Fungal-Bacterial Interactions in Health and Disease
Abstract
1. Introduction
1.1. Origins of Microbiota Research
1.2. Blooming Awareness of Cross-Kingdom Microbial Interactions
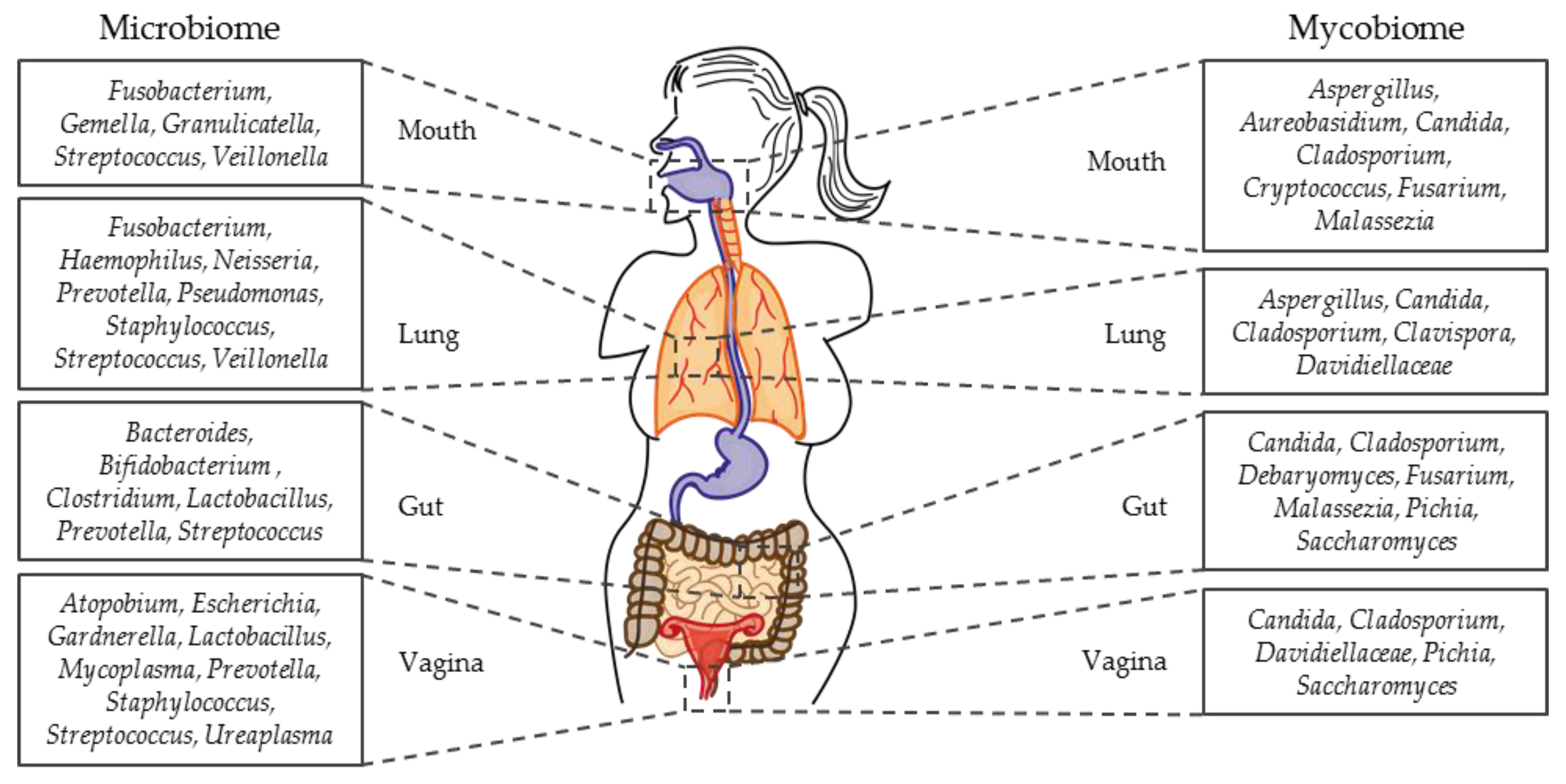
1.3. Micro- and Mycobiome Studies
1.4. Polymicrobial Interactions
1.5. Scope of This Review
| Mechanism | Fungi | Bacteria | Relationship | Study Setting | References |
|---|---|---|---|---|---|
| Physical Interaction | Aspergillus spp. | K. pneumoniae | antagonism | In vitro co-culture → prevention of spore germination and filamentation | [55] |
| A. fumigatus | P. aeruginosa | antagonism | In vitro co-culture → decreased filamentation, biofilm formation, and conidia biomass | [56] | |
| C. albicans | A. baumannii | antagonism | In vitro co-culture → induced fungal apoptosis | [57] | |
| F. nucleatum | antagonism | In vitro co-culture → inhibited growth and filamentation | [58] | ||
| Group B Streptococcus | synergism | In vitro: vaginal epithelial cells → enhanced fungal and bacterial adhesion | [59] | ||
| P. aeruginosa | antagonism | In vitro co-cultures → killing of filamentous fungus | [60,61] | ||
| S. aureus | synergism | Ex vivo mouse tongue infection [62]; in vivo oral mouse co-infection [63]; in vivo oral mouse infection [64] → promoted bacterial invasion | [62,63,64] | ||
| S. epidermidis | non-competitive | In vitro adhesion model → bacteria bind to fungal germtubes | [65] | ||
| S. gordonii | non-competitive | In vitro co-aggregation assays → C. albicans adhesin binds bacterial cell wall proteins | [66,67,68] | ||
| Chemical Interaction and Release of Metabolic Byproducts | A. fumigatus | A. baumannii | antagonism | Gliotoxin treated bacterial biofilm → decreased bacterial biomass | [69] |
| P. aeruginosa | antagonism | In vitro co-culture → inhibited fungal biofilm formation [56]; Gliotoxin treated bacterial biofilm → decreased bacterial biomass [69]; In vitro assay → inhibited fungal growth [70] | [56,69,70] | ||
| S. aureus | antagonism | Gliotoxin-treated bacterial biofilm → decreased bacterial biomass | [69] | ||
| C. albicans | A. actinomycetemcomitans | antagonism | In vitro co-culture → AI-2 inhibits fungal biofilm formation | [71] | |
| C. difficile | antagonism | In vitro assay → p-cresol involved in filamentation | [72] | ||
| E. coli | antagonism | In vitro biofilm assay → inhibited fungal biofilm formation [73]; In vitro assay → soluble factor kills C. albicans [74] | [73,74] | ||
| E. faecalis | antagonism | In vitro biofilm model, in vivo nematode model, in vivo murine candidiasis model [75]; In vivo nematode model, in vitro biofiolm model [76] → inhibition of filamentation and fungal virulence | [75,76] | ||
| Lactobacillus spp. | antagonism | In vitro: HeLa cells → reduced fungal adhesion [77]; In vitro C. albicans growth → stimulation of pseudohyphae and repression of growth [78]; In vitro model: vaginal epithelial cells → bactericidal mode against C. albicans [79]; In vitro co-culture → inhibition of filamentation [80] | [77,78,79,80] | ||
| P. aeruginosa | antagonism | In vitro assay → inhibition of fungal growth [70]; In vitro co-culture → decreased bacterial virulence [81]; In vitro co-culture → reduces fungal viability [82] | [70,81,82] | ||
| S. aureus | synergism | In vitro assay → enhanced tolerance to antimicrobial compounds | [83] | ||
| S. gordonii | synergism | In vitro assay → enhanced filamentation | [68] | ||
| S. mutans | synergism antagonism | In vitro assay → enhanced bacterial growth [84]; In vitro co-culture → inhibited filamentation [85,86] | [84] [85,86] | ||
| S. enterica serovar Typhimurium | antagonism | In vivo nematode model, in vitro co-culture → repressed filamentation | [87] | ||
| C. neoformans | K. aerogenes | synergism | In vitro co-culture → promoted fungal melanization | [88] | |
| S. cerevisiae | Acinetobacter spp. | synergism | In vitro co-culture, in vivo nematode model → enhanced bacterial growth and increased pathogenicity | [89] | |
| Influencing the Environment | C. albicans | B. fragilis | synergism | In vitro assay → protection of bacteria by fungal biofilm | [90] |
| C. difficile | synergism | In vitro co-culture → anaerobic growth of C. difficile | [72] | ||
| C. perfringens | synergism | In vitro assay → protection by fungal biofilm | [90] | ||
| Competition | C. albicans | Lactobacillus spp. | antagonism | In vitro model: vaginal epithelial cells → reduced bacterial adherence | [79,91,92,93] |
| S. mitis | antagonism | In vitro co-culture in a chemostat → competition for glucose | [94] | ||
| S. sobrinus | antagonism | In vitro co-culture in a chemostat → competition for glucose | [94] | ||
| Biofilm Formation | C. albicans | A. actinomycetemcomitans | antagonism | In vitro Bioflux assay → decreased fungal biofilm formation | [71] |
| C. freundii | non-competitive | In vitro co-culture → ability to form mixed biofilms | [95] | ||
| C. perfringens | synergism | In vitro assay → protection by fungal biofilm | [90] | ||
| E. coli | synergism | In vitro assay → increased mixed biofilm formation | [96] | ||
| E. faecalis | synergism | In vitro assay → increased mixed biofilm formation | [97] | ||
| K. pneumoniae | antagonism | In vitro assay → decreased fungal biofilm formation | [90,98] | ||
| P. aeruginosa | antagonism | In vitro assay → decreased fungal biofilm formation | [60,99] | ||
| P. gingivalis | synergism | In vitro assay → protection by fungal biofilm | [100] | ||
| S. aureus | synergism | In vitro assay → increased mixed biofilm formation | [83,101,102,103] | ||
| S. epidermidis | synergism | In vitro co-culture → increased mixed biofilm formation | [104,105] | ||
| Streptococcus spp. | synergism | In vitro model: oral epithelial cells [106]; In vivo oral mouse model [107]; In vitro assay, in vivo oral rat model [108,109] → increased mixed biofilm formation | [106,107,108,109] | ||
| C. tropicalis | E. coli | synergism | In vitro assay → increased mixed biofilm formation | [20] | |
| S. marcescens | synergism | In vitro assay → increased mixed biofilm formation | [20] | ||
| T. asahii | S. simulans | non-competitive | In vitro co-culture → ability to form mixed biofilms | [95] |
2. Fungal-Bacterial Interactions—Niche by Niche
2.1. Oral Cavity
2.1.1. Niche Landscape of the Human Mouth
2.1.2. Oral Micro- and Mycobiota in Health
2.1.3. Oral Dysbiosis
2.1.4. C. albicans and Bacteria in the Human Mouth
2.2. Vagina
2.2.1. Niche Landscape of the Human Vagina
2.2.2. Vaginal Micro- and Mycobiota in Health
2.2.3. C. albicans and Lactobacilli in the Human Vagina
2.2.4. C. albicans and Streptococci in the Human Vagina
2.3. Gut
2.3.1. Niche Landscape of the Human Gut
2.3.2. Gut Micro- and Mycobiota in Health and Disease
2.3.3. Candida spp. and Bacteria in the Human Gut
2.3.4. Saccharomyces spp. and Bacteria in the Human Gut
2.3.5. Yeasts and Clostridia spp. in the Human Gut
2.3.6. C. albicans and E. faecalis in the Human Gut
2.4. Lung
2.4.1. Niche Landscape of the Human Lung
2.4.2. Lung Micro- and Mycobiota in Health and Disease
2.4.3. Fungi and P. aeruginosa in the Human Lung
2.4.4. Fungi and Klebsiella spp. in the Human Lung
2.4.5. Mucorales and Bacteria in the Human Lung
2.5. Wound, Medical Device-Associated, and Systemic Infections
2.5.1. Niche Landscapes of Skin, Wound, and Bloodstream
2.5.2. Micro- and Mycobiota of Wounds
2.5.3. C. albicans and Staphylococci in Mixed Biofilms
2.5.4. C. albicans and E. coli in Mixed Biofilms
2.5.5. C. albicans and Staphylococci during Tissue Invasion and Systemic Infections
2.5.6. Mixed Bloodstream Infections in Patients
2.5.7. Mixed Systemic and Bloodstream Infections in Mouse Models
3. Conclusions
- Aspergilli are saprophytic molds, growing primarily on rotting biological material from where they spread via air as small and light spores. Unlike Candida spp., Aspergillus spp. are no natural colonizers of the human body. After inhalation, conidia can cause allergic reactions or severe diseases, like chronic pulmonary infections in patients with an impaired immune system. Major infective agents are A. fumigatus and A. nidulans. During infection, conidia swell, form germlings, and eventually long filaments [98,340,341].
