Role of Daptomycin in the Induction and Persistence of the Viable but Non-Culturable State of Staphylococcus Aureus Biofilms
Abstract
:1. Introduction
2. Results and Discussion
2.1. Biofilm Production, Stress Exposure and Non-Culturability
2.2. Persistence of the VBNC State
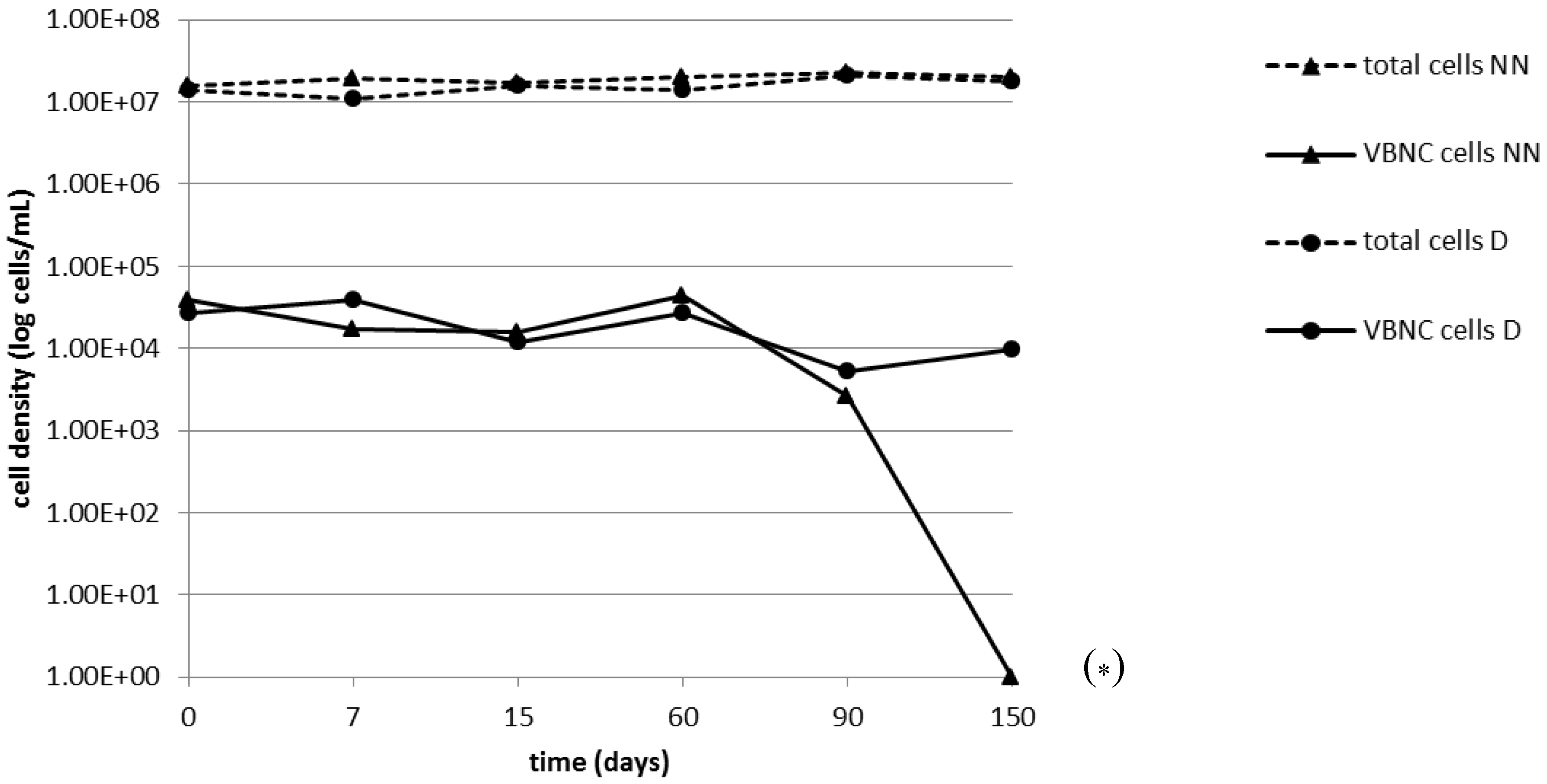
| Stress Condition | Days Since Achievement of Non-Culturability | Cells/mL | |
|---|---|---|---|
| Total | Viable | ||
| Nutrient depletion | 0 | 1.6×107 | 3.8 × 104 |
| 7 | 1.9 × 107 | 1.7 × 104 | |
| 15 | 1.7 × 107/1.5 × 108 * | 1.6 × 104/8.2 × 104 * | |
| 60 | 2.0 × 107 | 4.4 × 104 | |
| 90 | 2.3 × 107 | 2.6 × 103 | |
| 150 | 2.0 × 107 | <60 | |
| Nutrient depletion+ daptomycin | 0 | 1.4 × 107 | 2.7 × 104 |
| 7 | 1.1 × 107 | 3.9 × 104 | |
| 15 | 1.6 × 107/1.4 × 108 * | 1.2 × 104/7.6 × 104 * | |
| 60 | 1.4 × 107 | 2.7 × 104 | |
| 90 | 2.1 × 107 | 5.3 × 103 | |
| 150 | 1.8 × 107 | 9.8 × 103 | |
2.3. Gene Expression of VBNC Cells
| Stress Condition | Time from non Culturability | Gene Analysis | |||
|---|---|---|---|---|---|
| 16SrDNA | glt | nuc | mecA | ||
| Nutrient depletion | T0 | + | ND | ND | ND |
| T7 | + | - | - | + | |
| T15 | + | - | - | + | |
| T60 | + | - | - | + | |
| Nutrient depletion + daptomycin | T0 | + | ND | ND | ND |
| T7 | + | + | - | + | |
| T15 | + | + | - | + | |
| T60 | + | - | - | + | |
3. Experimental Section
3.1. Bacterial Strains, Media, Antibiotics and Enzymes
3.2. MIC Determination
3.3. Biofilm Production, Stress Exposure and Culturability Assays
3.4. Epifluorescence Microscopy and Flow Cytometry
3.5. Real-Time RT–PCR Assays
| Target Gene | Gene Function | Primer Pair (5ʹ-3ʹ) | Annealing Temperature (°C) | Product Size (bp) | Reference |
|---|---|---|---|---|---|
| 16S rDNA | Housekeeping | F-TGGAGCATGTGGTTTAATTCGA R-TGCGGGACTTAACCCAACA | 60 | 159 | [32] |
| glt | Species specific, coding for glutamate synthase | F-AATCTTTGTCGGTACACGATATTCTTCACG R-CGTAATGAGATTTCAGTAGATAATACAACA | 58 | 108 | [33] |
| nuc | Virulence factor, coding for thermonuclease | F-GACTATTATTGGTTGATCCACCTG R- GCCTTGACGAACTAAAGCTTCG | 60 | 218 | [34] |
| mecA | Methicillin resistance | F-TCCAGATTACAACTTCACCAGG R-CCACTTCATATCTTGTAACG | 57 | 162 | [35] |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Donlan, R.; Costerton, J. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Francolini, I.; Donelli, G. Prevention and control of biofilm-based medical- device-related infections. FEMS Immunol. Med. Microbiol. 2010, 59, 227–238. [Google Scholar] [PubMed]
