri, R. Deciphering the dark proteome of Chikungunya virus. Sci. Rep. 2018, 8, 5822. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, A.; Uversky, V.N.; Giri, R. Understanding the interactability of chikungunya virus proteins via molecular recognition feature analysis. RSC Adv. 2018, 8, 27293–27303. [Google Scholar] [CrossRef]
- Giri, R.; Kumar, D.; Sharma, N.; Uversky, V.N. Intrinsically Disordered Side of the Zika Virus Proteome. Front. Cell. Infect. Microbiol. 2016, 6, 144. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.M.; Uversky, V.N.; Giri, R. Molecular Recognition Features in Zika Virus Proteome. J. Mol. Biol. 2018, 430, 2372–2388. [Google Scholar] [CrossRef]
- Xue, B.; Blocquel, D.; Habchi, J.; Uversky, A.V.; Kurgan, L.; Uversky, V.N.; Longhi, S. Structural Disorder in Viral Proteins. Chem. Rev. 2014, 114, 6880–6911. [Google Scholar] [CrossRef]
- Toto, A.; Camilloni, C.; Giri, R.; Brunori, M.; Vendruscolo, M.; Gianni, S. Molecular Recognition by Templated Folding of an Intrinsically Disordered Protein. Sci. Rep. 2016, 6, 21994. [Google Scholar] [CrossRef]
- Wang, J.; Cao, Z.; Zhao, L.; Li, S. Novel strategies for drug discovery based on Intrinsically Disordered Proteins (IDPs). Int. J. Mol. Sci. 2011, 12, 3205–3219. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Sharma, N.; Giri, R. Therapeutic Interventions of Cancers Using Intrinsically Disordered Proteins as Drug Targets: C-Myc as Model System. Cancer Inform. 2017, 16, 1176935117699408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Liang, B.; Aarthy, M.; Singh, S.K.; Garg, N.; Mysorekar, I.U.; Giri, R. Hydroxychloroquine Inhibits Zika Virus NS2B-NS3 Protease. ACS Omega 2018, 3, 18132–18141. [Google Scholar] [CrossRef] [PubMed]
- Siklos, M.; BenAissa, M.; Thatcher, G.R.J. Cysteine proteases as therapeutic targets: Does selectivity matter? A systematic review of calpain and cathepsin inhibitors. Acta Pharm. Sin. B 2015, 5, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Trinchese, F.; Fa, M.; Liu, S.; Zhang, H.; Hidalgo, A.; Schmidt, S.D.; Yamaguchi, H.; Yoshii, N.; Mathews, P.M.; Nixon, R.A.; et al. Inhibition of calpains improves memory and synaptic transmission in a mouse model of Alzheimer disease. J. Clin. Investig. 2008, 118, 2796–2807. [Google Scholar] [CrossRef] [PubMed]