- Candida albicans is an opportunistic fungal pathogen that causes disease mostly in immunocompromised patients [342,343]. In healthy individuals, its major reservoir is the gut, but this yeast can be found in many niches of the human body, for instance throughout the entire GI tract [344,345]. The most relevant virulence trait of C. albicans is the ability to switch from yeast to hypha form and thereby either proliferate or adhere, penetrate tissues, and disseminate [346].
- Enterococci are opportunistic Gram-positive lactic acid producing bacteria commonly found as members of the microbiota of mammals and in a wide range of environmental niches. This wide distribution is due to their high tolerance against pH extremes, elevated temperatures, as well as salt concentrations. In humans, E. faecalis and E. faecium cause nosocomial infections like UTIs, bacteremia, and endocarditis [347].
- Escherichia coli is a Gram-negative bacterium that is the most prominent cause of infections in humans. As a common colonizer of the gut, it is also a widely-used indicator of fecal contaminations in food or water. E. coli shows a unique pathovariety from probiotic to life-threatening [329]. For instance, enterotoxigenic E. coli (ETEC), which cause moderate-to-severe diarrhea and can lead to malnutrition and death in young children, is one of the main health problems in developing countries [348].
- Klebsiella pneumoniae is a Gram-negative bacterium that can be found in the lung and throughout the human GI tract. Mostly harmless, it can cause a variety of infections. Especially nosocomially-acquired pneumonia can be problematic for immunocompromised patients in the hospital setting. K. pneumoniae possesses a thick capsule that protects it from the host immune system and other external threats and leads to characteristic slimy colonies.
- Lactobacillus spp. are Gram-positive, facultatively anaerobic bacteria that are also part of the gastrointestinal and vaginal flora [349]. They belong to a group of lactic acid producing bacteria (LAB) which ferment carbohydrates and produce lactic acid. Other genera of this group are, for example, Streptococcus, Lactococcus, and Enterococcus [349,350].
- Mucorales are ubiquitous molds that are able to cause infections opportunistically in immunocompromised and diabetic individuals, called mucormycosis [288,289,290]. The infections, so-called mucormycoses, are typically acquired via spores and filaments are formed within the infected organs. Mucormycosis often originates in the respiratory tract, but commonly disseminates into other organ systems [288].
- Pseudomonas aeruginosa is a Gram-negative bacterium that is well-adapted to many different niches. It survives in the environment but is also able to cause severe infections in humans. Especially in CF-patients, P. aeruginosa is feared for its ability to cause persistent lung infections. The ability of P. aeruginosa to form biofilms is of special importance in its virulence [351,352].
- Serratia marcescens is a Gram-negative opportunistic pathogen that forms characteristic red colonies when grown on culture media. It causes mainly UTIs, but also wound and bloodstream infections, mostly in neonates. It tolerates temperatures at 5–40 °C, is found ubiquitously in the environment, and is among the top ten isolated pathogens from hospitals all over the world [353,354].
- Staphylococci are Gram-positive bacteria. The two most relevant species in human disease are the coagulase-negative S. epidermidis and the coagulase-positive S. aureus. As commensals, S. epidermidis is a frequent colonizer of human skin whereas S. aureus can be found in the nasal cavities of roughly 30% of the human population. Staphylococci can cause a wide range of infections, from superficial to systemic and life-threatening [355,356].
- Streptococci are Gram-positive bacteria that include a wide range of opportunistic pathogens with various virulence factors. S. pyogenes (Group A Streptococci) is the causative agent of diseases such as scarlet fever, impetigo, and necrotizing fasciitis. S. pneumoniae (pneumococci) causes infections of the respiratory tract. Oral streptococci (group viridans) can cause local infections of oral mucosa and caries. Especially malignant are long-term complications such as endocarditis due to crossreactive antibodies produced during acute infection [357,358,359].
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
Glossary
| Polymicrobial | Referring to more than one microbe; can be poly-fungal, poly-bacterial, or fungal-bacterial |
| Monoinfections | Infections with one microbe; either fungal or bacterial |
| Co-infections | Infections with more than one microbe; can be poly-fungal, poly-bacterial, or fungal-bacterial |
| Mixed biofilms or infections | Biofilms or infections with at least one bacterium and one fungus |
| Microbiota/microbiome | Entity of bacteria/bacterial genes in a certain niche or sample |
| Mycobiota/mycobiome | Entity of fungi/fungal genes in a certain niche or sample |
| Microbes/microbial: | In general, referring to bacteria and fungi |
References
- Albert, M.J.; Mathan, V.I.; Baker, S.J. Vitamin B12 synthesis by human small intestinal bacteria. Nature 1980, 283, 781–782. [Google Scholar] [CrossRef]
- Bruce, A.W.; Chadwick, P.; Hassan, A.; VanCott, G.F. Recurrent urethritis in women. Can. Med. Assoc. J. 1973, 108, 973–976. [Google Scholar] [PubMed]
- Enaud, R.; Vandenborght, L.-E.; Coron, N.; Bazin, T.; Prevel, R.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Lamireau, T.; Delhaes, L. The mycobiome: A neglected component in the microbiota-gut-brain axis. Microorganisms 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207. [Google Scholar]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Hermann, C.; Hermann, J.; Munzel, U.; Ruchel, R. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 1999, 42, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yoon, Y.K.; Kim, M.J.; Sohn, J.W. Risk factors for and clinical implications of mixed Candida/bacterial bloodstream infections. Clin. Microbiol. Infect. 2013, 19, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Chasin, B.S.; Powell, B.; Gaur, N.K.; Lipke, P.N. Polymicrobial bloodstream infections involving Candida species: Analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 2007, 59, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Reno, J.; Doshi, S.; Tunali, A.K.; Stein, B.; Farley, M.M.; Ray, S.M.; Jacob, J.T. Epidemiology of methicillin-resistant Staphylococcus aureus bloodstream coinfection among adults with candidemia in Atlanta (GA) 2008-2012. Infect. Control Hosp. Epidemiol. 2015, 36, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Kett, D.H.; Azoulay, E.; Echeverria, P.M.; Vincent, J.L. Candida bloodstream infections in intensive care units: Analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 2011, 39, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Chotirmall, S.H.; Greene, C.M.; McElvaney, N.G. Candida species in cystic fibrosis: A road less travelled. Med. Mycol. J. 2010, 48, 114–124. [Google Scholar] [CrossRef][Green Version]
- Amin, R.; Dupuis, A.; Aaron, S.D.; Ratjen, F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 2010, 137, 171–176. [Google Scholar] [CrossRef]
- Drell, T.; Lillsaar, T.; Tummeleht, L.; Simm, J.; Aaspõllu, A.; Väin, E.; Saarma, I.; Salumets, A.; Donders, G.G.; Metsis, M. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS ONE 2013, 8, e54379. [Google Scholar] [CrossRef] [PubMed]
- Charlson, E.S.; Diamond, J.M.; Bittinger, K.; Fitzgerald, A.S.; Yadav, A.; Haas, A.R.; Bushman, F.D.; Collman, R.G. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012, 186, 536–545. [Google Scholar] [CrossRef]
- Botterel, F.; Angebault, C.; Cabaret, O.; Stressmann, F.A.; Costa, J.M.; Wallet, F.; Wallaert, B.; Bruce, K.; Delhaes, L. Fungal and bacterial diversity of airway microbiota in adults with cystic fibrosis: Concordance between conventional methods and ultra-deep sequencing, and their practical use in the clinical laboratory. Mycopathologia 2018, 183, 171–183. [Google Scholar] [CrossRef]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio 2016, 7, e01250-16. [Google Scholar] [CrossRef]
- Saxena, R.; Mittal, P.; Clavaud, C.; Dhakan, D.B.; Hegde, P.; Veeranagaiah, M.M.; Saha, S.; Souverain, L.; Roy, N.; Breton, L.; et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell. Infect. Microbiol. 2018, 8, 346. [Google Scholar] [CrossRef]
- Vesty, A.; Gear, K.; Biswas, K.; Radcliff, F.J.; Taylor, M.W.; Douglas, R.G. Microbial and inflammatory-based salivary biomarkers of head and neck squamous cell carcinoma. Clin. Exp. Dent. Res. 2018, 4, 255–262. [Google Scholar] [CrossRef]
- Fukui, Y.; Aoki, K.; Ishii, Y.; Tateda, K. The palatine tonsil bacteriome, but not the mycobiome, is altered in HIV infection. BMC Microbiol. 2018, 18, 127. [Google Scholar] [CrossRef]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G.; et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef]
- Jurdi, N.E.; Filali-Mouhim, A.; Salem, I.; Retuerto, M.; Dambrosio, N.M.; Baer, L.; Lazarus, H.M.; Caimi, P.; Cooper, B.; Tomlinson, B.; et al. Gastrointestinal microbiome and mycobiome changes during autologous transplantation for multiple myeloma: Results of a prospective pilot study. Biol. Blood Marrow Transplant. 2019. [Google Scholar] [CrossRef][Green Version]
- Kragelund, C.; Keller, M.K. The oral microbiome in oral lichen planus during a 1-year randomized clinical trial. Oral Dis. 2018, 25, 327–338. [Google Scholar] [CrossRef]
- Dollive, S.; Chen, Y.-Y.; Grunberg, S.; Bittinger, K.; Hoffmann, C.; Vandivier, L.; Cuff, C.; Lewis, J.D.; Wu, G.D.; Bushman, F.D. Fungi of the murine gut: Episodic variation and proliferation during antibiotic treatment. PLoS ONE 2013, 8, e71806. [Google Scholar] [CrossRef] [PubMed]
- Vesty, A.; Biswas, K.; Taylor, M.W.; Gear, K.; Douglas, R.G. Evaluating the impact of DNA extraction method on the representation of human oral bacterial and fungal communities. PLoS ONE 2017, 12, e0169877. [Google Scholar] [CrossRef]
- Wesolowska-Andersen, A.; Bahl, M.I.; Carvalho, V.; Kristiansen, K.; Sicheritz-Pontén, T.; Gupta, R.; Licht, T.R. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014, 2, 19. [Google Scholar] [CrossRef]