- Zandri, G.; Pasquaroli, S.; Vignaroli, C.; Talevi, S.; Manso, E.; Donelli, G.; Biavasco, F. Detection of viable but non-culturable staphylococci in biofilms from central venous catheters negative on standard microbiological assays. Clin. Microbiol. Infect. 2012, 18, 259–261. [Google Scholar] [CrossRef]
- Chambers, H.F. Methicillin resistance in staphylococci: Molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 1997, 10, 781–791. [Google Scholar] [PubMed]
- Enoch, D.A.; Bygott, J.M.; Daly, M.L.; Karas, J.A. Daptomycin. J. Infect. 2007, 55, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Tarai, B.; Das, P.; Kumar, D. Recurrent challenges for clinicians: Emergence of methicillin-resistant Staphylococcus aureus, vancomycin resistance, and current treatment options. J. Lab. Phys. 2013, 5, 71–78. [Google Scholar]
- Popiel, K.Y.; Miller, M.A. Evaluation of Vancomycin-Resistant Enterococci (VRE)-associated morbidity following relaxation of VRE screening and isolation precautions in a tertiary care hospital. Infect. Control Hosp. Epidemiol. 2014, 35, 818–825. [Google Scholar] [CrossRef] [PubMed]
- Carugati, M.; Bayer, A.S.; Miró, J.M.; Park, L.P.; Guimarães, A.C.; Skoutelis, A.; Fortes, C.Q.; Durante-Mangoni, E.; Hannan, M.M.; Nacinovich, F.; et al. High-Dose daptomycin therapy for left-sided infective endocarditis: A prospective study from the international collaboration on endocarditis. Antimicrob. Agents Chemother. 2013, 57, 6213–6222. [Google Scholar] [CrossRef] [PubMed]
- Robbel, L.; Marahiel, M.A. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 2010, 285, 27501–27508. [Google Scholar] [CrossRef] [PubMed]
- Raad, I.; Hanna, H.; Jiang, Y.; Dvorak, T.; Reitzel, R.; Chaiban, G.; Sherertz, R.; Hachem, R. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob. Agents Chemother. 2007, 51, 1656–1660. [Google Scholar] [CrossRef] [PubMed]
- Barber, K.E.; Werth, B.J.; McRoberts, J.P.; Rybak, M.J. A novel approach utilizing biofilm time-kill curves to assess the bactericidal activity of ceftaroline combinations against biofilm-producing methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 2989–2992. [Google Scholar] [CrossRef] [PubMed]
- Len, O.; Montejo, M.; Cervera, C.; Fariñas, M.C.; Sabé, N.; Ramos, A.; Cordero, E.; Torre-Cisneros, J.; Martín-Dávila, P.; Azanza, J.R.; et al. Daptomycin is safe and effective for the treatment of gram-positive cocci infections in solid organ transplantation. Transpl. Infect. Dis. 2014. [Google Scholar] [CrossRef]
