Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures
Abstract
1. Introduction
2. Results
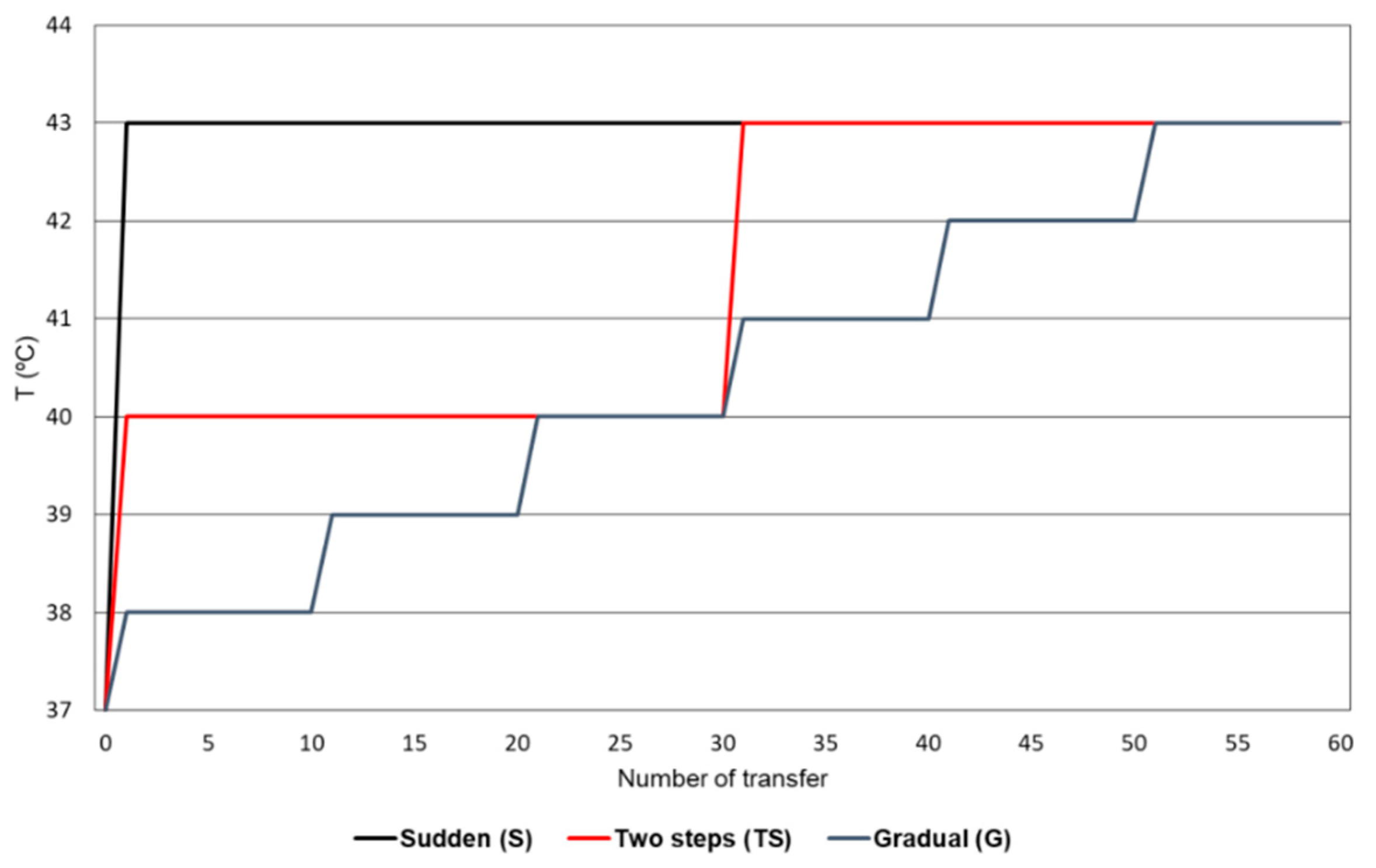
2.1. Evolution of Bacteriophage Qβ Under Different Patterns of Temperature Increase
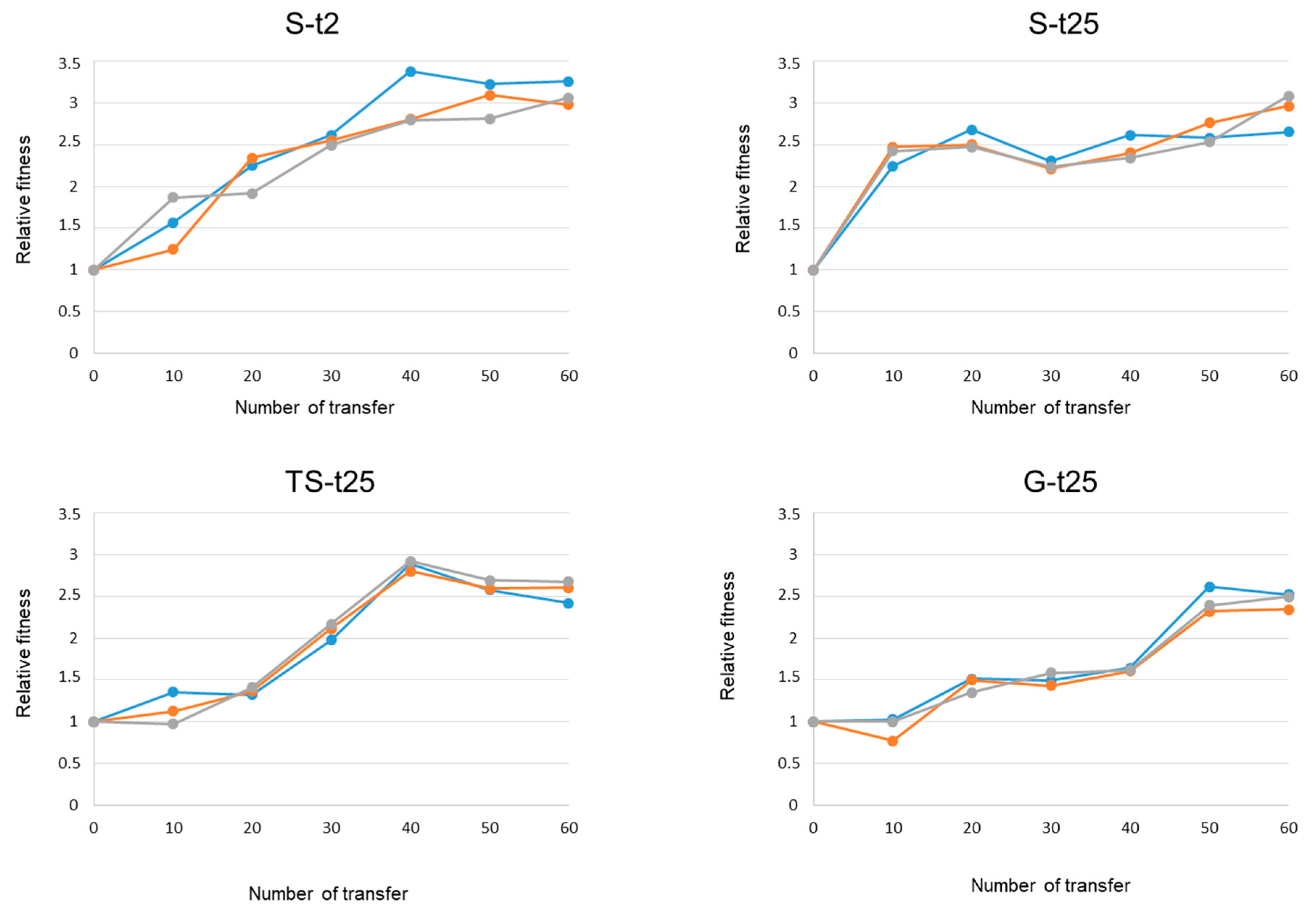
2.2. Dynamics of Adaptation to 43 °C
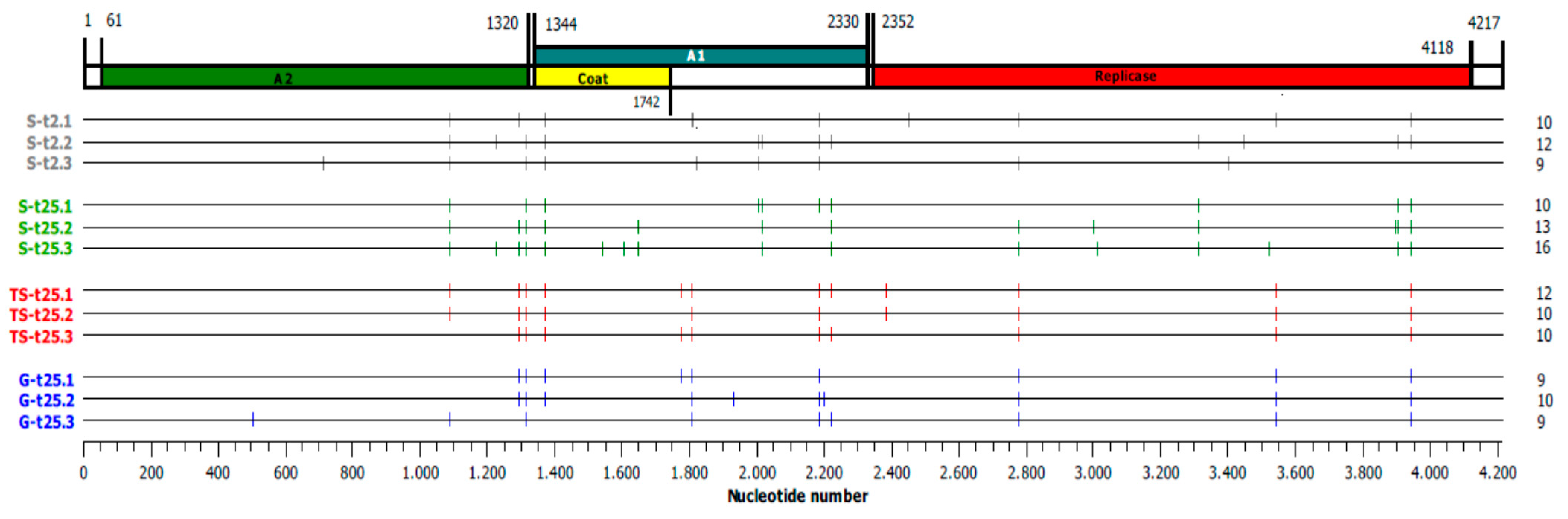
2.3. Analysis of the Consensus Sequences of Final Qβ Populations
2.4. Evolutionary Convergence among Lineages and Treatments
2.5. Evolution of the Consensus Sequences through Adaptation
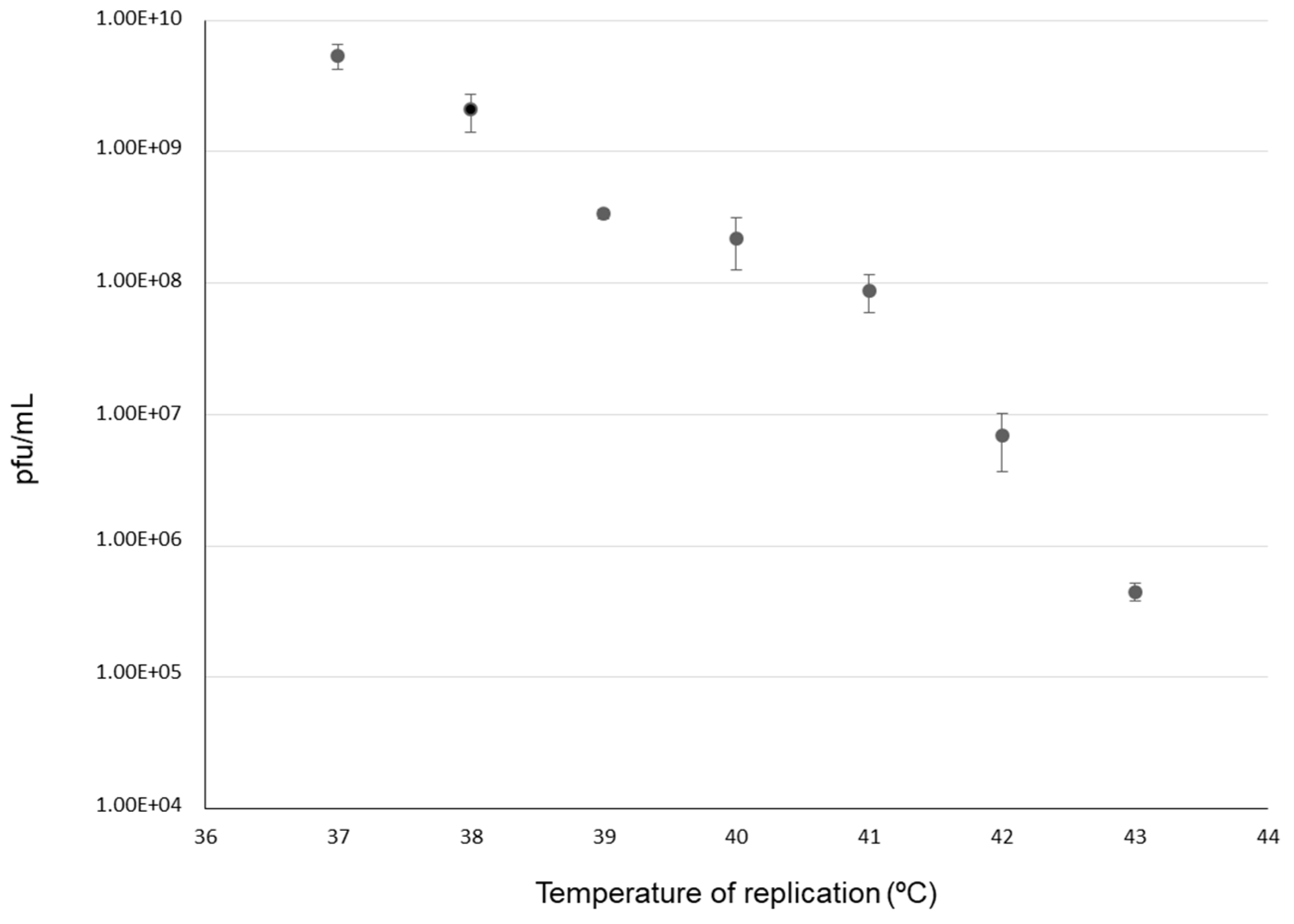
2.6. Fixation of Mutations and Temperature of Evolution
3. Discussion
4. Materials and Methods
4.1. Virus Populations, Bacteria, and Standard Procedures for Infection
4.2. Fitness Determinations
4.3. Determination of the Consensus Sequences of Virus Populations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elena, S.F.; Lenski, R.E. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003, 4, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, T.J.; Lenski, R.E.; Ebert, D.; Hollis, B.; Olivieri, I.; Whitlock, M.C. Experimental evolution. Trends Ecol. Evol. 2012, 27, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.I.; Desai, M.M. The spectrum of adaptive mutations in experimental evolution. Genomics 2014, 104, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Jerison, E.R.; Desai, M.M. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Curr. Opin. Genet. Dev. 2015, 35, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.F.; Bataillon, T. Can the experimental evolution programme help us elucidate the genetic basis of adaptation in nature? Mol. Ecol. 2016, 25, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Sanjuán, R. Virus Evolution: Insights from an Experimental Approach. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 27–52. [Google Scholar] [CrossRef]
- Collins, S. Many Possible Worlds: Expanding the Ecological Scenarios in Experimental Evolution. Evol. Biol. 2011, 38, 3–14. [Google Scholar] [CrossRef]
- Barrick, J.E.; Lenski, R.E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 2013, 14, 827–839. [Google Scholar] [CrossRef]