- Underhill, D.M.; Iliev, I.D. The mycobiota: Interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 2014, 14, 405–416. [Google Scholar] [CrossRef]
- Tang, J.; Iliev, I.D.; Brown, J.; Underhill, D.M.; Funari, V.A. Mycobiome: Approaches to analysis of intestinal fungi. J. Immunol. Methods 2015, 421, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Z.; Guo, R.; Chen, N.; Lu, H.; Huang, S.; Wang, J.; Li, L. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn. Microbiol. Infect. Dis. 2011, 70, 492–498. [Google Scholar] [CrossRef]
- Brogden, K.A.; Guthmiller, J.M.; Taylor, C.E. Human polymicrobial infections. Lancet 2005, 365, 253–255. [Google Scholar] [CrossRef]
- Allison, D.L.; Willems, H.M.; Jayatilake, J.A.; Bruno, V.M.; Peters, B.M.; Shirtliff, M.E. Candida-bacteria interactions: Their impact on human disease. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Förster, T.M.; Mogavero, S.; Dräger, A.; Graf, K.; Polke, M.; Jacobsen, I.D.; Hube, B. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell. Microbiol. 2016, 18, 1709–1715. [Google Scholar] [CrossRef] [PubMed]
- Kapitan, M.; Niemiec, M.J.; Steimle, A.; Frick, J.S.; Jacobsen, I.D. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 2018, 1, 1–37. [Google Scholar]
- Wargo, M.J.; Hogan, D.A. Fungal-bacterial interactions: A mixed bag of mingling microbes. Curr. Opin. Microbiol. 2006, 9, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Leclair, L.W.; Hogan, D.A. Mixed bacterial-fungal infections in the CF respiratory tract. Med. Mycol. 2010, 48, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Hogan, D.A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef] [PubMed]
- Rall, G.; Knoll, L.J. Development of complex models to study co- and polymicrobial infections and diseases. PLoS Pathog. 2016, 12, e1005858. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial–fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340. [Google Scholar] [CrossRef]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009, 299, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Ye, Y.; Zhou, Y.; Fodor, A.A. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE 2012, 7, e34242. [Google Scholar] [CrossRef] [PubMed]
- Klimesova, K.; Jiraskova Zakostelska, Z.; Tlaskalova-Hogenova, H. Oral bacterial and fungal microbiome impacts colorectal carcinogenesis. Front. Microbiol. 2018, 9, 774. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, A.K.; David, M.S.; Li, L.; Heider, T.N.; Peterson, J.D.; Montano, E.A.; Dongari-Bagtzoglou, A.; Diaz, P.I.; Strausbaugh, L.D. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: Discovery of Malassezia as a prominent commensal. PLoS ONE 2014, 9, e90899. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Charlson, E.S.; Bittinger, K.; Haas, A.R.; Fitzgerald, A.S.; Frank, I.; Yadav, A.; Bushman, F.D.; Collman, R.G. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011, 184, 957–963. [Google Scholar] [CrossRef]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011, 6, e16384. [Google Scholar] [CrossRef]
- Van Woerden, H.C.; Gregory, C.; Brown, R.; Marchesi, J.R.; Hoogendoorn, B.; Matthews, I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: A community based case control study. BMC Infect. Dis. 2013, 13, 69. [Google Scholar] [CrossRef]
- Mailhe, M.; Ricaboni, D.; Vitton, V.; Gonzalez, J.-M.; Bachar, D.; Dubourg, G.; Cadoret, F.; Robert, C.; Delerce, J.; Levasseur, A. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol. 2018, 18, 157. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A. The gut mycobiome of the human microbiome project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, M.; Pereira, L.; Jenull, S.; Kuchler, K.; Lion, T. Klebsiella pneumoniae prevents spore germination and hyphal development of Aspergillus species. Sci. Rep. 2019, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 2010, 313, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef]
- Bor, B.; Cen, L.; Agnello, M.; Shi, W.; He, X. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci. Rep. 2016, 6, 27956. [Google Scholar] [CrossRef]
- Pidwill, G.R.; Rego, S.; Jenkinson, H.F.; Lamont, R.J.; Nobbs, A.H. Coassociation between Group B Streptococcus and Candida albicans promotes interactions with vaginal epithelium. Infect. Immun. 2018, 86, e00669-17. [Google Scholar] [CrossRef]
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef]
- Brand, A.; Barnes, J.D.; Mackenzie, K.S.; Odds, F.C.; Gow, N.A. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2008, 287, 48–55. [Google Scholar] [CrossRef]
- Peters, B.M.; Ovchinnikova, E.S.; Krom, B.P.; Schlecht, L.M.; Zhou, H.; Hoyer, L.L.; Busscher, H.J.; van der Mei, H.C.; Jabra-Rizk, M.A.; Shirtliff, M.E. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 2012, 158, 2975–2986. [Google Scholar] [CrossRef]
- Schlecht, L.M.; Peters, B.M.; Krom, B.P.; Freiberg, J.A.; Hansch, G.M.; Filler, S.G.; Jabra-Rizk, M.A.; Shirtliff, M.E. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 2015, 161, 168–181. [Google Scholar] [CrossRef]
- Kong, E.F.; Kucharikova, S.; Van Dijck, P.; Peters, B.M.; Shirtliff, M.E.; Jabra-Rizk, M.A. Clinical implications of oral candidiasis: Host tissue damage and disseminated bacterial disease. Infect. Immun. 2015, 83, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Beaussart, A.; Herman, P.; El-Kirat-Chatel, S.; Lipke, P.N.; Kucharikova, S.; Van Dijck, P.; Dufrene, Y.F. Single-cell force spectroscopy of the medically important Staphylococcus epidermidis-Candida albicans interaction. Nanoscale 2013, 5, 10894–10900. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Oh, S.-H.; Jones, R.; Cota, E. A proposed mechanism for the interaction between the Candida albicans Als3 adhesin and streptococcal cell wall proteins. Front. Microbiol. 2014, 5, 564. [Google Scholar] [CrossRef]
- Silverman, R.J.; Nobbs, A.H.; Vickerman, M.M.; Barbour, M.E.; Jenkinson, H.F. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 2010, 78, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Bamford, C.V.; D’mello, A.; Nobbs, A.H.; Dutton, L.C.; Vickerman, M.M.; Jenkinson, H.F. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 2009, 77, 3696–3704. [Google Scholar] [CrossRef] [PubMed]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus fumigatus inhibits Pseudomonas aeruginosa in co-culture: Implications of a mutually antagonistic relationship on virulence and inflammation in the CF airway. Front. Microbiol. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Kerr, J.; Taylor, G.; Rutman, A.; Høiby, N.; Cole, P.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef] [PubMed]
- Bachtiar, E.W.; Bachtiar, B.M.; Jarosz, L.M.; Amir, L.R.; Sunarto, H.; Ganin, H.; Meijler, M.M.; Krom, B.P. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front. Cell. Infect. Microbiol. 2014, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, P.T.; van der Peet, J.M.; Bikker, F.J.; Hoogenkamp, M.A.; Paiva, A.M.O.; Kostidis, S.; Mayboroda, O.A.; Smits, W.K.; Krom, B.P. Interspecies interactions between Clostridium difficile and Candida albicans. MSphere 2016, 1, e00187-16. [Google Scholar] [CrossRef]
- Bandara, H.M.; Cheung, B.P.K.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Secretory products of Escherichia coli biofilm modulate Candida biofilm formation and hyphal development. J. Investig. Clin. Dent. 2013, 4, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Cabral, D.J.; Penumutchu, S.; Norris, C.; Morones-Ramirez, J.R.; Belenky, P. Microbial competition between Escherichia coli and Candida albicans reveals a soluble fungicidal factor. Microb. Cell 2018, 5, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef]
- Parolin, C.; Marangoni, A.; Laghi, L.; Foschi, C.; Ñahui Palomino, R.A.; Calonghi, N.; Cevenini, R.; Vitali, B. Isolation of vaginal lactobacilli and characterization of anti-Candida activity. PLoS ONE 2015, 10, e0131220. [Google Scholar] [CrossRef]
- Okkers, D.J.; Dicks, L.M.T.; Silvester, M.; Joubert, J.J.; Odendaal, H.J. Characterization of pentocin TV35b, a bacteriocin-like peptide isolated from Lactobacillus pentosus with a fungistatic effect on Candida albicans. J. Appl. Microbiol. 1999, 87, 726–734. [Google Scholar] [CrossRef]
- Kaewsrichan, J.; Peeyananjarassri, K.; Kongprasertkit, J. Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol. Med. Microbiol. 2006, 48, 75–83. [Google Scholar] [CrossRef]
- Noverr, M.C.; Huffnagle, G.B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 2004, 72, 6206–6210. [Google Scholar] [CrossRef]
- Cugini, C.; Calfee, M.W.; Farrow, J.M.; Morales, D.K.; Pesci, E.C.; Hogan, D.A. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 896–906. [Google Scholar] [CrossRef]
- Gibson, J.; Sood, A.; Hogan, D.A. Pseudomonas aeruginosa-Candida albicans interactions: Localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 2009, 75, 504–513. [Google Scholar] [CrossRef]
- Kong, E.F.; Tsui, C.; Kucharikova, S.; Van Dijck, P.; Jabra-Rizk, M.A. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob. Agents Chemother. 2017, 61, e01573-17. [Google Scholar] [CrossRef]
- Kim, D.; Sengupta, A.; Niepa, T.H.R.; Lee, B.-H.; Weljie, A.; Freitas-Blanco, V.S.; Murata, R.M.; Stebe, K.J.; Lee, D.; Koo, H. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 2017, 7, 41332. [Google Scholar] [CrossRef]
- Vílchez, R.; Lemme, A.; Ballhausen, B.; Thiel, V.; Schulz, S.; Jansen, R.; Sztajer, H.; Wagner-Döbler, I. Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). Chembiochem 2010, 11, 1552–1562. [Google Scholar] [CrossRef]
- Jarosz, L.M.; Deng, D.M.; van der Mei, H.C.; Crielaard, W.; Krom, B.P. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell 2009, 8, 1658–1664. [Google Scholar] [CrossRef]
- Kim, Y.; Mylonakis, E. Killing of Candida albicans filaments by Salmonella enterica serovar Typhimurium is mediated by SopB effectors, parts of a type III secretion system. Eukaryot Cell 2011, 10, 782–790. [Google Scholar] [CrossRef]
- Frases, S.; Chaskes, S.; Dadachova, E.; Casadevall, A. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl. Environ. Microbiol. 2006, 72, 1542–1550. [Google Scholar] [CrossRef]