- Vilhena, C.; Bettencourt, A. Daptomycin: A review of properties, clinical use, drug delivery and resistance. Mini Rev. Med. Chem. 2012, 12, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Kelesidis, T. The interplay between daptomycin and the immune system. Front. Immunol. 2014, 5, 52. [Google Scholar] [PubMed]
- Agarwal, A.; Singh, K.P.; Jain, A. Medical significance and management of staphylococcal biofilm. FEMS Immunol. Med. Microbiol. 2010, 58, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Meije, Y.; Almirante, B.; del Pozo, J.L.; Martín, M.T.; Fernández-Hidalgo, N.; Shan, A.; Basas, J.; Pahissa, A.; Gavaldà, J. Daptomycin is effective as antibiotic-lock therapy in a model of Staphylococcus aureus catheter-related infection. J. Infect. 2014, 68, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, A.; Rasmussen, M. Antibiotic regimens with rifampicin for treatment of Enterococcus faecium in biofilms. Int. J. Antimicrob. Agents 2014, 44, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. Recent findings on the viable but non-culturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2009, 34, 415–425. [Google Scholar] [PubMed]
- Nowakowska, J.; Oliver, J.D. Resistance to environmental stresses by Vibrio vulnificus in the viable but non-culturable state. FEMS Microbiol. Ecol. 2013, 84, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Pasquaroli, S.; Zandri, G.; Vignaroli, C.; Vuotto, C.; Donelli, G.; Biavasco, F. Antibiotic pressure can induce the viable but non-culturable state in Staphylococcus aureus growing in biofilms. J. Antimicrob. Chemother. 2013, 68, 1812–1817. [Google Scholar]
- Pascoe, B.; Dams, L.; Wilkinson, T.S.; Harris, L.G.; Bodger, O.; Mack, D.; Davies, A.P. Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS One 2014, 9, e85998. [Google Scholar] [CrossRef] [PubMed]
- El-Azizi, M.; Rao, S.; Kanchanapoom, T.; Khardori, N. In vitro activity of vancomycin, quinupristin/dalfopristin, and linezolid against intact and disrupted biofilms of staphylococci. Ann. Clin. Microbiol. Antimicrob. 2005, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donelli, G.; Francolini, I.; Romoli, D.; Guaglianone, E.; Piozzi, A.; Ragunath, C.; Kaplan, J.B. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob. Agents Chemother. 2007, 51, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Lahtinen, S.J.; Ahokoski, H.; Reinikainen, J.P.; Gueimonde, M.; Nurmi, J.; Ouwehand, A.C.; Salminen, S.J. Degradation of 16S rRNA and attributes of viability of viable but non-culturable probiotic bacteria. Lett. Appl. Microbiol. 2008, 46, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Wiltshire, M.D.; Foster, S.J. Identification and analysis of Staphylococcus aureus components expressed by a model system of growth in serum. Infect. Immun. 