- Collins, S.; de Meaux, J. Adaptation to different rates of environmental change in chlamydomonas. Evolution 2009, 63, 2952–2965. [Google Scholar] [CrossRef]
- Toprak, E.; Veres, A.; Michel, J.B.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012, 44, 101–105. [Google Scholar] [CrossRef]
- Gonzalez, A.; Bell, G. Evolutionary rescue and adaptation to abrupt environmental change depends upon the history of stress. Philos. Trans. R. Soc. Lond. B 2012, 368, 20120079. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, H.A.; Gallie, J.; Taylor, S.; Kerr, B. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature 2013, 494, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Morley, V.J.; Mendiola, S.Y.; Turner, P.E. Rate of novel host invasion affects adaptability of evolving RNA virus lineages. Proc. R. Soc. B 2015, 282, 20150801. [Google Scholar] [CrossRef] [PubMed]
- Gorter, F.A.; Aarts, M.M.; Zwaan, B.J.; de Visser, J.A.G.M. Dynamics of Adaptation in Experimental Yeast Populations Exposed to Gradual and Abrupt Change in Heavy Metal Concentration. Am. Nat. 2016, 187, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Morley, V.J.; Turner, P.E. Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change. Evolution 2017, 71, 872–883. [Google Scholar] [CrossRef] [PubMed]
- Gorter, F.A.; Derks, M.F.L.; van den Heuvel, J.; Aarts, M.G.M.; Zwaan, B.J.; de Ridder, D.; de Visser, J.A.G.M. Genomics of Adaptation Depends on the Rate of Environmental Change in Experimental Yeast Populations. Mol. Biol. Evol. 2017, 34, 2613–2626. [Google Scholar] [CrossRef] [PubMed]
- Bello, Y.; Waxman, D. Near-periodic substitution and the genetic variance induced by environmental change. J. Theor. Biol. 2006, 239, 152–160. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kopp, M.; Hermisson, J. Adaptation of a Quantitative Trait to a Moving Optimum. Genetics 2007, 176, 715–719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kopp, M.; Hermisson, J. The Genetic Basis of Phenotypic Adaptation II: The Distribution of Adaptive Substitutions in the Moving Optimum Model. Genetics 2009, 183, 1453–1476. [Google Scholar] [CrossRef]
