Can Oral Fluids Replace Nasal Swabs in Swine Influenza A Virus (swIAV) PCR Diagnostics?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. swIAV Detection and Molecular Subtyping
2.3. Statistical Analyses
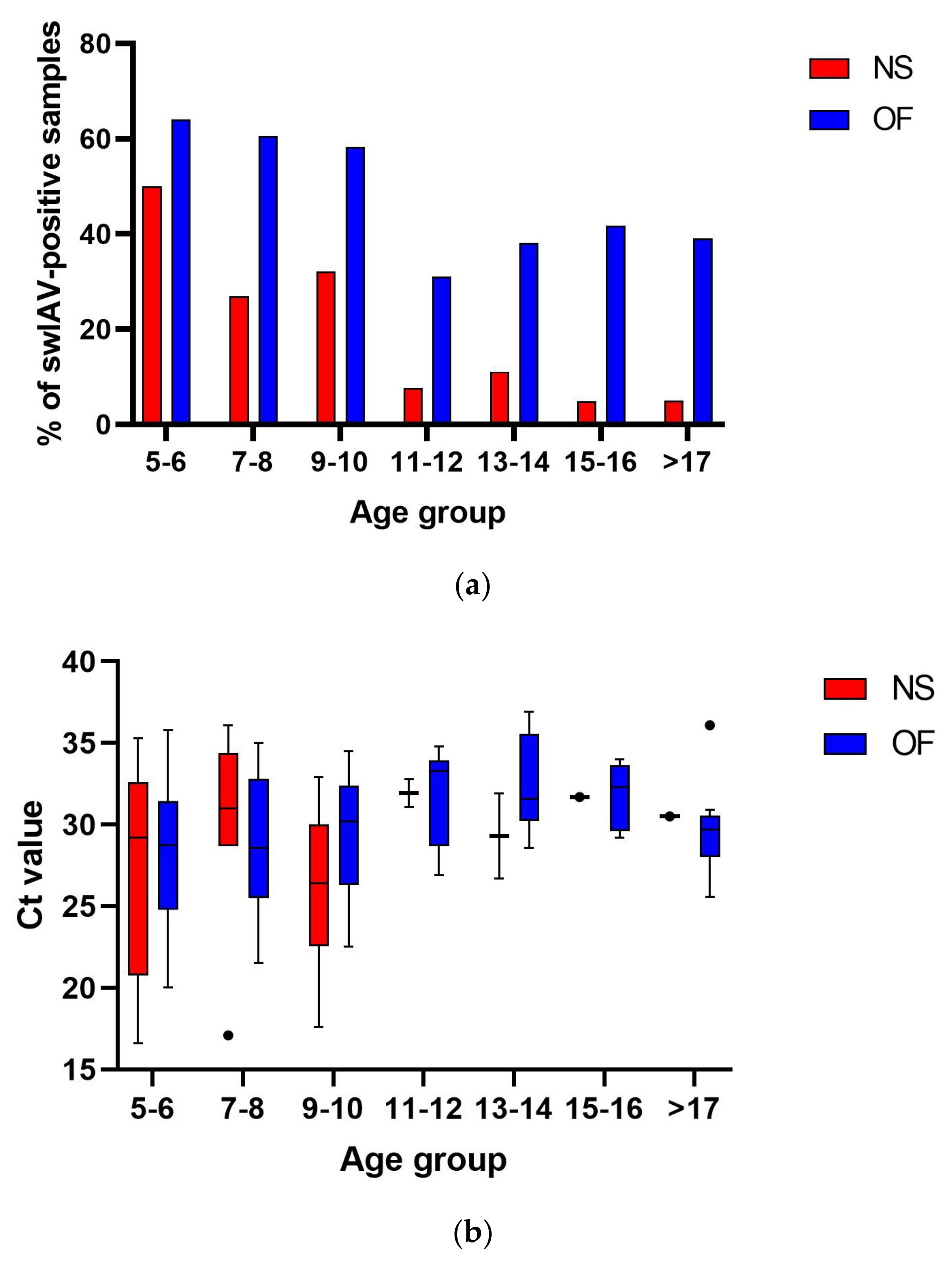
3. Results
3.1. The Prevalence of swIAV in Herds with Influenza-like Clinical Signs
3.2. Subtyping of swIAV from Polish Pig Herds
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IAV | influenza A virus |
| swIAV | swine influenza A virus |
| NS | nasal swab |
| OF | oral fluid |
| HA | hemagglutinin |
| NA | neuraminidase |
| PRDC | porcine respiratory disease complex |
| MDA | maternally derived antibody |
| PRRSV | porcine reproductive and respiratory syndrome |
| PPIV-1 | porcine parainfluenza virus 1 |
References
- Van Reeth, K.; Vincent, A.L. Influenza viruses. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Willey and Sons Inc.: Ames, IA, USA, 2019; pp. 480–504. [Google Scholar]
- Diaz, A.; Perez, A.; Sreevatsan, S.; Davies, P.; Culhane, M.; Torremorell, M. Association between influenza A virus infection and pigs subpopulations in endemically infected breeding herds. PLoS ONE 2015, 10, e0129213. [Google Scholar] [CrossRef] [PubMed]
- Mancera Gracia, J.C.; Pearce, D.S.; Masic, A.; Balasch, M. Influenza A Virus in Swine: Epidemiology, Challenges and Vaccination Strategies. Front. Vet. Sci. 2020, 7, 647. [Google Scholar] [CrossRef] [PubMed]
- Ryt-Hansen, P.; Pedersen, A.G.; Larsen, I.; Krog, J.S.; Kristensen, C.S.; Larsen, L.E. Acute Influenza A virus outbreak in an enzootic infected sow herd: Impact on viral dynamics, genetic and antigenic variability and effect of maternally derived antibodies and vaccination. PLoS ONE 2019, 14, e0224854. [Google Scholar] [CrossRef] [PubMed]