- Smith, M.G.; Des Etages, S.G.; Snyder, M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004, 24, 3874–3884. [Google Scholar] [CrossRef]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Cell. Microbiol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef]
- Boris, S.; Suárez, J.E.; Vázquez, F.; Barbés, C. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 1998, 66, 1985–1989. [Google Scholar] [PubMed]
- Donnarumma, G.; Molinaro, A.; Cimini, D.; De Castro, C.; Valli, V.; De Gregorio, V.; De Rosa, M.; Schiraldi, C. Lactobacillus crispatus L1: High cell density cultivation and exopolysaccharide structure characterization to highlight potentially beneficial effects against vaginal pathogens. BMC Microbiol. 2014, 14, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Losacco, A.; Carratelli, C.R. Lactobacillus crispatus modulates epithelial cell defense against Candida albicans through Toll-like receptors 2 and 4, interleukin 8 and human β-defensins 2 and 3. Immun. Lett. 2013, 156, 102–109. [Google Scholar] [CrossRef]
- Basson, N.J. Competition for glucose between Candida albicans and oral bacteria grown in mixed culture in a chemostat. J. Med. Microbiol. 2000, 49, 969–975. [Google Scholar] [CrossRef]
- Kalan, L.; Loesche, M.; Hodkinson, B.P.; Heilmann, K.; Ruthel, G.; Gardner, S.E.; Grice, E.A. Redefining the chronic-wound microbiome: Fungal communities are prevalent, dynamic, and associated with delayed healing. MBio 2016, 7, e01058-16. [Google Scholar] [CrossRef] [PubMed]
- De Brucker, K.; Tan, Y.; Vints, K.; De Cremer, K.; Braem, A.; Verstraeten, N.; Michiels, J.; Vleugels, J.; Cammue, B.P.; Thevissen, K. Fungal β-1, 3-glucan increases ofloxacin-tolerance of Escherichia coli in a polymicrobial E. coli–Candida albicans biofilm. Antimicrob. Agents Chemother. 2015, 59, 3052–3058. [Google Scholar] [CrossRef]
- Adair, C.G.; Gorman, S.P.; Feron, B.M.; Byers, L.M.; Jones, D.S.; Goldsmith, C.E.; Moore, J.E.; Kerr, J.R.; Curran, M.D.; Hogg, G. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 1999, 25, 1072–1076. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.M.; Lam, O.L.T.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Bacterial lipopolysaccharides variably modulate in vitro biofilm formation of Candida species. J. Med. Microbiol. 2010, 59, 1225–1234. [Google Scholar] [CrossRef]
- Bandara, H.; K Cheung, B.; Watt, R.; Jin, L.; Samaranayake, L. Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol. Oral Microbiol. 2013, 28, 54–69. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Bartnicka, D.; Zawrotniak, M.; Zielinska, G.; Kierońska, A.; Bochenska, O.; Ciaston, I.; Koziel, J.; Potempa, J.; Baster, Z.; et al. The activity of bacterial peptidylarginine deiminase is important during formation of dual-species biofilm by periodontal pathogen Porphyromonas gingivalis and opportunistic fungus Candida albicans. Pathog. Dis. 2018, 76, fty033. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharikova, S.; Andes, D.; Van Dijck, P.; Jabra-Rizk, M.A. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio 2016, 7, e01365-16. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; Scheper, M.A.; Leid, J.G.; Costerton, J.W.; Shirtliff, M.E. Microbial interactions and differential protein expression in Staphylococcus aureus—Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Adam, B.; Baillie, G.S.; Douglas, L.J. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 2002, 51, 344–349. [Google Scholar] [CrossRef]
- Pammi, M.; Liang, R.; Hicks, J.; Mistretta, T.A.; Versalovic, J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013, 13, 257. [Google Scholar] [CrossRef] [PubMed]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microb. 2013, 16, 214–231. [Google Scholar] [CrossRef]
- Kim, D.; Liu, Y.; Benhamou, R.I.; Sanchez, H.; Simón-Soro, Á.; Li, Y.; Hwang, G.; Fridman, M.; Andes, D.R.; Koo, H. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 2018, 12, 1427–1442. [Google Scholar] [CrossRef] [PubMed]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.-H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilm in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Arweiler, N.B.; Netuschil, L. The oral microbiota. In Microbiota of the Human Body: Implications in Health and Disease; Schwiertz, A., Ed.; Springer International Publishing: Cham, Germany, 2016; pp. 45–60. [Google Scholar]
- Marsh, P.D.; Do, T.; Beighton, D.; Devine, D.A. Influence of saliva on the oral microbiota. Periodontol 2000 2016, 70, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hebraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Mylonakis, E. Characteristics, clinical relevance, and the role of echinocandins in fungal-bacterial interactions. Clin. Infect. Dis. 2015, 61, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Demuyser, L.; Jabra-Rizk, M.A.; Van Dijck, P. Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog. Dis. 2014, 70, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef]
- Utter, D.R.; Mark Welch, J.L.; Borisy, G.G. Individuality, stability, and variability of the plaque microbiome. Front. Microbiol. 2016, 7, 564. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Stahringer, S.S.; Clemente, J.C.; Corley, R.P.; Hewitt, J.; Knights, D.; Walters, W.A.; Knight, R.; Krauter, K.S. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 2012, 22, 2146–2152. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, 89. [Google Scholar] [CrossRef]
- Cameron, S.J.; Huws, S.A.; Hegarty, M.J.; Smith, D.P.; Mur, L.A. The human salivary microbiome exhibits temporal stability in bacterial diversity. FEMS Microbiol. Ecol. 2015, 91, 91. [Google Scholar] [CrossRef] [PubMed]
- Belstrom, D.; Holmstrup, P.; Bardow, A.; Kokaras, A.; Fiehn, N.E.; Paster, B.J. Temporal stability of the salivary microbiota in oral health. PLoS ONE 2016, 11, e0147472. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Bio.l 2011, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Flores, G.E.; Caporaso, J.G.; Henley, J.B.; Rideout, J.R.; Domogala, D.; Chase, J.; Leff, J.W.; Vazquez-Baeza, Y.; Gonzalez, A.; Knight, R.; et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014, 15, 531. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-da-Silva, F.; Araujo, R.; Sampaio-Maia, B. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med. Mycol. 2014, 52, 498–505. [Google Scholar] [CrossRef]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef]
- Costa, A.C.; Pereira, C.A.; Junqueira, J.C.; Jorge, A.O. Recent mouse and rat methods for the study of experimental oral candidiasis. Virulence 2013, 4, 391–399. [Google Scholar] [CrossRef]
- Millsop, J.W.; Fazel, N. Oral candidiasis. Clin. Dermatol. 2016, 34, 487–494. [Google Scholar] [CrossRef]
- Chanda, W.; Joseph, T.P.; Wang, W.; Padhiar, A.A.; Zhong, M. The potential management of oral candidiasis using anti-biofilm therapies. Med. Hypotheses 2017, 106, 15–18. [Google Scholar] [CrossRef]
- Jain, A.; Jain, S.; Rawat, S. Emerging fungal infections among children: A review on its clinical manifestations, diagnosis, and prevention. J. Pharm Bioallied Sci. 2010, 2, 314–320. [Google Scholar] [CrossRef]
- Dickstein, B. Neonatal oral candidiasis: Evaluation of a new chemotherapeutic agent. Drug Des. Dev. Ther. 1964, 3, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Dzidic, M.; Collado, M.C.; Abrahamsson, T.; Artacho, A.; Stensson, M.; Jenmalm, M.C.; Mira, A. Oral microbiome development during childhood: An ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 2018, 12, 2292–2306. [Google Scholar] [CrossRef] [PubMed]
- Hoepelman, I.M.; Dupont, B. Oral candidiasis: The clinical challenge of resistance and management. Int. J. Antimicrob. Agents 1996, 6, 155–159. [Google Scholar] [CrossRef]
- Sampaio-Maia, B.; Monteiro-Silva, F. Acquisition and maturation of oral microbiome throughout childhood: An update. Dent. Res. J. 2014, 11, 291–301. [Google Scholar]
- Hajishengallis, G. The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol. 2014, 29, 248–257. [Google Scholar] [CrossRef]
- Canabarro, A.; Valle, C.; Farias, M.R.; Santos, F.B.; Lazera, M.; Wanke, B. Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J. Periodontal Res. 2013, 48, 428–432. [Google Scholar] [CrossRef] [PubMed]
- De-La-Torre, J.; Quindós, G.; Marcos-Arias, C.; Marichalar-Mendia, X.; Gainza, M.L.; Eraso, E.; Acha-Sagredo, A.; Aguirre-Urizar, J.M. Oral Candida colonization in patients with chronic periodontitis. Is there any relationship? Rev. Iberoam Micol. 2018, 35, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Bachtiar, E.W.; Bachtiar, B.M. Relationship between Candida albicans and Streptococcus mutans in early childhood caries, evaluated by quantitative PCR. F1000Res 2018, 7, 1645–1660. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, F.G.; Silva, D.S.; Hebling, J.; Spolidorio, L.C.; Spolidorio, D.M.P. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 2006, 51, 1024–1028. [Google Scholar]
- Raja, M.; Hannan, A.; Ali, K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010, 44, 272–276. [Google Scholar] [CrossRef]
- Yang, X.Q.; Zhang, Q.; Lu, L.Y.; Yang, R.; Liu, Y.; Zou, J. Genotypic distribution of Candida albicans in dental biofilm of chinese children associated with severe early childhood caries. Arch. Oral Biol. 2012, 57, 1048–1053. [Google Scholar] [CrossRef]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond Streptococcus mutans: Dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef]
- Metwalli, K.H.; Khan, S.A.; Krom, B.P.; Jabra-Rizk, M.A. Streptococcus mutans, Candida albicans, and the human mouth: A sticky situation. PLoS Pathog. 2013, 9, e1003616. [Google Scholar] [CrossRef]
- Forssten, S.D.; Björklund, M.; Ouwehand, A.C. Streptococcus mutans, caries and simulation models. Nutrients 2010, 2, 290–298. [Google Scholar] [CrossRef]
- Falsetta, M.L.; Klein, M.I.; Lemos, J.A.; Silva, B.B.; Agidi, S.; Scott-Anne, K.K.; Koo, H. Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo. Antimicrob. Agents Chemother. 2012, 56, 6201–6211. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Liu, Y.; Kim, D.; Li, Y.; Krysan, D.J.; Koo, H. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017, 13, e1006407. [Google Scholar] [CrossRef]