- Rashad, A.A.; Mahalingam, S.; Keller, P.A. Chikungunya virus: Emerging targets and new opportunities for medicinal chemistry. J. Med. Chem. 2014, 57, 1147–1166. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.-J.; Delang, L.; Ng, L.F.P.; Priego, E.-M. Chikungunya virus drug discovery: Still a long way to go? Expert Opin. Drug Discov. 2019, 14, 855–866. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Yu, H.; Keller, P.A. Identification of chikungunya virus nsP2 protease inhibitors using structure-base approaches. J. Mol. Graph. Model. 2015, 57, 1–8. [Google Scholar] [CrossRef]
- Sajid, M.; McKerrow, J.H. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 2002, 120, 1–21. [Google Scholar] [CrossRef]
- Russo, A.T.; White, M.A.; Watowich, S.J. The Crystal Structure of the Venezuelan Equine Encephalitis Alphavirus nsP2 Protease. Structure 2006, 14, 1449–1458. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.L.; Shen, G.; Zhong, H.F. Pharmacokinetics of ribostamycin in paediatric patients. Clin. Pharmacokinet. 1992, 22, 144–151. [Google Scholar] [PubMed]
- Ennifar, E. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: From crystal to cell. Nucleic Acids Res. 2006, 34, 2328–2339. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.J.; Kembhavi, A.A.; Brown, M.A.; Kirschke, H.; Knight, C.G.; Tamai, M.; Hanada, K. L-trans-Epoxysuccinyl-leucylamido (4-guanidino) butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 1982, 201, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Pintér, M.; Aszódi, A.; Friedrich, P.; Ginzburg, I. Calpeptin, a calpain inhibitor, promotes neurite elongation in differentiating PC12 cells. Neurosci. Lett. 1994, 170, 91–93. [Google Scholar] [CrossRef]
- Salawu, E.O. In Silico Study Reveals How E64 Approaches, Binds to, and Inhibits Falcipain-2 of Plasmodium falciparum that Causes Malaria in Humans. Sci. Rep. 2018, 8, 16380. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Mizoue, K.; Kitamura, K.; Tse, W.C.; Huber, C.P.; Ishida, T. Structural basis of inhibition of cysteine proteases by E-64 and its derivatives. Biopolymers 1999, 51, 99–107. [Google Scholar] [CrossRef]