- Kopp, M.; Hermisson, J. The Genetic Basis of Phenotypic Adaptation I: Fixation of Beneficial Mutations in the Moving Optimum Model. Genetics 2009, 182, 233–249. [Google Scholar] [CrossRef][Green Version]
- Rokyta, D.R.; Joyce, P.; Caudle, S.B.; Wichman, H.A. An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat. Genet. 2005, 37, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Eyre-Walker, A.; Keightley, P.D. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007, 8, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Bons, E.; Bertels, F.; Regoes, R.R. Estimating the mutational fitness effects distribution during early HIV infection. Virus Evol. 2018, 4, vey029. [Google Scholar] [CrossRef] [PubMed]
- Bürger, R.; Lynch, M. Evolution and extinction in a changing environment: A quantitative-genetic analysis. Evolution 1995, 49, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.; Collins, S. Adaptation, extinction and global change: Adaption, extinction and global change. Evol. Appl. 2008, 1, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Gomulkiewicz, R.; Houle, D. Demographic and Genetic Constraints on Evolution. Am. Nat. 2009, 174, E218–E229. [Google Scholar] [CrossRef] [PubMed]
- Kopp, M.; Matuszewski, S. Rapid evolution of quantitative traits: Theoretical perspectives. Evol. Appl. 2014, 7, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Yubero, P.; Manrubia, S.; Aguirre, J. The space of genotypes is a network of networks: Implications for evolutionary and extinction dynamics. Sci. Rep. 2017, 7, 13813. [Google Scholar] [CrossRef] [PubMed]
- Schenk, M.F.; Szendro, I.G.; Salverda, M.L.M.; Krug, J.; de Visser, J.A.G.M. Patterns of Epistasis between Beneficial Mutations in an Antibiotic Resistance Gene. Mol. Biol. Evol. 2013, 30, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Schoustra, S.; Hwang, S.; Krug, J.; de Visser, J.A.G.M. Diminishing-returns epistasis among random beneficial mutations in a multicellular fungus. Proc. R. Soc. B 2016, 283, 20161376. [Google Scholar] [CrossRef] [PubMed]