- Willems, M.; Kos, K.; Jabra-Rizk, M.A.; Krom, B. Candida albicans in oral biofilms could prevent caries. Pathog. Dis. 2016, 74, ftw039. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.O.; Rossoni, R.D.; Vilela, S.F.; de Alvarenga, J.A.; Velloso, M.; Prata, M.C.; Jorge, A.O.; Junqueira, J.C. Streptococcus mutans can modulate biofilm formation and attenuate the virulence of Candida albicans. PLoS ONE 2016, 11, e0150457. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Sobue, T.; Bertolini, M.; Thompson, A.; Dongari-Bagtzoglou, A. Streptococcus oralis and Candida albicans synergistically activate μ-calpain to degrade e-cadherin from oral epithelial junctions. J. Infect. Dis. 2016, 214, 925–934. [Google Scholar] [CrossRef]
- Tamai, R.; Sugamata, M.; Kiyoura, Y. Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb. Pathog. 2011, 51, 250–254. [Google Scholar] [CrossRef]
- Janus, M.M.; Crielaard, W.; Volgenant, C.M.; van der Veen, M.H.; Brandt, B.W.; Krom, B.P. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017, 9, 1270613. [Google Scholar] [CrossRef]
- Fuochi, V.; Li Volti, G.; Furneri, P.M. Commentary: Lactobacilli dominance and vaginal pH: Why is the human vaginal microbiome unique? Front. Microbiol. 2017, 8, 1815. [Google Scholar] [CrossRef]
- Greenbaum, S.; Greenbaum, G.; Moran-Gilad, J.; Weintruab, A.Y. Ecological dynamics of the vaginal microbiome in relation to health and disease. Am. J. Obstet. Gynecol. 2018, 18, 114–118. [Google Scholar] [CrossRef]
- Vaneechoutte, M. The human vaginal microbial community. Res. Microbiol. 2017, 168, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Bradford, L.L.; Ravel, J. The vaginal mycobiome: A contemporary perspective on fungi in women’s health and diseases. Virulence 2017, 8, 342–351. [Google Scholar] [CrossRef]
- Sobel, J.D.; Faro, S.; Force, R.W.; Foxman, B.; Ledger, W.J.; Nyirjesy, P.R.; Reed, B.D.; Summers, P.R. Vulvovaginal candidiasis: Epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 1998, 178, 203–211. [Google Scholar] [CrossRef]
- De Bernardis, F.; Graziani, S.; Tirelli, F.; Antonopoulou, S. Candida vaginitis: Virulence, host response and vaccine prospects. Med. Mycol. 2018, 56, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; Alves, C.T.; Henriques, M.; Azeredo, J.; Silva, S. Vulvovaginal candidiasis: Epidemiology, microbiology and risk factors. Crit. Rev. Microbiol. 2016, 42, 905–927. [Google Scholar]
- Mendling, W. Vaginal microbiota. In Microbiota of the Human Body: Implications in Health and Disease; Schwiertz, A., Ed.; Springer International Publishing: Cham, Germany, 2016; pp. 83–93. [Google Scholar]
- Köhler, G.A.; Assefa, S.; Reid, G. Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infect. Dis. Obstet. Gynecol. 2012, 2012, 1–12. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Lopes, L.C.L.; Cordero, R.J.B.; Nosanchuk, J.D. Sodium butyrate inhibits pathogenic yeast growth and enhances the functions of macrophages. J. Antimicrob. Chemother. 2011, 66, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Buffo, J.; Herman, M.A.; Soll, D.R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 1984, 85, 21–30. [Google Scholar] [CrossRef] [PubMed]
- De Barros, P.P.; Scorzoni, L.; Ribeiro, F.; Fugisaki, L.R.; Fuchs, B.B.; Mylonakis, E.; Jorge, A.O.; Junqueira, J.C.; Rossoni, R.D. Lactobacillus paracasei 28.4 reduces in vitro hyphae formation of Candida albicans and prevents the filamentation in an experimental model of Caenorhabditis elegans. Microb. Pathog. 2018, 117, 80–87. [Google Scholar]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef]
- De Oliveira, F.E.; Rossoni, R.D.; de Barros, P.P.; Begnini, B.E.; Junqueira, J.C.; Jorge, A.O.C.; Leão, M.V.P.; de Oliveira, L.D. Immunomodulatory effects and anti-Candida activity of lactobacilli in macrophages and in invertebrate model of Galleria mellonella. Microb. Pathog. 2017, 110, 603–611. [Google Scholar] [CrossRef]
- Ribeiro, F.C.; de Barros, P.P.; Rossoni, R.D.; Junqueira, J.C.; Jorge, A.O.C. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 2017, 122, 201–211. [Google Scholar] [CrossRef]
- Rossoni, R.D.; Fuchs, B.B.; de Barros, P.P.; Velloso, M.d.S.; Jorge, A.O.C.; Junqueira, J.C.; Mylonakis, E. Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLoS ONE 2017, 12, e0173332. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.A.; Pires, M.C.V.; Leão, T.L.; Silva, A.K.S.; Miranda, L.S.; Martins, F.S.; Silva, A.M.; Nicoli, J.R. Anti-inflammatory effect of two Lactobacillus strains during infection with Gardnerella vaginalis and Candida albicans in a HeLa cell culture model. Microbiology 2018, 164, 349–358. [Google Scholar] [CrossRef]
- Ene, I.V.; Cheng, S.-C.; Netea, M.G.; Brown, A.J.P. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect. Immun. 2013, 81, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef]
- Zhang, Z.; Lv, J.; Pan, L.; Zhang, Y. Roles and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 2018, 102, 8135–8143. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V. From yaks to yogurt: The history, development, and current use of probiotics. Clin. Infect. Dis. 2015, 60, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Merenstein, D.J.; Wang, C.; Hamilton, P.R.; Blackmon, M.L.; Chen, H.; Calderone, R.A.; Li, D. Impact of eating probiotic yogurt on colonization by Candida species of the oral and vaginal mucosa in HIV-infected and HIV-uninfected women. Mycopathologia 2013, 176, 175–181. [Google Scholar] [CrossRef]
- Mezzasalma, V.; Manfrini, E.; Ferri, E.; Boccarusso, M.; Di Gennaro, P.; Schiano, I.; Michelotti, A.; Labra, M. Orally administered multispecies probiotic formulations to prevent uro-genital infections: A randomized placebo-controlled pilot study. Arch. Gynecol. Obstet. 2017, 295, 163–172. [Google Scholar] [CrossRef]
- De Alberti, D.; Russo, R.; Terruzzi, F.; Nobile, V.; Ouwehand, A. Lactobacilli vaginal colonisation after oral consumption of Respecta® complex: A randomised controlled pilot study. Arch. Gynecol. Obstet. 2015, 292, 861–867. [Google Scholar] [CrossRef]
- Reid, G.; Bruce, A.W.; Fraser, N.; Heinemann, C.; Owen, J.; Henning, B. Oral probiotics can resolve urogenital infections. FEMS Immunol. Med. Microbiol. 2001, 30, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Verdenelli, M.C.; Cecchini, C.; Coman, M.M.; Silvi, S.; Orpianesi, C.; Coata, G.; Cresci, A.; Di Renzo, G.C. Impact of probiotic SYNBIO® administered by vaginal suppositories in promoting vaginal health of apparently healthy women. Curr. Microbiol. 2016, 73, 483–490. [Google Scholar] [CrossRef]
- Kovachev, S.M.; Vatcheva-Dobrevska, R.S. Local probiotic therapy for vaginal Candida albicans infections. Probiotics Antimicrob. Proteins 2015, 7, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Pendharkar, S.; Brandsborg, E.; Hammarström, L.; Marcotte, H.; Larsson, P.-G. Vaginal colonisation by probiotic lactobacilli and clinical outcome in women conventionally treated for bacterial vaginosis and yeast infection. BMC Infect. Dis. 2015, 15, 255. [Google Scholar] [CrossRef] [PubMed]
- De Seta, F.; Parazzini, F.; De Leo, R.; Banco, R.; Maso, G.P.; De Santo, D.; Sartore, A.; Stabile, G.; Inglese, S.; Tonon, M.; et al. Lactobacillus plantarum P17630 for preventing Candida vaginitis recurrence: A retrospective comparative study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 182, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Cools, P.; Jespers, V.; Hardy, L.; Crucitti, T.; Delany-Moretlwe, S.; Mwaura, M.; Ndayisaba, G.F.; van de Wijgert, J.H.H.M.; Vaneechoutte, M. A multi-country cross-sectional study of vaginal carriage of Group B Streptococci (GBS) and Escherichia coli in resource-poor settings: Prevalences and risk factors. PLoS ONE 2016, 11, e0148052. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Y.; Fu, F.; Kong, W.-N.; Xuan, Q.-K.; Wen, D.-H.; Chen, X.-Q.; He, Y.-M.; He, L.-H.; Guo, J.; Zhou, A.-P.; et al. Streptococcus agalactiae inhibits Candida albicans hyphal development and diminishes host vaginal mucosal TH17 response. Front. Microbiol. 2018, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef]
- Steimle, A.; Frick, J.S. Molecular mechanisms of induction of tolerant and tolerogenic intestinal dendritic cells in mice. J. Immunol. Res. 2016, 2016. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 2017, 15, 55–64. [Google Scholar] [CrossRef]
- Witherden, E.A.; Moyes, D. Mycobiome and gut inflammation: Implications in gut disease. In Immunity and Inflammation in Health and Disease; Chatterjee, S., Jungriathmyr, W., Bagchi, D., Eds.; Elsevier: New York, NY, USA, 2017. [Google Scholar]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Strati, F.; Di Paola, M.; Stefanini, I.; Albanese, D.; Rizzetto, L.; Lionetti, P.; Calabrò, A.; Jousson, O.; Donati, C.; Cavalieri, D. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 2016, 7, 1227. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59. [Google Scholar] [CrossRef]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef]
- Durban, A.; Abellan, J.J.; Jimenez-Hernandez, N.; Ponce, M.; Ponce, J.; Sala, T.; D’Auria, G.; Latorre, A.; Moya, A. Assessing gut microbial diversity from feces and rectal mucosa. Microb. Ecol. 2011, 61, 123–133. [Google Scholar] [CrossRef]
- Altomare, A.; Putignani, L.; Del Chierico, F.; Cocca, S.; Angeletti, S.; Ciccozzi, M.; Tripiciano, C.; Dalla Piccola, B.; Cicala, M.; Guarino, M.P.L. Gut mucosal-associated microbiota better discloses Inflammatory Bowel Disease differential patterns than faecal microbiota. Dig. Liver Dis. 2018, 18. [Google Scholar] [CrossRef]
- Carstens, A.; Roos, A.; Andreasson, A.; Magnuson, A.; Agreus, L.; Halfvarson, J.; Engstrand, L. Differential clustering of fecal and mucosa-associated microbiota in ‘healthy’ individuals. J. Dig. Dis. 2018, 19, 745–752. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Rajilic-Stojanovic, M.; de Vos, W.M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef]
- Seed, P.C. The human mycobiome. Cold Spring Harb. Perspect. Med. 2014, 5, a019810. [Google Scholar] [CrossRef]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastrol. 2008, 43, 831–841. [Google Scholar] [CrossRef]