- Gastaminza, P.; Whitten-Bauer, C.; Chisari, F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA 2010, 107, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Thuaud, F.; Basmadjian, C.; Roytrakul, S.; Ubol, S.; Désaubry, L.; Smith, D.R. Assessment of flavaglines as potential chikungunya virus entry inhibitors. Microbiol. Immunol. 2015, 59, 129–141. [Google Scholar] [CrossRef]
- Lin, L.-T.; Chen, T.-Y.; Lin, S.-C.; Chung, C.-Y.; Lin, T.-C.; Wang, G.-H.; Anderson, R.; Lin, C.-C.; Richardson, C.D. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol. 2013, 13, 187. [Google Scholar] [CrossRef]
- Sharma, N.; Murali, A.; Singh, S.K.; Giri, R. Epigallocatechin gallate, an active green tea compound inhibits the Zika virus entry into host cells via binding the envelope protein. Int. J. Biol. Macromol. 2017, 104, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Sharma, N.; Aarthy, M.; Singh, S.; Giri, R. Mechanistic insights into Zika virus NS3 helicase inhibition by Epigallocatechin-3-gallate. bioRxiv 2019, 530600. [Google Scholar] [CrossRef]
- Rothan, H.A.; Bahrani, H.; Mohamed, Z.; Teoh, T.C.; Shankar, E.M.; Rahman, N.A.; Yusof, R. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS ONE 2015, 10, e0126360. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Dhanwani, R.; Patro, I.K.; Rao, P.V.L.; Parida, M.M. Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antivir. Res. 2011, 89, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Briolant, S.; Garin, D.; Scaramozzino, N.; Jouan, A.; Crance, J.M. In vitro inhibition of Chikungunya and Semliki Forest viruses replication by antiviral compounds: Synergistic effect of interferon-alpha and ribavirin combination. Antivir. Res. 2004, 61, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Albulescu, I.C.; van Hoolwerff, M.; Wolters, L.A.; Bottaro, E.; Nastruzzi, C.; Yang, S.C.; Tsay, S.-C.; Hwu, J.R.; Snijder, E.J.; van Hemert, M.J. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antivir. Res. 2015, 121, 39–46. [Google Scholar] [CrossRef]