- Perron, G.G.; Gonzalez, A.; Buckling, A. The rate of environmental change drives adaptation to an antibiotic sink. J. Evol. Biol. 2008, 21, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.; Gonzalez, A. Adaptation and Evolutionary Rescue in Metapopulations Experiencing Environmental Deterioration. Science 2011, 332, 1327–1330. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.-Q.; Brockhurst, M.A.; Petchey, O.L.; Zhang, Q.-G. Evolutionary rescue can be impeded by temporary environmental amelioration. Ecol. Lett. 2015, 18, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, A.; Sugawara, R.; Sano Tsushima, F.; Kumagai, T.; Yomo, T. Contribution of Silent Mutations to Thermal Adaptation of RNA Bacteriophage Qβ. J. Virol. 2014, 88, 11459–11468. [Google Scholar] [CrossRef] [PubMed]
- Baym, M.; Lieberman, T.D.; Kelsic, E.D.; Chait, R.; Gross, R.; Yelin, I.; Kishony, R. Spatiotemporal microbial evolution on antibiotic landscapes. Science 2016, 353, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.; Leon Guerrero, C.M.; Whang, S.G.; McClure, E.M.; Busch, H.G.; Kerr, B. Adaptations of an RNA virus to increasing thermal stress. PLoS ONE 2017, 12, e0189602. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.; van Duin, J. Bacteriophages with ssRNA. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Bradwell, K.; Combe, M.; Domingo-Calap, P.; Sanjuán, R. Correlation Between Mutation Rate and Genome Size in Riboviruses: Mutation Rate of Bacteriophage Qβ. Genetics 2013, 195, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Moya, A.; Holmes, E.C.; González-Candelas, F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004, 2, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, E. RNA Viruses: Control, Mutagenesis and Extinction. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2011; p. a0023276. ISBN 978-0-470-01617-6. [Google Scholar]
- Domingo, E.; Sheldon, J.; Perales, C. Viral Quasispecies Evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed]
- Guzella, T.S.; Dey, S.; Chelo, I.M.; Pino-Querido, A.; Pereira, V.F.; Proulx, S.R.; Teotónio, H. Slower environmental change hinders adaptation from standing genetic variation. PLoS Genet. 2018, 14, e1007731. [Google Scholar] [CrossRef]