- Lepage, P.; Seksik, P.; Sutren, M.; de la Cochetiere, M.F.; Jian, R.; Marteau, P.; Dore, J. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 2005, 11, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Sepehri, S.; Kotlowski, R.; Bernstein, C.N.; Krause, D.O. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.P.; Schippa, S.; Zamboni, I.; Penta, M.; Chiarini, F.; Seganti, L.; Osborn, J.; Falconieri, P.; Borrelli, O.; Cucchiara, S. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006, 55, 1760–1767. [Google Scholar] [CrossRef]
- Libertucci, J.; Young, V.B. The role of the microbiota in infectious diseases. Nat. Microbiol. 2019, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Djukovic, A.; Isaac, S. Roles of the intestinal microbiota in pathogen protection. Clin. Transl. Immun. 2017, 6, e128. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Dongari-Bagtzoglou, A. Tipping the balance: C. albicans adaptation in polymicrobial environments. JoF 2018, 4, 112. [Google Scholar]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Noverr, M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef]
- Ianiro, G.; Tilg, H.; Gasbarrini, A. Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut 2016, 65, 1906–1915. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Angebault, C.; Ghozlane, A.; Volant, S.; Botterel, F.; D’enfert, C.; Bougnoux, M.E. Combined bacterial and fungal intestinal microbiota analyses: Impact of storage conditions and DNA extraction protocols. PLoS ONE 2018, 13, e0201174. [Google Scholar] [CrossRef]
- Skalski, J.H.; Limon, J.J.; Sharma, P.; Gargus, M.D.; Nguyen, C.; Tang, J.; Coelho, A.L.; Hogaboam, C.M.; Crother, T.R.; Underhill, D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018, 14. [Google Scholar] [CrossRef]
- Scupham, A.J.; Presley, L.L.; Wei, B.; Bent, E.; Griffith, N.; McPherson, M.; Zhu, F.; Oluwadara, O.; Rao, N.; Braun, J.; et al. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006, 72, 793–801. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Ranjith, K.; Dutta, A.; Pinna, N.K.; Mande, S.S.; Sharma, S.; Garg, P.; Murthy, S.I.; Shivaji, S. Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Jiang, T.T.; Shao, T.-Y.; Ang, W.G.; Kinder, J.M.; Turner, L.H.; Pham, G.; Whitt, J.; Alenghat, T.; Way, S.S. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe 2017, 22, 809–816. [Google Scholar] [CrossRef]
- Mason, K.L.; Downward, J.R.E.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during re-colonization following broad spectrum antibiotic therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef]
- Kennedy, M.J.; Volz, P.A. Effect of various antibiotics on gastrointestinal colonization and dissemination by Candida albicans. J. Med. Vet. Mycol. 1985, 23, 265–273. [Google Scholar] [CrossRef]
- Noverr, M.C.; Falkowski, N.R.; McDonald, R.A.; McKenzie, A.N.; Huffnagle, G.B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: Role of host genetics, antigen, and interleukin-13. Infect. Immun. 2005, 73, 30–38. [Google Scholar] [CrossRef]
- Koh, A.Y. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot. Cell 2013, 12, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Coughlin, L.A.; Neubauer, M.M.; Kim, J.; Kim, M.S.; Zhan, X.; Simms-Waldrip, T.R.; Xie, Y.; Hooper, L.V.; Koh, A.Y. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 2015, 21, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Fidel, P.L., Jr.; Odds, F.C. Animal models of mucosal Candida infection. FEMS Microbiol. Lett. 2008, 283, 129–139. [Google Scholar] [CrossRef]
- Erb-Downward, J.R.; Falkowski, N.R.; Mason, K.L.; Muraglia, R.; Huffnagle, G.B. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci. Rep. 2013, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- López-García, B.; Lee, P.H.; Yamasaki, K.; Gallo, R.L. Anti-fungal activity of cathelicidins and their potential role in Candida albicans skin infection. J. Investig. Dermatol. 2005, 125, 108–115. [Google Scholar] [CrossRef]
- Charlet, R.; Pruvost, Y.; Tumba, G.; Istel, F.; Poulain, D.; Kuchler, K.; Sendid, B.; Jawhara, S. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci. Rep. 2018, 8, 3316. [Google Scholar] [CrossRef] [PubMed]
- Charlet, R.; Bortolus, C.; Barbet, M.; Sendid, B.; Jawhara, S. A decrease in anaerobic bacteria promotes Candida glabrata overgrowth while beta-glucan treatment restores the gut microbiota and attenuates colitis. Gut Pathog. 2018, 10, 50–59. [Google Scholar] [CrossRef]
- Murphy, A.; Kavanagh, K. Emergence of Saccharomyces cerevisiae as a human pathogen: Implications for biotechnology. Enzym. Microb. Technol. 1999, 25, 551–557. [Google Scholar] [CrossRef]
- De Llanos, R.; Querol, A.; Pemán, J.; Gobernado, M.; Fernández-Espinar, M.T. Food and probiotic strains from the Saccharomyces cerevisiae species as a possible origin of human systemic infections. Int. J. Food Microbiol. 2006, 110, 286–290. [Google Scholar] [CrossRef]
- Byron, J.K.; Clemons, K.V.; McCusker, J.H.; Davis, R.W.; Stevens, D.A. Pathogenicity of Saccharomyces cerevisiae in complement factor five-deficient mice. Infect. Immun. 1995, 63, 478–485. [Google Scholar]
- Munoz, P.; Bouza, E.; Cuenca-Estrella, M.; Eiros, J.M.; Pérez, M.J.; Sánchez-Somolinos, M.; Rincón, C.; Hortal, J.; Peláez, T. Saccharomyces cerevisiae fungemia: An emerging infectious disease. Clin. Infect. Dis. 2005, 40, 1625–1634. [Google Scholar] [CrossRef]
- Massot, J.; Sanchez, O.; Couchy, R.; Astoin, J.; Parodi, A.L. Bacterio-pharmacological activity of Saccharomyces boulardii in clindamycin-induced colitis in the hamster. Arzneimittel-Forschung 1984, 34, 794–797. [Google Scholar]
- Kelesidis, T.; Pothoulakis, C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap. Adv. Gastroenterol. 2012, 5, 111–125. [Google Scholar] [CrossRef]
- Lessard, M.; Dupuis, M.; Gagnon, N.; Nadeau, E.; Matte, J.; Goulet, J.; Fairbrother, J. Administration of Pediococcus acidilactici or Saccharomyces cerevisiae boulardii modulates development of porcine mucosal immunity and reduces intestinal bacterial translocation after Escherichia coli challenge. J. Anim. Sci. 2009, 87, 922–934. [Google Scholar] [CrossRef]
- Roussel, C.; Sivignon, A.; de Vallée, A.; Garrait, G.; Denis, S.; Tsilia, V.; Ballet, N.; Vandekerckove, P.; Van de Wiele, T.; Barnich, N. Anti-infectious properties of the probiotic Saccharomyces cerevisiae CNCM I-3856 on enterotoxigenic E. coli (ETEC) strain H10407. Appl. Microbiol. Biotechnol. 2018, 102, 6175–6189. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.C.; Owings, M.; Jernigan, D.B. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg. Infect. Dis. 2006, 12, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Corthier, G.; Dubos, F.; Ducluzeau, R. Prevention of Clostridium difficile induced mortality in gnotobiotic mice by Saccharomyces boulardii. Can. J. Microbiol. 1986, 32, 894–896. [Google Scholar] [CrossRef]
- Zuo, T.; Wong, S.H.; Cheung, C.P.; Lam, K.; Lui, R.; Cheung, K.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Wu, J.C.Y.; et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat. Commun. 2018, 9, 3663. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Palasuk, M.; Hiengrach, P.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019, 14, e0210798. [Google Scholar] [CrossRef]
- Markey, L.; Shaban, L.; Green, E.R.; Lemon, K.P.; Mecsas, J.; Kumamoto, C.A. Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes 2018, 9, 497–509. [Google Scholar] [CrossRef]
- Mitchell, A.B.; Glanville, A.R. The human respiratory microbiome: Implications and impact. Semin. Respir. Crit. Care Med. 2018, 39, 199–212. [Google Scholar] [CrossRef]
- Ingenito, E.; Solway, J.; McFadden, E., Jr.; Pichurko, B.; Bowman, H.; Michaels, D.; Drazen, J. Indirect assessment of mucosal surface temperatures in the airways: Theory and tests. J. Appl. Physiol. 1987, 63, 2075–2083. [Google Scholar] [CrossRef] [PubMed]
- West, J.B. Regional differences in the lung. Chest 1978, 74, 426–437. [Google Scholar]
- Marsland, B.J.; Gollwitzer, E.S. Host–microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014, 14, 827. [Google Scholar] [CrossRef]
- Huang, L.; Cattamanchi, A.; Davis, J.L.; Boon, S.; Kovacs, J.; Meshnick, S.; Miller, R.F.; Walzer, P.D.; Worodria, W.; Masur, H. HIV-associated Pneumocystis pneumonia. Proc. Am. Thorac. Soc. 2011, 8, 294–300. [Google Scholar] [CrossRef]
- Sethi, S.; Murphy, T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 2008, 359, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Montes-Cano, M.; De La Horra, C.; Dapena, F.; Mateos, I.; Friaza, V.; Respaldiza, N.; Muñoz-Lobato, F.; Medrano, F.; Calderon, E.; Varela, J. Dynamic colonisation by different Pneumocystis jirovecii genotypes in cystic fibrosis patients. Clin. Microbiol. Infect. 2007, 13, 1008–1011. [Google Scholar] [CrossRef]
- Chabé, M.; Aliouat-Denis, C.-M.; Delhaes, L.; Aliouat, E.M.; Viscogliosi, E.; Dei-Cas, E. Pneumocystis: From a doubtful unique entity to a group of highly diversified fungal species. FEMS Yeast Res. 2011, 11, 2–17. [Google Scholar] [CrossRef]
- Peterson, J.C.; Cushion, M.T. Pneumocystis: Not just pneumonia. Curr. Opin. Microbiol. 2005, 8, 393–398. [Google Scholar] [CrossRef]
- Medrano, F.J.; Montes-Cano, M.; Conde, M.; de la Horra, C.; Respaldiza, N.; Gasch, A.; Perez-Lozano, M.J.; Varela, J.M.; Calderon, E.J. Pneumocystis jirovecii in general population. Emerg. Infect. Dis. 2005, 11, 245–250. [Google Scholar] [CrossRef]
- Ponce, C.A.; Gallo, M.; Bustamante, R.; Vargas, S.L. Pneumocystis colonization is highly prevalent in the autopsied lungs of the general population. Clin. Infect. Dis. 2010, 50, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E. The airway microbiota in cystic fibrosis: A complex fungal and bacterial community—Implications for therapeutic management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef] [PubMed]
- Limper, A.H.; Thomas, C.F., Jr.; Anders, R.A.; Leof, E.B. Interactions of parasite and host epithelial cell cycle regulation during Pneumocystis carinii pneumonia. J. Lab. Clin. Med. 1997, 130, 132–138. [Google Scholar] [CrossRef]
- Cui, L.; Lucht, L.; Tipton, L.; Rogers, M.B.; Fitch, A.; Kessinger, C.; Camp, D.; Kingsley, L.; Leo, N.; Greenblatt, R.M. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am. J. Respir. Crit. Care Med. 2015, 191, 932–942. [Google Scholar] [CrossRef]