2001, 69, 5198–5202. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I.; Shahamat, M.; Kirchman, P.A.; Russek-Cohen, E.; Colwell, R.R. Methionine uptake and cytopathogenicity of viable but non-culturable Shigella dysenteriae type 1. Appl. Environ. Microb. 1994, 60, 3573–3578. [Google Scholar]
- Kiedrowski, M.R.; Kavanaugh, J.S.; Malone, C.L.; Mootz, J.M.; Voyich, J.M.; Smeltzer, M.S.; Bayles, K.W.; Horswill, A.R. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 2011, 6, e26714. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Spiegelman, G.B.; Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006, 9, 445–453. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 4.0. 2014. Available online: http://www.eucast.org (accessed on 17 September 2014).
- Warwick, S.; Wilks, M.; Hennessy, E.; Powell-Tuck, J.; Small, M.; Sharp, J.; Millar, M.R. Use of quantitative 16S ribosomal DNA detection for diagnosis of central vascular catheter-associated bacterial infection. J. Clin. Microbiol. 2004, 42, 1402–1408. [Google Scholar] [CrossRef] [PubMed]
- Martineau, F.; Picard, F.J.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Species-Specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 1998, 36, 618–623. [Google Scholar] [PubMed]
- Depardieu, F.; Perichon, B.; Courvalin, P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 2004, 42, 5857–5860. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; Milheiric, C.; de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3374–3377. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pasquaroli, S.; Citterio, B.; Cesare, A.D.; Amiri, M.; Manti, A.; Vuotto, C.; Biavasco, F. Role of Daptomycin in the Induction and Persistence of the Viable but Non-Culturable State of Staphylococcus Aureus Biofilms. Pathogens 2014, 3, 759-768. https://doi.org/10.3390/pathogens3030759
Pasquaroli S, Citterio B, Cesare AD, Amiri M, Manti A, Vuotto C, Biavasco F. Role of Daptomycin in the Induction and Persistence of the Viable but Non-Culturable State of Staphylococcus Aureus Biofilms. Pathogens. 2014; 3(3):759-768. https://doi.org/10.3390/pathogens3030759
Chicago/Turabian StylePasquaroli, Sonia, Barbara Citterio, Andrea Di Cesare, Mehdi Amiri, Anita Manti, Claudia Vuotto, and Francesca Biavasco. 2014. "Role of Daptomycin in the Induction and Persistence of the Viable but Non-Culturable State of Staphylococcus Aureus Biofilms" Pathogens 3, no. 3: 759-768. https://doi.org/10.3390/pathogens3030759