- Arribas, M.; Kubota, K.; Cabanillas, L.; Lázaro, E. Adaptation to Fluctuating Temperatures in an RNA Virus Is Driven by the Most Stringent Selective Pressure. PLoS ONE 2014, 9, e100940. [Google Scholar] [CrossRef] [PubMed]
- Arribas, M.; Cabanillas, L.; Kubota, K.; Lázaro, E. Impact of increased mutagenesis on adaptation to high temperature in bacteriophage Qβ. Virology 2016, 497, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, E.; Arribas, M.; Cabanillas, L.; Román, I.; Acosta, E. Evolutionary adaptation of an RNA bacteriophage to the simultaneous increase in the within-host and extracellular temperatures. Sci. Rep. 2018, 8, 8080. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Palmieri, M.; Weissmann, C. Qβ DNA-containing hybrid plasmids giving rise to Qβ phage formation in the bacterial host. Nature 1978, 274, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Barrera, I.; Schuppli, D.; Sogo, J.M.; Weber, H. Different Mechanisms of Recognition of Bacteriophage Qβ Plus and Minus Strand RNAs by Qβ Replicase. J. Mol. Biol. 1993, 232, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Inomata, T.; Kimura, H.; Hayasaka, H.; Shiozaki, A.; Fujita, Y.; Kashiwagi, A. Quantitative comparison of the RNA bacteriophage Qβ infection cycle in rich and minimal media. Arch. Virol. 2012, 157, 2163–2169. [Google Scholar] [CrossRef] [PubMed]
- Callanan, J.; Stockdale, S.; Shkoporov, A.; Draper, L.; Ross, R.; Hill, C. RNA Phage Biology in a Metagenomic Era. Viruses 2018, 10, 386. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Watson, R.A.; Chao, L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution 2005, 59, 1165–1174. [Google Scholar] [CrossRef]
- Parera, M.; Martinez, M.A. Strong Epistatic Interactions within a Single Protein. Mol. Biol. Evol. 2014, 31, 1546–1553. [Google Scholar] [CrossRef] [PubMed]
- Lyons, D.; Lauring, A. Mutation and Epistasis in Influenza Virus Evolution. Viruses 2018, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Bernet, G.P.; Elena, S.F. Distribution of mutational fitness effects and of epistasis in the 5′ untranslated region of a plant RNA virus. BMC Evol. Biol. 2015, 15, 274. [Google Scholar] [CrossRef] [PubMed]