- Jin, J.; Simmons, G. Antiviral functions of monoclonal antibodies against chikungunya virus. Viruses 2019, 11, 305. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. In Protein dynamics; Humana Press: Totowa, NJ, USA, 2014; Volume 1084, pp. 198–226. ISBN 978162703. [Google Scholar]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinforma. 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Aarthy, M.; Kumar, D.; Giri, R.; Singh, S.K. E7 oncoprotein of human papillomavirus: Structural dynamics and inhibitor screening study. Gene 2018, 658, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2009, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins Struct. Funct. Genet. 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Desdouits, N.; Nilges, M.; Blondel, A. Principal Component Analysis reveals correlation of cavities evolution and functional motions in proteins. J. Mol. Graph. Model. 2015, 55, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Shukla, H.; Tripathi, T. Structural and energetic understanding of novel natural inhibitors of Mycobacterium tuberculosis malate synthase. J. Cell. Biochem. 2018, 120, 2469–2482. [Google Scholar] [CrossRef] [PubMed]
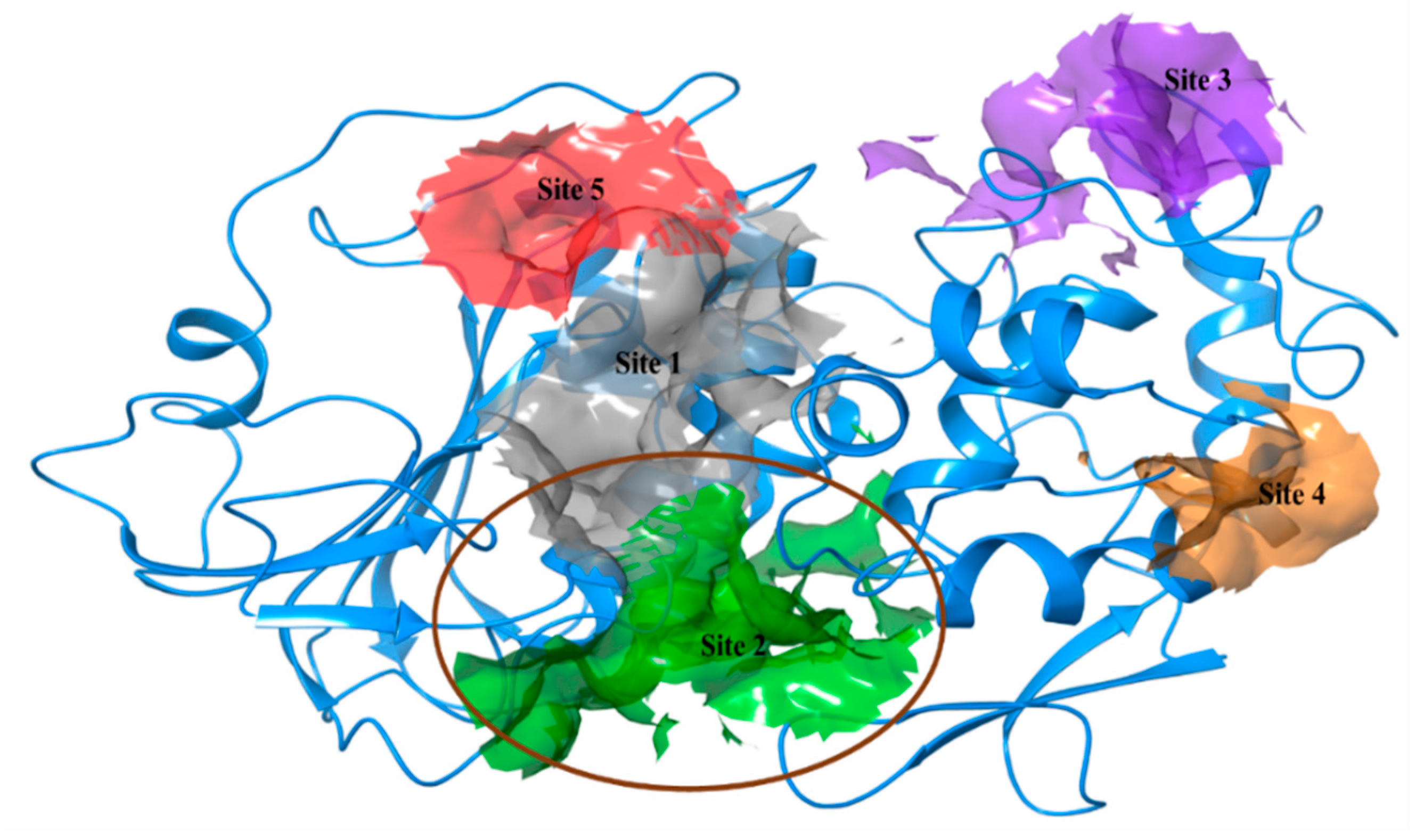
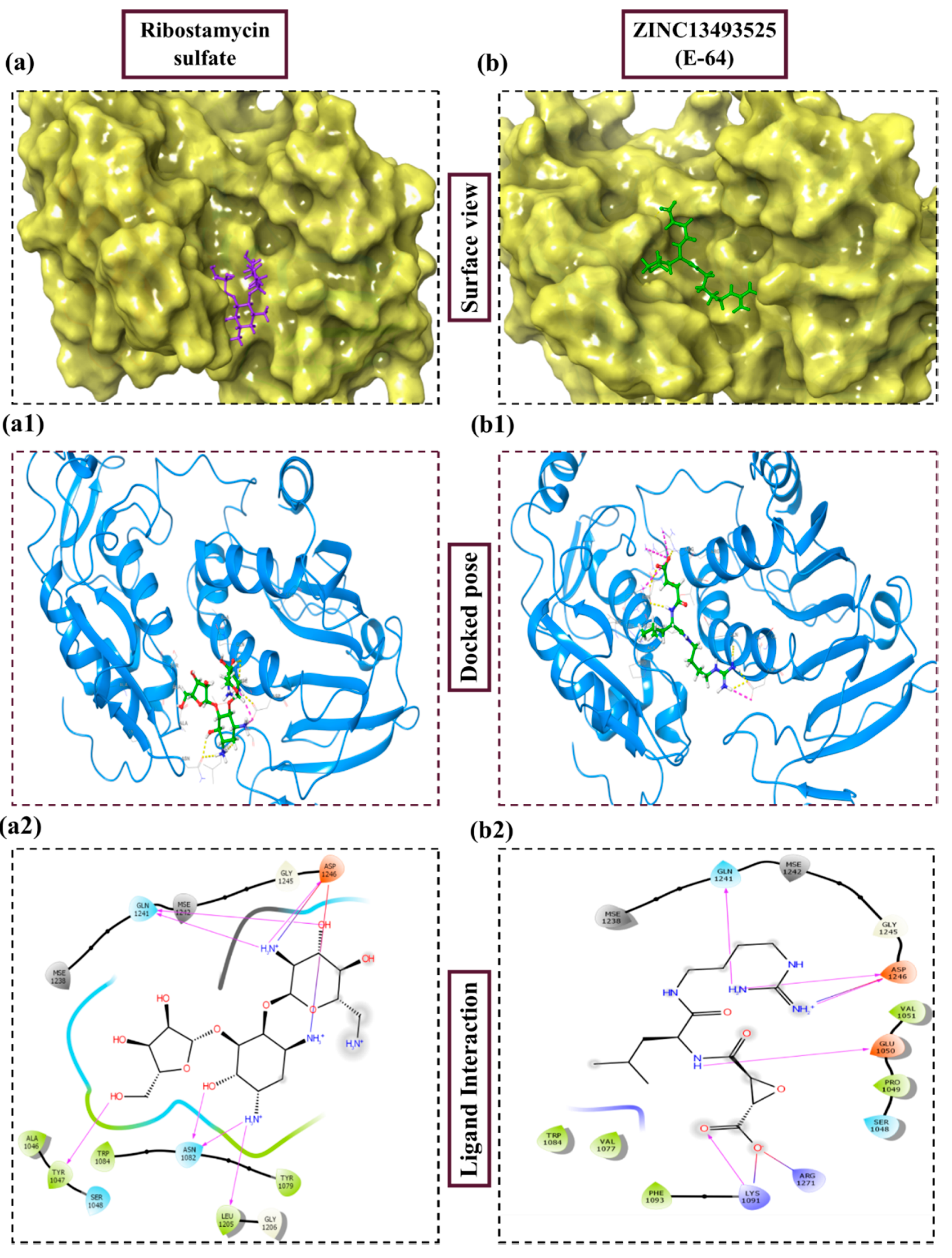
| Predicted Active Sites | Site Score | Dscore | Volume | Phobic | Philic | Residues |
|---|---|---|---|---|---|---|
| Site 1 | 1.007 | 0.998 | 215.747 | 0.475 | 1.127 | I1038, Q1039, A1040, 1042, E1043, K1045, A1046, Y1047, E1055, K1191, MSE1192, N1202, L1203, E1204, I1221, T1223, P1224, R1226, V1234, D1235, MSE1238, K1239, MSE1242, L1243 |
| Site 2 | 0.98 | 1.016 | 311.101 | 0.561 | 0.95 | N1011, C1013, W1014, A1046, Y1047, S1048, E1050, V1051, N1054, L1065, Y1079, N1082, W1084, L1192, Y1201, N1202, E1204, L1205, G1206, L1207, P1208, Q1241, MSE1242, G1245, D1246, L1248, R1267, T1268, R1271, V1272, V1275, L1276 |
| Site 3 | 0.791 | 0.761 | 134.456 | 0.421 | 1.058 | G1062, Q1119, C1121, V1122, T1123, T1124, R1126, I1127, E1128, D1129, N1131, T1133, T1134, N1135, I1136, I1137, P1138, V1139 |
| Site 4 | 0.728 | 0.492 | 38.416 | 0.834 | 1.48 | N1004, F1006, V1019, L1022, E1023, I1027, K1028, L1029, W1034 |
| Site 5 | 0.533 | 0.455 | 58.653 | 0.144 | 0.951 | H1151, H1222, T1223, P1224, F1225, Q1232, Y1262, T1292, S1293, E1296 |
| Sr. No. | Compounds | Structure | Docking Score (kcal/mol) | MMGBSA dG Bind (kcal/mol) | Hydrogen Bond and Other Non-Covalent Interactions |
|---|---|---|---|---|---|
| Docking with FDA drugs | |||||