- Decorte, I.; Steensels, M.; Lambrecht, B.; Cay, A.B.; De Regge, N. Detection and Isolation of Swine Influenza A Virus in Spiked Oral Fluid and Samples from Individually Housed, Experimentally Infected Pigs: Potential Role of Porcine Oral Fluid in Active Influenza A Virus Surveillance in Swine. PLoS ONE 2015, 10, e0139586. [Google Scholar] [CrossRef] [PubMed]
- Pardo, F.O.C.; Wayne, S.; Culhane, M.R.; Perez, A.; Allerson, M.; Torremorell, M. Effect of strain-specific maternally-derived antibodies on influenza A virus infection dynamics in nursery pigs. PLoS ONE 2019, 14, e0210700. [Google Scholar]
- Simon, G.; Larsen, L.E.; Dürrwald, R.; Foni, E.; Harder, T.; Van Reeth, K.; Markowska-Daniel, I.; Reid, S.M.; Dan, A.; Maldonado, J.; et al. European surveillance network for influenza in pigs: Surveillance programs, diagnostic tools and Swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS ONE 2014, 9, e115815. [Google Scholar] [CrossRef] [PubMed]
- Henritzi, D.; Petric, P.P.; Lewis, N.S.; Graaf, A.; Pessia, A.; Starick, E.; Breithaupt, A.; Strebelow, G.; Luttermann, C.; Parker, L.M.K.; et al. Surveillance of European Domestic Pig Populations Identifies an Emerging Reservoir of Potentially Zoonotic Swine Influenza A Viruses. Cell Host Microbe 2020, 28, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Byrne, A.; European Swine Influenza Network. European Swine Influenza Network-swIAV Evolution and Diversity in Europe Report #1: October 2022–September 2023. Available online: https://www.openagrar.de/servlets/MCRFileNodeServlet/openagrar_derivate_00057345/ESFLU_WG2_Report_1.pdf (accessed on 7 July 2025).
- Gautier, R.; Ryt-Hansen, P.; Byrne, A.; European Swine Influenza Network. European Swine Influenza Network Report #2 on Swine Influenza A Viruses Evolution and Diversity in Europe from October 2022 to September 2024. Available online: https://zenodo.org/records/15673794 (accessed on 7 July 2025).