- Beck, J.M.; Schloss, P.D.; Venkataraman, A.; Twigg III, H.; Jablonski, K.A.; Bushman, F.D.; Campbell, T.B.; Charlson, E.S.; Collman, R.G.; Crothers, K. Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am. J. Respir. Crit. Care Med. 2015, 192, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med. 2013, 7, 245–257. [Google Scholar] [CrossRef]
- Cui, L.; Morris, A.; Huang, L.; Beck, J.M.; Twigg III, H.L.; von Mutius, E.; Ghedin, E. The microbiome and the lung. Ann. Am. Thorac. Soc. 2014, 11, 227–232. [Google Scholar] [CrossRef]
- Segal, L.N.; Clemente, J.C.; Tsay, J.-C.J.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 16031. [Google Scholar] [CrossRef]
- Robinson, M.; Daviskas, E.; Eberl, S.; Baker, J.; Chan, H.; Anderson, S.; Bye, P. The effect of inhaled mannitol on bronchial mucus clearance in cystic fibrosis patients: A pilot study. Eur. Respir. J. 1999, 14, 678–685. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hauser, A.R.; Jain, M.; Bar-Meir, M.; McColley, S.A. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 2011, 24, 29–70. [Google Scholar] [CrossRef] [PubMed]
- Harrison, F. Microbial ecology of the cystic fibrosis lung. J. Microbiol. 2007, 153, 917–923. [Google Scholar] [CrossRef]
- Gordee, R.S.; Matthews, T.R. Systemic antifungal activity of pyrrolnitrin. J. Appl. Microbiol. 1969, 17, 690–694. [Google Scholar]
- Kerr, J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J. Infect. 1994, 28, 305–310. [Google Scholar] [CrossRef]
- Hogan, D.A.; Vik, Å.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef]
- Briard, B.; Heddergott, C.; Latgé, J.-P. Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus. MBio 2016, 7, e00219-16. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.A.; Penner, J.C.; Moss, R.B.; Haagensen, J.A.; Clemons, K.V.; Spormann, A.M.; Nazik, H.; Cohen, K.; Banaei, N.; Carolino, E. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS ONE 2015, 10, e0134692. [Google Scholar] [CrossRef]
- Kaur, J.; Pethani, B.P.; Kumar, S.; Kim, M.; Sunna, A.; Kautto, L.; Penesyan, A.; Paulsen, I.T.; Nevalainen, H. Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 2015, 6, 866. [Google Scholar] [CrossRef]
- Sedlacek, L.; Graf, B.; Schwarz, C.; Albert, F.; Peter, S.; Würstl, B.; Wagner, S.; Klotz, M.; Becker, A.; Haase, G. Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J. Cyst. Fibros. 2015, 14, 237–241. [Google Scholar] [CrossRef]
- Chen, S.C.-A.; Patel, S.; Meyer, W.; Chapman, B.; Yu, H.; Byth, K.; Middleton, P.G.; Nevalainen, H.; Sorrell, T.C. Pseudomonas aeruginosa inhibits the growth of Scedosporium and Lomentospora in vitro. Mycopathologia 2018, 183, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Gaudry, S.; Khoy-Ear, L.; Aloulou, M.; Phillips-Houlbracq, M.; Bex, J.; Skurnik, D.; Denamur, E.; Monteiro, R.C.; Dreyfuss, D. Airway fungal colonization compromises the immune system allowing bacterial pneumonia to prevail. Crit. Care Med. 2013, 41, 191–199. [Google Scholar] [CrossRef]
- Ader, F.; Jawhara, S.; Nseir, S.; Kipnis, E.; Faure, K.; Vuotto, F.; Chemani, C.; Sendid, B.; Poulain, D.; Guery, B. Short term Candida albicans colonization reduces Pseudomonas aeruginosa-related lung injury and bacterial burden in a murine model. J. Crit. Care 2011, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Mylonakis, E. Fungal–bacterial interactions and their relevance in health. Cell Microbiol. 2015, 17, 1442–1446. [Google Scholar] [CrossRef] [PubMed]
- Azoulay, E.; Timsit, J.-F.; Tafflet, M.; de Lassence, A.; Darmon, M.; Zahar, J.-R.; Adrie, C.; Garrouste-Orgeas, M.; Cohen, Y.; Mourvillier, B. Candida colonization of the respiratory tract and subsequent Pseudomonas ventilator-associated pneumonia. Chest 2006, 129, 110–117. [Google Scholar] [CrossRef]
- Kollef, M.H.; Shorr, A.; Tabak, Y.P.; Gupta, V.; Liu, L.Z.; Johannes, R. Epidemiology and outcomes of health-care–associated pneumonia: Results from a large US database of culture-positive pneumonia. Chest 2005, 128, 3854–3862. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, 3574–3581. [Google Scholar] [CrossRef] [PubMed]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [PubMed]
- Zowawi, H.M.; Forde, B.M.; Alfaresi, M.; Alzarouni, A.; Farahat, Y.; Chong, T.-M.; Yin, W.-F.; Chan, K.-G.; Li, J.; Schembri, M.A. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci. Rep. 2015, 5, 15082. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-J.; Lin, T.-L.; Chen, C.-T.; Chen, Y.-Y.; Hsieh, P.-F.; Hsu, C.-R.; Wu, M.-C.; Wang, J.-T. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015, 5, 15573. [Google Scholar] [CrossRef]
- Naparstek, L.; Carmeli, Y.; Navon-Venezia, S.; Banin, E. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014, 69, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef]
- Richards, A.M.; Abu Kwaik, Y.; Lamont, R.J. Code blue: Acinetobacter baumannii, a nosocomial pathogen with a role in the oral cavity. Mol. Oral Microbiol. 2015, 30, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, R.; Zhu, S.; Wang, H.; Yan, D.; Zhang, X.; Farmakiotis, D.; Mylonakis, E. Candida albicans airway colonization facilitates subsequent Acinetobacter baumannii pneumonia in a rat model. Antimicrob. Agents Chemother. 2016, 60, 3348–3354. [Google Scholar] [CrossRef]
- Mendoza, L.; Vilela, R.; Voelz, K.; Ibrahim, A.S.; Voigt, K.; Lee, S.C. Human fungal pathogens of Mucorales and Entomophthorales. Cold Spring Harb. Perspect. Med. 2014, 5, a019562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hassan, M.I.A.; Voigt, K. Pathogenicity patterns of mucormycosis: Epidemiology, interaction with immune cells and virulence factors. Med. Mycol. 2019, 57, 245–256. [Google Scholar] [CrossRef]
- Morace, G.; Borghi, E. Invasive mold infections: Virulence and pathogenesis of Mucorales. Int. J. Med. Microbiol. 2012, 2012, 349278. [Google Scholar] [CrossRef] [PubMed]
- Hover, T.; Maya, T.; Ron, S.; Sandovsky, H.; Shadkchan, Y.; Kijner, N.; Mitiagin, Y.; Fichtman, B.; Harel, A.; Shanks, R.M.; et al. Mechanisms of bacterial (Serratia marcescens) attachment to migration along, and killing of fungal hyphae. Appl. Environ. Microbiol. 2016, 82, 2585–2594. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Gebremariam, T.; Liu, M.; Chamilos, G.; Kontoyiannis, D.; Mink, R.; Kwon-Chung, K.J.; Fu, Y.; Skory, C.D.; Edwards, J.E., Jr.; et al. Bacterial endosymbiosis is widely present among zygomycetes but does not contribute to the pathogenesis of mucormycosis. J. Infect. Dis. 2008, 198, 1083–1090. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Crouch, J.A. Bacterial/Fungal interactions: From pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 2009, 47, 63–82. [Google Scholar] [CrossRef]
- Church, D.; Elsayed, S.; Reid, O.; Winston, B.; Lindsay, R. Burn wound infections. Clin. Microbiol. Rev. 2006, 19, 403–434. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Murphy, K.P.; Janeway, C.A.; Travers, P.; Walport, M.; Ehrenstein, M. The Complement System and Innate Immunity. In Janeway’s Immunobiology; Schank, D., Ed.; Garland Science: New York, NY, USA, 2008. [Google Scholar]
- Macfarlane, S.; Dillon, J.F. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 2007, 102, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Ward, R.M.; Rane, H.S.; Lee, S.A.; Noverr, M.C. Efficacy of ethanol against Candida albicans and Staphylococcus aureus polymicrobial biofilms. Antimicrob. Agents Chemother. 2013, 57, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob. Agents Chemother. 2010, 54, 3746–3755. [Google Scholar] [CrossRef]
- Siala, W.; Kucharikova, S.; Braem, A.; Vleugels, J.; Tulkens, P.M.; Mingeot-Leclercq, M.P.; Van Dijck, P.; Van Bambeke, F. The antifungal caspofungin increases fluoroquinolone activity against Staphylococcus aureus biofilms by inhibiting N-acetylglucosamine transferase. Nat. Commun. 2016, 7, 13286. [Google Scholar] [CrossRef]
- Rogiers, O.; Holtappels, M.; Siala, W.; Lamkanfi, M.; Van Bambeke, F.; Lagrou, K.; Van Dijck, P.; Kucharikova, S. Anidulafungin increases the antibacterial activity of tigecycline in polymicrobial Candida albicans/Staphylococcus aureus biofilms on intraperitoneally implanted foreign bodies. J. Antimicrob. Chemother. 2018, 73, 2806–2814. [Google Scholar] [CrossRef]
- Budič, M.; Rijavec, M.; Petkovšek, Ž.; Žgur-Bertok, D. Escherichia coli bacteriocins: Antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS ONE 2011, 6, e28769. [Google Scholar] [CrossRef]
- Bell, T.; O’Grady, N.P. Prevention of central line-associated bloodstream infections. Infect. Dis. Clin. N. Am. 2017, 31, 551–559. [Google Scholar] [CrossRef]
- Dyess, D.L.; Garrison, R.N.; Fry, D.E. Candida sepsis. Implications of polymicrobial blood-borne infection. Arch. Surg. 1985, 120, 345–348. [Google Scholar] [CrossRef]
- Pletz, M.W.; Weis, S.; Forstner, C.; Wagenlehner, F. Urosepsis. Der Urologe 2018, 57, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Gouba, N.; Drancourt, M. Digestive tract mycobiota: A source of infection. Med. Mal Infect. 2015, 45, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Inweregbu, K. Nosocomial infections. CEACCP 2005, 5, 14–17. [Google Scholar] [CrossRef]
- Orsini, J.; Mainardi, C.; Muzylo, E.; Karki, N.; Cohen, N.; Sakoulas, G. Microbiological profile of organisms causing bloodstream infection in critically ill patients. J. Clin. Med. Res. 2012, 4, 371–377. [Google Scholar] [CrossRef]
- Bouza, E.; Burillo, A.; Munoz, P.; Guinea, J.; Marin, M.; Rodriguez-Creixems, M. Mixed bloodstream infections involving bacteria and Candida spp. J. Antimicrob. Chemother. 2013, 68, 1881–1888. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klingspor, L.; Muhammed, S.A.; Ozenci, V. Comparison of the two blood culture systems, Bactec 9240 and BacT/Alert 3D, in the detection of Candida spp. and bacteria with polymicrobial sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2983–2987. [Google Scholar] [CrossRef]
- Kaufman, D.A.; Brown, A.T.; Eisenhuth, K.K.; Yue, J.; Grossman, L.B.; Hazen, K.C. More serious infectious morbidity and mortality associated with simultaneous candidemia and coagulase-negative staphylococcal bacteremia in neonates and in vitro adherence studies between Candida albicans and Staphylococcus epidermidis. Early Hum. Dev. 2014, 90, 66–70. [Google Scholar] [CrossRef]