- Wiser, M.J.; Ribeck, N.; Lenski, R.E. Long-Term Dynamics of Adaptation in Asexual Populations. Science 2013, 342, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- Perfeito, L.; Sousa, A.; Bataillon, T.; Gordo, I. Rates of fitness decline and rebound suggest pervasive epistasis: Pervasive epistasis in fitness decline and rebound. Evolution 2014, 68, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Kryazhimskiy, S.; Rice, D.P.; Jerison, E.R.; Desai, M.M. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 2014, 344, 1519–1522. [Google Scholar] [CrossRef] [PubMed]
- Couce, A.; Tenaillon, O.A. The rule of declining adaptability in microbial evolution experiments. Front. Genet. 2015, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-H.; Chiu, H.-C.; Delaney, N.F.; Segre, D.; Marx, C.J. Diminishing Returns Epistasis Among Beneficial Mutations Decelerates Adaptation. Science 2011, 332, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.I.; Dinh, D.M.; Schneider, D.; Lenski, R.E.; Cooper, T.F. Negative Epistasis Between Beneficial Mutations in an Evolving Bacterial Population. Science 2011, 332, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, A.; Kadoya, T.; Kumasaka, N.; Kumagai, T.; Tsushima, F.S.; Yomo, T. Influence of adaptive mutations, from thermal adaptation experiments, on the infection cycle of RNA bacteriophage Qβ. Arch. Virol. 2018, 163, 2655–2662. [Google Scholar] [CrossRef] [PubMed]
- Rumnieks, J.; Tars, K. Crystal Structure of the Maturation Protein from Bacteriophage Qβ. J. Mol. Biol. 2017, 429, 688–696. [Google Scholar] [CrossRef] [PubMed]
- García-Villada, L.; Drake, J.W. Experimental selection reveals a trade-off between fecundity and lifespan in the coliphage Qß. Open Biol. 2013, 3, 130043. [Google Scholar] [CrossRef]
- Rumnieks, J.; Tars, K. Crystal structure of the read-through domain from bacteriophage Qβ A1 protein. Protein Sci. 2011, 20, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Rumnieks, J.; Tars, K. Crystal structure of the bacteriophage Qβ coat protein in complex with the RNA operator of the replicase gene. J. Mol. Biol. 2014, 426, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, D.; Tomita, K. Assembly of Qβ viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. USA 2010, 107, 15733–15738. [Google Scholar] [CrossRef] [PubMed]