| 1. | Ribostamycin Sulfate | 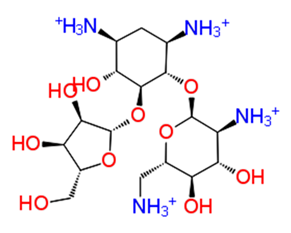 | −12.085 | −30.997 | H-bond: Y1047, N1082 (2), L1205, Q1241 (2), D1246 Salt bridge: D1246 (2) |
| 2. | Kanamycin sulfate | 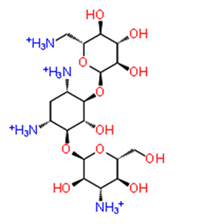 | −10.864 | −24.635 | H-bond: Y1079, N1082 (2), Q1241 (2), D1246 (2) Salt bridge: D1246 (2) |
| 3. | Sennoside A |  | −10.404 | −47.652 | H-bond: G1090, Q1241, D1246 (2), R1271 Salt bridge: K1091 |
| 4. | Amikacin sulfate |  | −10.389 | −34.798 | H-bond: W1084, E1204, G1245 D1246 Salt bridge: D1246 |
| 5. | Dihydrostreptomycin sulfate |  | −10.36 | −34.720 | H-bond: N1082, L1205, D1246 (2) Salt bridge: D1246 (2) |
| 6. | Acarbose |  | −10.2 | −44.927 | H-bond: Y1079, L1205, G1206, Q1241, D1246 (2) |
| 7. | Iron sucrose |  | −10.13 | 3.473 | H-bond: K1045, A1046 (2), L1203, K1239 Salt bridge: K1045, K1239 |
| 8. | Pemetrexed disodium hydrate |  | −10.077 | −33.812 | H-bond: D1246, R1267 Pi-Pi stacking: Y1079, Salt bridge: K1091, R1267 |
| 9. | Ibandronate sodium |  | −10.045 | −37.746 | H-bond: E1050, R1267 (2), R1271 (2) Salt bridge: E1050, K1091 (2), R1267, R1271 |
| 10. | Etidronate |  | −10.014 | −15.141 | H-bond: E1050, K1091 (2), R1271 Salt bridge: K1091, R1267 (2), R1271 (2) |
| Docking with cysteine protease inhibitors | |||||
| 11. | E-64 (ZINC13493525) |  | −8.738 | −31.659 | H-bond: E1050, K1091, Q1241, D1246 (2) Salt bridge: K1091, D1246, R1271 |
| 12. | Leupeptin hemisulfate |  | −7.08 | −44.641 | H-bond: Q1241, D1246, W1084 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, P.; Kumar, D.; Giri, R. Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors. Pathogens 2019, 8, 128. https://doi.org/10.3390/pathogens8030128
Kumar P, Kumar D, Giri R. Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors. Pathogens. 2019; 8(3):128. https://doi.org/10.3390/pathogens8030128
Chicago/Turabian StyleKumar, Prateek, Deepak Kumar, and Rajanish Giri. 2019. "Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors" Pathogens 8, no. 3: 128. https://doi.org/10.3390/pathogens8030128
APA StyleKumar, P., Kumar, D., & Giri, R. (2019). Targeting the nsp2 Cysteine Protease of Chikungunya Virus Using FDA Approved Library and Selected Cysteine Protease Inhibitors. Pathogens, 8(3), 128. https://doi.org/10.3390/pathogens8030128