- Pammi, M.; Zhong, D.; Johnson, Y.; Revell, P.; Versalovic, J. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: A case-control study. BMC Infect. Dis. 2014, 14, 390. [Google Scholar] [CrossRef]
- Cateau, E.; Cognee, A.S.; Tran, T.C.; Vallade, E.; Garcia, M.; Belaz, S.; Kauffmann-Lacroix, C.; Rodier, M.H. Impact of yeast-bacteria coinfection on the detection of Candida spp. in an automated blood culture system. Diagn. Microbiol. Infect. Dis. 2012, 72, 328–331. [Google Scholar] [CrossRef]
- Hockey, L.J.; Fujita, N.K.; Gibson, T.R.; Rotrosen, D.; Montgomerie, J.Z.; Edwards, J.E., Jr. Detection of fungemia obscured by concomitant bacteremia: In vitro and in vivo studies. J. Clin. Microbiol. 1982, 16, 1080–1085. [Google Scholar]
- Pulimood, S.; Ganesan, L.; Alangaden, G.; Chandrasekar, P. Polymicrobial candidemia. Diagn. Microbiol Infect. Dis. 2002, 44, 353–357. [Google Scholar] [CrossRef]
- Nair, N.; Biswas, R.; Gotz, F.; Biswas, L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 2014, 82, 2162–2169. [Google Scholar] [CrossRef]
- Luo, Y.; Cui, M.Y.; Liao, B.; Pui, M.H.; Dong, Z.; Li, X.H.; Sun, C.H.; Peng, Z.P.; Li, Z.P.; Feng, S.T. Diagnostic and post-treatment CT appearance of biopsy proven mixed Cryptococcus and Candida cholangitis. J. Xray Sci. Technol. 2014, 22, 727–733. [Google Scholar]
- Sulis, G.; Villanacci, V.; Missale, G.; Salemme, M.; Castelli, F.; Caligaris, S. Whipple’s disease concomitant with Candida esophagitis and subsequent Giardia lamblia coinfection. Eur. J. Gastroenterol. Hepatol. 2014, 26, 1181–1185. [Google Scholar] [CrossRef]
- Skedros, J.G.; Keenan, K.E.; Updike, W.S.; Oliver, M.R. Failed reverse total shoulder arthroplasty caused by recurrent Candida glabrata infection with prior Serratia marcescens coinfection. Case Rep. Infect. Dis. 2014, 14. [Google Scholar] [CrossRef]
- Carlson, E. Enhancement by Candida albicans of Staphylococcus aureus, Serratia marcescens, and Streptococcus faecalis in the establishment of infection in mice. Infect. Immun. 1983, 39, 193–197. [Google Scholar]
- Carlson, E. Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect. Immun. 1983, 42, 285–292. [Google Scholar]
- Peters, B.M.; Noverr, M.C. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect. Immun. 2013, 81, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Nash, E.E.; Peters, B.M.; Palmer, G.E.; Fidel, P.L.; Noverr, M.C. Morphogenesis is not required for Candida albicans-Staphylococcus aureus intra-abdominal infection-mediated dissemination and lethal sepsis. Infect. Immun. 2014, 82, 3426–3435. [Google Scholar] [CrossRef]
- Ikeh, M.A.C.; Fidel, P.L., Jr.; Noverr, M.C. Identification of specific components of the eicosanoid biosynthetic and signaling pathway involved in pathological inflammation during intra-abdominal infection with Candida albicans and Staphylococcus aureus. Infect. Immun. 2018, 86, e00144-18. [Google Scholar] [CrossRef] [PubMed]
- Nash, E.E.; Peters, B.M.; Fidel, P.L.; Noverr, M.C. Morphology-independent virulence of Candida species during polymicrobial intra-abdominal infections with Staphylococcus aureus. Infect. Immun. 2016, 84, 90–98. [Google Scholar] [CrossRef]
- Lilly, E.A.; Ikeh, M.; Nash, E.E.; Fidel, P.L., Jr.; Noverr, M.C. Immune protection against lethal fungal-bacterial intraabdominal infections. MBio 2018, 9, e01472-17. [Google Scholar] [CrossRef]
- Sawyer, R.G.; Adams, R.B.; Rosenlof, L.K.; May, A.K.; Pruett, T.L. The role of Candida albicans in the pathogenesis of experimental fungal/bacterial peritonitis and abscess formation. Am. Surg. 1995, 61, 726–731. [Google Scholar]
- Klaerner, H.G.; Uknis, M.E.; Acton, R.D.; Dahlberg, P.S.; Carlone-Jambor, C.; Dunn, D.L. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J. Surg. Res. 1997, 70, 161–165. [Google Scholar] [CrossRef]
- Stenutz, R.; Weintraub, A.; Widmalm, G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 2006, 30, 382–403. [Google Scholar] [CrossRef] [PubMed]
- Akagawa, G.; Abe, S.; Yamaguchi, H. Mortality of Candida albicans-infected mice is facilitated by superinfection of Escherichia coli or administration of its lipopolysaccharide. J. Infect. Dis. 1995, 171, 1539–1544. [Google Scholar] [CrossRef]
- Toscano, M.G.; Ganea, D.; Gamero, A.M. Cecal ligation puncture procedure. J. Vis. Exp. 2011, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.; Traeger, T.; Entleutner, M.; Westerholt, A.; Kleist, B.; Huser, N.; Holzmann, B.; Stier, A.; Pfeffer, K.; Heidecke, C.D. Cecal ligation and puncture versus colon ascendens stent peritonitis: Two distinct animal models for polymicrobial sepsis. Shock 2004, 21, 505–511. [Google Scholar] [CrossRef]
- Davis, C.G.; Chang, K.; Osborne, D.; Walton, A.H.; Dunne, W.M.; Muenzer, J.T. Increased susceptibility to Candida infection following cecal ligation and puncture. Biochem. Biophys. Res. Commun. 2011, 414, 37–43. [Google Scholar] [CrossRef]
- Chen, X.P.; Zheng, H.; Li, W.G.; Chen, G.D.; Lu, J.X. Bacteria-induced susceptibility to Candida albicans super-infection in mice via monocyte methyltransferase Setdb2. Cell Microbiol. 2018, 20, e12860. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Finkelman, M.; Worasilchai, N.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1-->3)-beta-D-glucan. PLoS ONE 2017, 12, e0181439. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Worasilchai, N.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Gastrointestinal colonization of Candida albicans increases serum (1-->3)-beta-D-glucan, without candidemia, and worsens cecal ligation and puncture sepsis in murine model. Shock 2018, 49, 62–70. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Worasilchai, N.; Wannalerdsakun, S.; Jutivorakool, K.; Somparn, P.; Issara-Amphorn, J.; Tachaboon, S.; Srisawat, N.; Finkelman, M.; Chindamporn, A. Gastrointestinal leakage detected by serum (1-->3)-beta-D-glucan in mouse models and a pilot study in patients with sepsis. Shock 2016, 46, 506–518. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Hang, J.P.; Zhang, L.; Wang, F.; Zhang, D.C.; Gong, F.H. A systematic review and meta-analysis of diagnostic accuracy of serum 1,3-beta-D-glucan for invasive fungal infection: Focus on cutoff levels. J. Microbiol. Immunol. Infect. 2015, 48, 351–361. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Wolf, A.J.; Underhill, D.M. Beta-glucan recognition by the innate immune system. Immunol. Rev. 2009, 230, 38–50. [Google Scholar] [CrossRef]
- Askew, D.S. Aspergillus fumigatus: Virulence genes in a street-smart mold. Curr. Opin. Microbiol. 2008, 11, 331–337. [Google Scholar] [CrossRef]
- Latgé, J.-P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef]
- Rueping, M.J.; Vehreschild, J.J.; Cornely, O.A. Invasive candidiasis and candidemia: From current opinions to future perspectives. Expert Opin. Investig. Drugs 2009, 18, 735–748. [Google Scholar] [CrossRef]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef]
- Niemiec, M.J.; Kapitan, M.; Polke, M.; Jacobsen, I.D. Commensal to Pathogen Transition of Candida albicans. In Reference Module in Life Sciences; Elsevier: New York, NY, USA, 2017. [Google Scholar]
- Prieto, D.; Correia, I.; Pla, J.; Roman, E. Adaptation of Candida albicans to commensalism in the gut. Future Microbiol. 2016, 11, 567–583. [Google Scholar] [CrossRef]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Ch’ng, J.-H.; Chong, K.K.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2018, 1, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Vidal, R.M.; Muhsen, K.; Tennant, S.M.; Svennerholm, A.-M.; Sow, S.O.; Sur, D.; Zaidi, A.K.; Faruque, A.S.; Saha, D.; Adegbola, R. Colonization factors among enterotoxigenic Escherichia coli isolates from children with moderate-to-severe diarrhea and from matched controls in the Global Enteric Multicenter Study (GEMS). PLoS Negl. Trop. Dis. 2019, 13, e0007037. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.C.; Tyrrell, K.L.; Citron, D.M. Lactobacillus species: Taxonomic complexity and controversial susceptibilities. Clin. Infect. Dis. 2015, 60, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Gonzalez, D.; Mavridou, D.A.; Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Hejazi, A.; Falkiner, F.R. Serratia marcescens. J. Med. Microbiol. 1997, 46, 903–912. [Google Scholar] [CrossRef]
- Sandner-Miranda, L.; Vinuesa, P.; Cravioto, A.; Morales-Espinosa, R. The genomic basis of intrinsic and acquired antibiotic resistance in the genus Serratia. Front. Microbiol. 2018, 9, 828. [Google Scholar] [CrossRef]
- Krismer, B.; Weidenmaier, C.; Zipperer, A.; Peschel, A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 2017, 15, 675–687. [Google Scholar] [CrossRef]
- Sause, W.E.; Buckley, P.T.; Strohl, W.R.; Lynch, A.S.; Torres, V.J. Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trend Pharmacol. Sci. 2016, 37, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Engholm, D.H.; Kilian, M.; Goodsell, D.S.; Andersen, E.S.; Kjaergaard, R.S. A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 2017, 41, 854–879. [Google Scholar] [CrossRef] [PubMed]
- Sitkiewicz, I. How to become a killer, or is it all accidental? Virulence strategies in oral streptococci. Mol. Oral Microbiol. 2018, 33, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wollein Waldetoft, K.; Raberg, L. To harm or not to harm? On the evolution and expression of virulence in group A streptococci. Trends Microbiol. 2014, 22, 7–13. [Google Scholar]
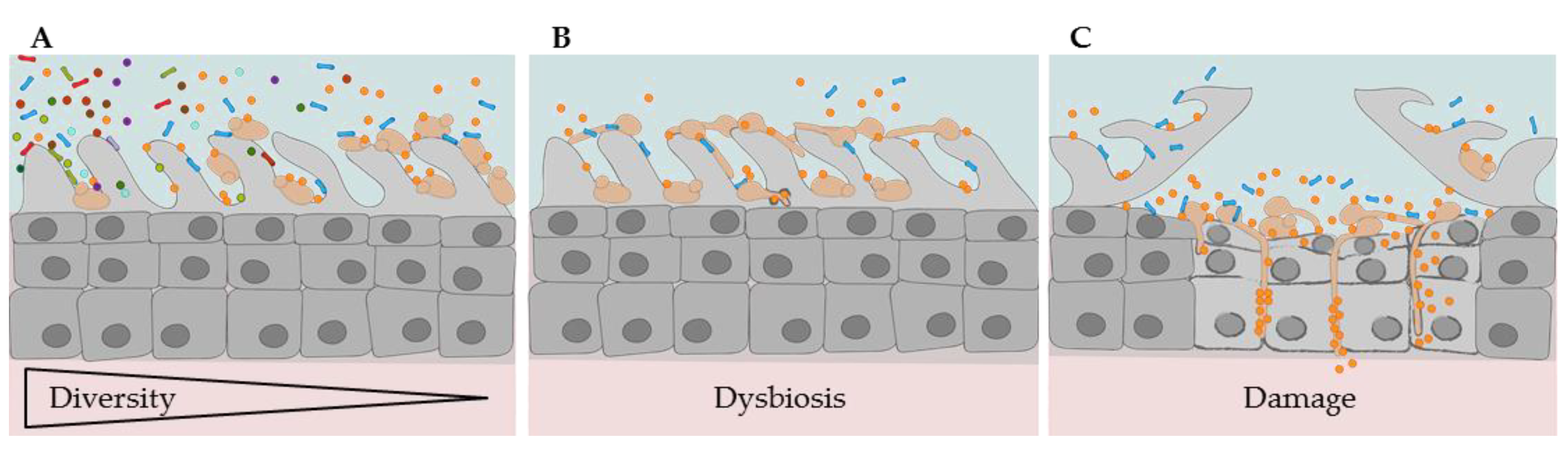
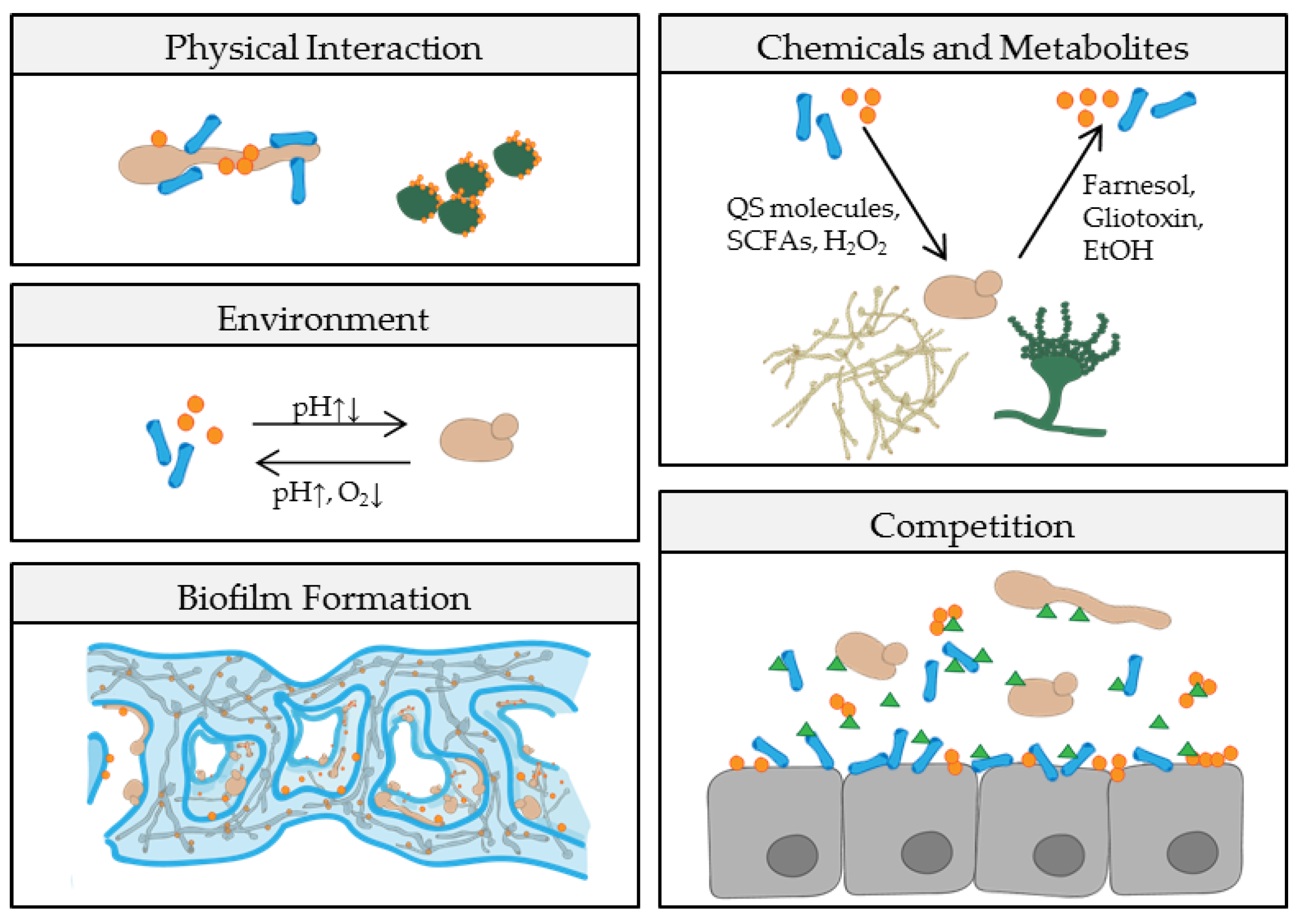
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. https://doi.org/10.3390/pathogens8020070
Krüger W, Vielreicher S, Kapitan M, Jacobsen ID, Niemiec MJ. Fungal-Bacterial Interactions in Health and Disease. Pathogens. 2019; 8(2):70. https://doi.org/10.3390/pathogens8020070
Chicago/Turabian StyleKrüger, Wibke, Sarah Vielreicher, Mario Kapitan, Ilse D. Jacobsen, and Maria Joanna Niemiec. 2019. "Fungal-Bacterial Interactions in Health and Disease" Pathogens 8, no. 2: 70. https://doi.org/10.3390/pathogens8020070
APA StyleKrüger, W., Vielreicher, S., Kapitan, M., Jacobsen, I. D., & Niemiec, M. J. (2019). Fungal-Bacterial Interactions in Health and Disease. Pathogens, 8(2), 70. https://doi.org/10.3390/pathogens8020070