- Kidmose, R.T.; Vasiliev, N.N.; Chetverin, A.B.; Andersen, G.R.; Knudsen, C.R. Structure of the Qβ replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 10884–10889. [Google Scholar] [CrossRef] [PubMed]
- Arribas, M.; Cabanillas, L.; Lázaro, E. Identification of mutations conferring 5-azacytidine resistance in bacteriophage Qβ. Virology 2011, 417, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, L.; Arribas, M.; Lázaro, E. Evolution at increased error rate leads to the coexistence of multiple adaptive pathways in an RNA virus. BMC Evol. Biol. 2013, 13, 11. [Google Scholar] [CrossRef] [PubMed]

| Evolutionary Lineage | Transfer Number (Relative Fitness = 2) 1 | Relative Fitness at Transfer 60 | Fitness Difference (First 10 Transfers at 43 °C) |
|---|---|---|---|
| S-t2.1 | 16.3 | 3.3 | 0.6 |
| S-t2.2 | 16.9 | 3.0 | 0.3 |
| S-t2.3 | 21.5 | 3.1 | 0.9 |
| S-t2 (Mean ± SD) | 18.2 ± 2.8 | 3.1 ± 0.1 | 0.6 ± 0.3 |
| S-t25.1 | 8.1 | 2.7 | 1.2 |
| S-t25.2 | 6.8 | 3.0 | 1.5 |
| S-t25.3 | 7.0 | 3.1 | 1.4 |
| S-t25 (Mean ± SD) | 7.3 ± 0.7 | 2.9 ± 0.2 | 1.4 ± 0.1 |
| TS-t25.1 | 30.3 | 2.4 | 0.9 |
| TS-t25.2 | 28.5 | 2.6 | 0.7 |
| TS-t25.3 | 27.8 | 2.7 | 0.8 |
| TS-t25 (Mean ± SD) | 28.9 ± 1.3 | 2.6 ± 0.1 | 0.8 ± 0.1 |
| G-t25.1 | 43.7 | 2.5 | -0.1 |
| G-t25.2 | 45.5 | 2.3 | 0.02 |
| G-t25.3 | 45.0 | 2.5 | 0.1 |
| G-t25 (Mean ± SD) | 44.7 ± 0.9 | 2.5 ± 0.1 | 0.01 ± 0.1 |
| Mutation 1 | Gene 2 | Number of Lineages with the Mutation Per Treatment 3 | |||
|---|---|---|---|---|---|
| S-t2 | S-t25 | TS-t25 | G-t25 | ||
| G502C (G147R) | A2 | 0/1 | |||
| C714U (S) | A2 | 0/1 | |||
| A1088G (D342G) | A2 | 2/1 | 3/0 | 0/2 | 0/1 |
| C1228U (L389F) | A2 | 0/1 | 1/0 | ||
| U1295(C/G) (F411S/C) | A2 | 1/0 | 2/0 | 1/2 | 1/1 |
| G1312A (V417I) | A2 | 2/0 | 3/0 | 3/0 | 3/0 |
| G1371A (G9S) | C | 3/0 | 3/0 | 0/3 | 0/2 |
| U1544C (S) | C | 1/0 | |||
| G1607U (Q87H) | C | 1/0 | |||
| C1649U (S) | C | 2/0 | |||
| A1778G (S) | A1 | 1/1 | 1/0 | ||
| C1806U (P154S) | A1 | 3/0 | 2/1 | ||