- Panyasing, Y.; Goodell, C.; Kittawornrat, A.; Wang, C.; Levis, I.; Desfresne, L.; Rauh, R.; Gauger, P.C.; Zhang, J.; Lin, X.; et al. Influenza A Virus Surveillance Based on Pre-Weaning Piglet Oral Fluid Samples. Transbound. Emerg. Dis. 2016, 63, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Biernacka, K.; Karbowiak, P.; Wróbel, P.; Charęza, T.; Czopowicz, M.; Balka, G.; Goodell, C.; Rauh, R.; Stadejek, T. Detection of porcine reproductive and respiratory syndrome virus (PRRSV) and influenza A virus (IAV) in oral fluid of pigs. Res. Vet. Sci. 2016, 109, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Detmer, S.E.; Patnayak, D.P.; Jiang, Y.; Gramer, M.R.; Goyal, S.M. Detection of Influenza A virus in porcine oral fluid samples. J. Vet. Diagn. Investig. 2011, 23, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Woźniak, A.; Pauwels, A.; Coppens, S.; Nauwynck, H.; Cybulski, P.; Theuns, S.; Stadejek, T. Successful Whole Genome Nanopore Sequencing of Swine Influenza A Virus (swIAV) Directly from Oral Fluids Collected in Polish Pig Herds. Viruses 2023, 15, 435. [Google Scholar] [CrossRef] [PubMed]
- Price, R.H.M.; Graham, C.; Ramalingam, S. Association between viral seasonality and meteorological factors. Sci. Rep. 2019, 9, 929. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Chan, M.; Vander Zaag, A. Inactivation of avian influenza viruses on porous and non-porous surfaces is enhanced by elevating absolute humidity. Transbound. Emerg. Dis. 2017, 64, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Pardo, F.O.C.; Alba-Casals, A.; Nerem, J.; Morrison, R.B.; Puig, P.; Torremorell, M. Influenza herd-level prevalence and seasonality in breed-to-wean pig farms in the Midwestern United States. Front. Vet. Sci. 2017, 4, 167. [Google Scholar] [CrossRef]
- Ryt-Hansen, P.; Pedersen, A.; Larsen, I.; Kristensen, C.S.; Krog, J.; Wacheck, S.; Larsen, L.E. Substantial Antigenic Drift in the Hemagglutinin Protein of Swine Influenza A Viruses. Viruses 2020, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Henritzi, D.; Zhao, N.; Starick, E.; Simon, G.; Krog, J.S.; Larsen, L.E.; Reid, S.M.; Brown, I.H.; Chiapponi, C.; Foni, E.; et al. Rapid detection and subtyping of European swine influenza viruses in porcine clinical samples by haemagglutininand neuraminidase-specific tetra- and triplex real-time RT-PCRs. Influenza Other Respir. Viruses 2016, 6, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Cador, C.; Arnaud, M.; Willem, L.; Rose, N. Control of endemic swine flu persistence in farrow– to-finish pig farms: A stochastic metapopulation medeling assessment. Vet. Res. 2017, 48, 58. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, T.R.; Ryt-Hansen, P.; Nielsen, J.P.; Larsen, L.E.; Larsen, I.; Chamings, A.; Goecke, N.B.; Alexandersen, S. Infection Dynamics of Swine Influenza Virus in a Danish Pig Herd Reveals Recurrent Infections with Different Variants of the H1N2 Swine Influenza A Virus Subtype. Viruses 2020, 12, 1013. [Google Scholar] [CrossRef] [PubMed]
- Pardo, F.O.C.; Schelkopf, A.; Allerson, M.; Morrison, R.; Culhane, M.; Perez, A.; Torremorell, M. Breed-to-wean farm factors associated with influenza A virus infection in piglets at weaning. Prev. Vet. Med. 2018, 161, 33–40. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woźniak, A.; Cybulski, P.; Ryt-Hansen, P.; Larsen, L.E.; Biernacka, K.; Miłek, D.; Stadejek, T. Can Oral Fluids Replace Nasal Swabs in Swine Influenza A Virus (swIAV) PCR Diagnostics? Pathogens 2025, 14, 808. https://doi.org/10.3390/pathogens14080808
Woźniak A, Cybulski P, Ryt-Hansen P, Larsen LE, Biernacka K, Miłek D, Stadejek T. Can Oral Fluids Replace Nasal Swabs in Swine Influenza A Virus (swIAV) PCR Diagnostics? Pathogens. 2025; 14(8):808. https://doi.org/10.3390/pathogens14080808
Chicago/Turabian StyleWoźniak, Aleksandra, Piotr Cybulski, Pia Ryt-Hansen, Lars Erik Larsen, Kinga Biernacka, Dagmara Miłek, and Tomasz Stadejek. 2025. "Can Oral Fluids Replace Nasal Swabs in Swine Influenza A Virus (swIAV) PCR Diagnostics?" Pathogens 14, no. 8: 808. https://doi.org/10.3390/pathogens14080808
APA StyleWoźniak, A., Cybulski, P., Ryt-Hansen, P., Larsen, L. E., Biernacka, K., Miłek, D., & Stadejek, T. (2025). Can Oral Fluids Replace Nasal Swabs in Swine Influenza A Virus (swIAV) PCR Diagnostics? Pathogens, 14(8), 808. https://doi.org/10.3390/pathogens14080808