| C1807U (P154L) | A1 | 1/0 | |||
| U1810C (I155T) | A1 | 1/0 | |||
| G1820A (S) | A1 | 1/0 | |||
| A1930G (Q195R) | A1 | 0/1 | |||
| U2006G (S220R) | A1 | 0/2 | 0/1 | ||
| U2016C (F224L) | A1 | 1/0 | 2/1 | ||
| A2187C (S281R) | A1 | 0/3 | 0/1 | 3/0 | 2/1 |
| C2201U (S) | A1 | 1/0 | |||
| A2222C (S) | A1 | 1/0 | 3/0 | ||
| G2223A (V293I) | A1 | 1/1 | 0/1 | ||
| C2384U (S) | R | 0/2 | |||
| C2452U (A33V) | R | 1/0 | |||
| U2776C (V141A) | R | 2/0 | 1/1 | 3/0 | 3/0 |
| G3002A (S) | R | 1/0 | |||
| U3011C (S) | R | 1/0 | |||
| U3311G (I320M) | R | 1/0 | 3/0 | ||
| U3402C (S350P) | R | 1/0 | |||
| U3447C (C365R) | R | 0/1 | |||
| G3521U (E389D) | R | 1/0 | |||
| C3545U (S) | R | 0/1 | 3/0 | 2/1 | |
| G3896A (S) | R | 0/1 | |||
| C3903U (L517F) | R | 0/1 | 2/1 | ||
| G3945A (G531S) | R | 2/0 | 3/0 | 3/0 | 1/2 |
| Lineage | First Fixed Mutations | Number of Transfer |
|---|---|---|
| S-t2.1 | A1088G, G1371A, C2452U, U2776C | 30 |
| S-t2.2 | A1088G, G1371A, A2222C, U3311G | 20 |
| S-t2.3 | G1312A, U3402C | 10 |
| S-t25.1 | A1088G, C1649U, U2016C, A2222C, U2460C, U3311G, G3945A | 10 |
| S-t25.2 | A1088G, A2222C, U3311G | 10 |
| S-t25.3 | A1088G, C1649U, A2222C, U3311G | 10 |
| TS-t25.1 | G1312A | 20 |
| TS-t25.2 | G1312A, A1930G | 20 |
| TS-t25.3 | G1312A | 30 |
| G-t25.1 | G1312A | 20 |
| G-t25.2 | A1930G | 20 |
| G-t25.3 | G1312A | 20 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somovilla, P.; Manrubia, S.; Lázaro, E. Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures. Pathogens 2019, 8, 80. https://doi.org/10.3390/pathogens8020080
Somovilla P, Manrubia S, Lázaro E. Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures. Pathogens. 2019; 8(2):80. https://doi.org/10.3390/pathogens8020080
Chicago/Turabian StyleSomovilla, Pilar, Susanna Manrubia, and Ester Lázaro. 2019. "Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures" Pathogens 8, no. 2: 80. https://doi.org/10.3390/pathogens8020080
APA StyleSomovilla, P., Manrubia, S., & Lázaro, E. (2019). Evolutionary Dynamics in the RNA Bacteriophage Qβ Depends on the Pattern of Change in Selective Pressures. Pathogens, 8(2), 80. https://doi.org/10.3390/pathogens8020080





