Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Material and Fusarium Inoculation Under Controlled Conditions
2.2. Material and Fusarium Genotype Analysis
Evaluation of the Species Affiliation of the Pathogenic Fungus
2.3. Selection of Candidate Genes and Identification of Associated Molecular Markers
2.4. DNA Isolation
2.5. PCR Conditions
2.6. Electrophoresis
2.7. Quantitative (Real-Time) PCR
2.7.1. RNA Isolation
2.7.2. Primer Design and Conditions
2.7.3. Selection of Reference Genes for Quantitative PCR. Development of Standard Curves Genes
2.8. Transcriptomics Analysis
2.9. Statistical Analysis
3. Results
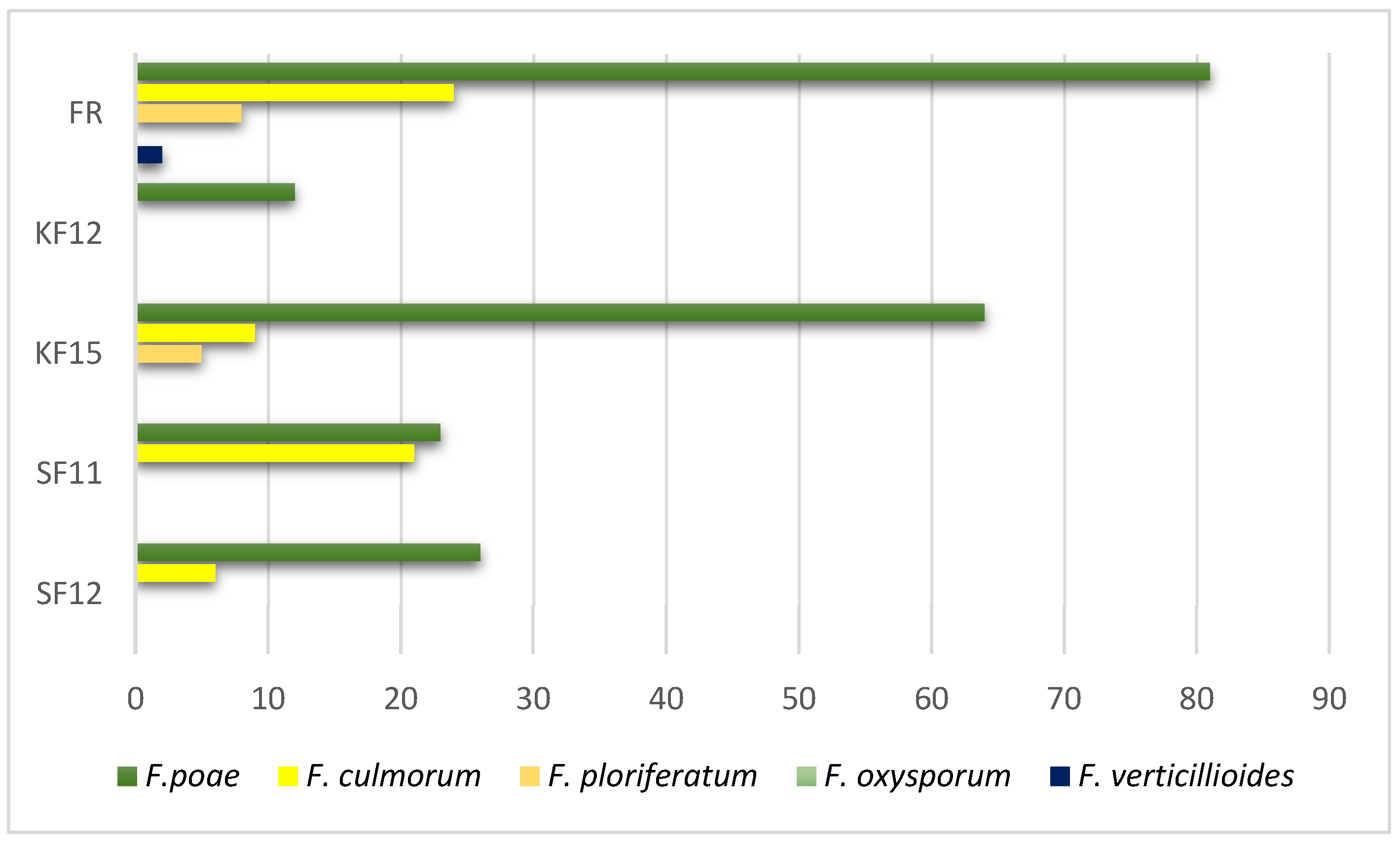
3.1. Colonization of Different Maize Varieties by Five Fusarium Species
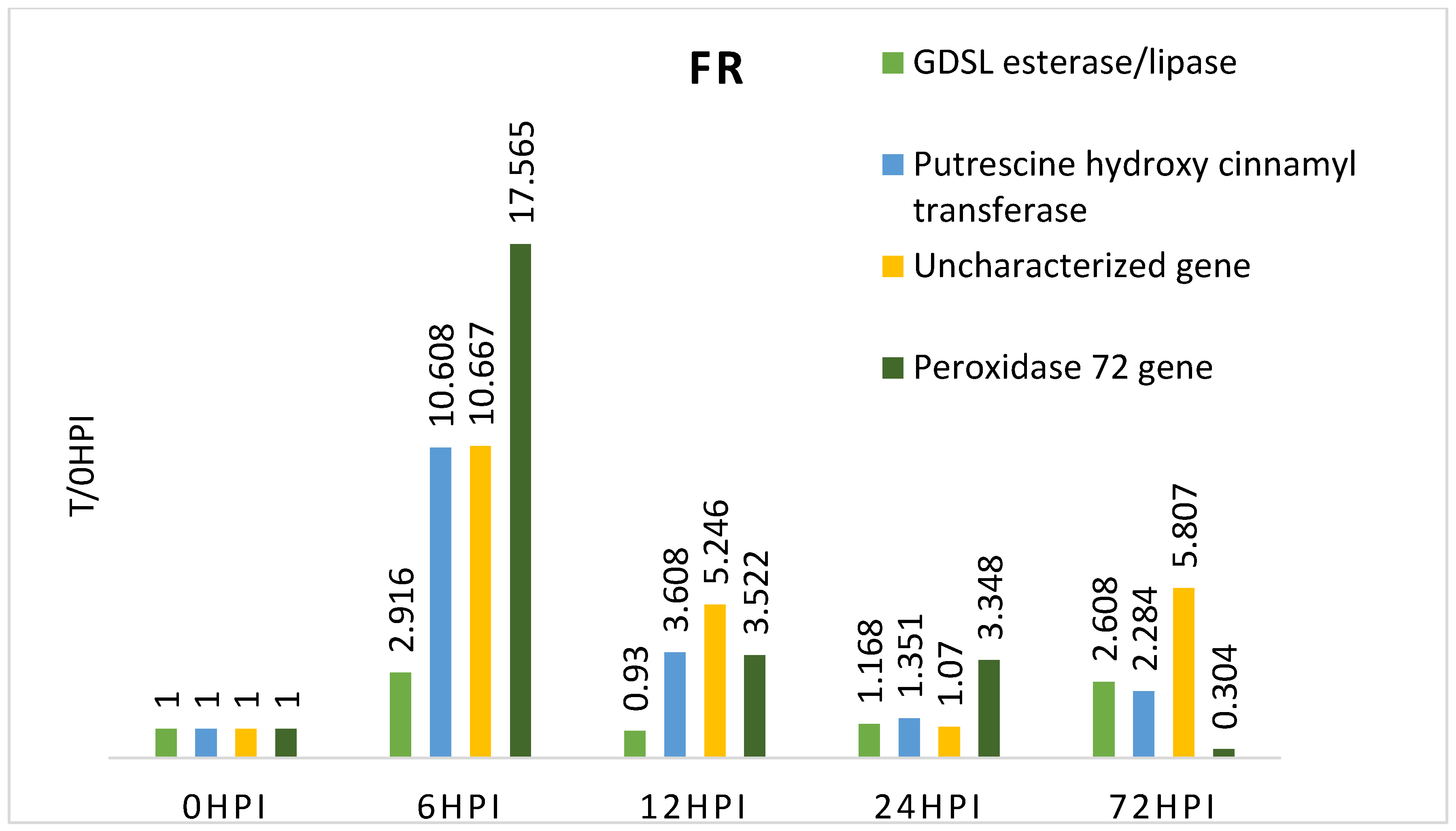
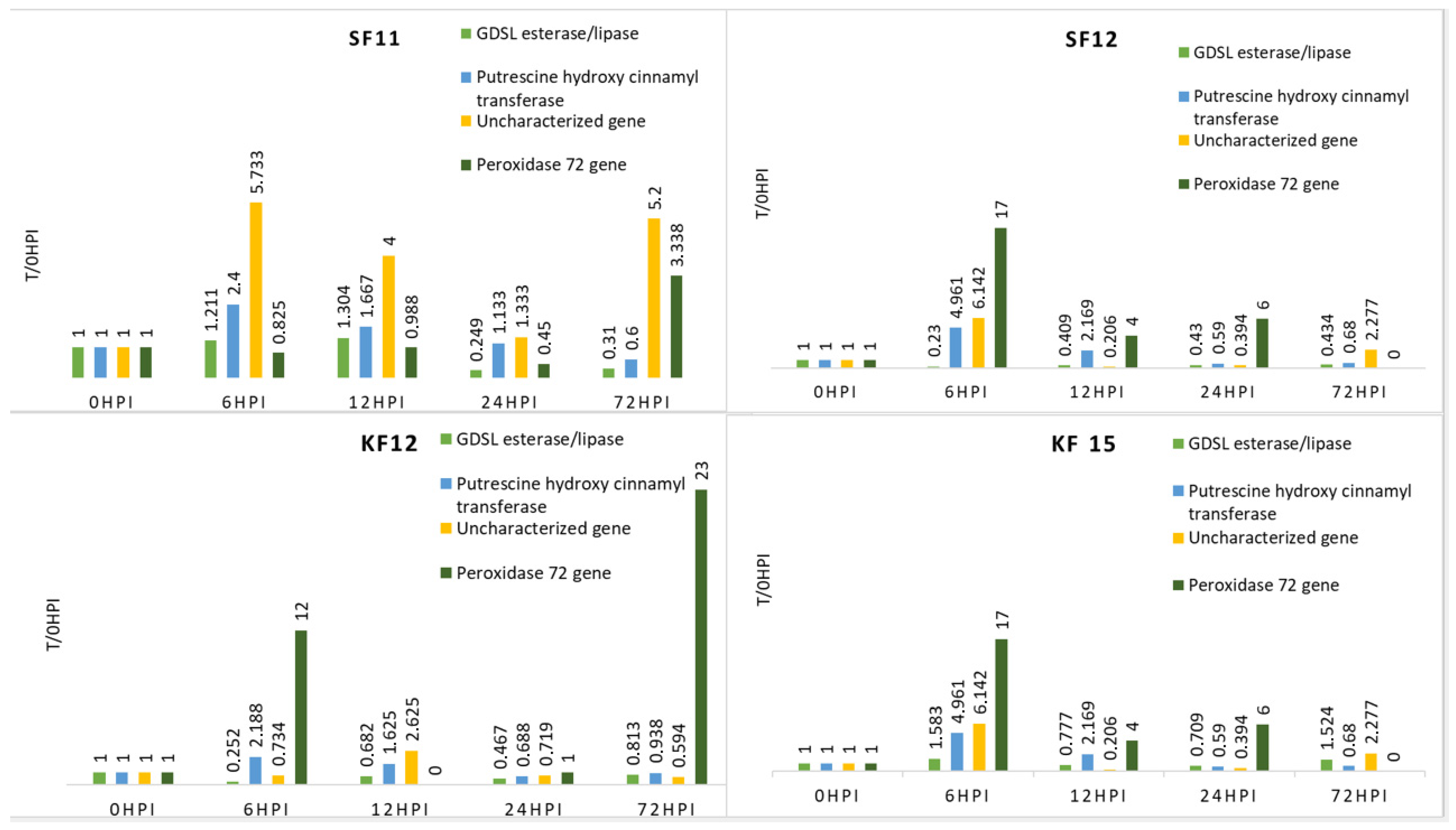
3.2. Expression of Candidate Genes Linked to Fusiarium Resistance
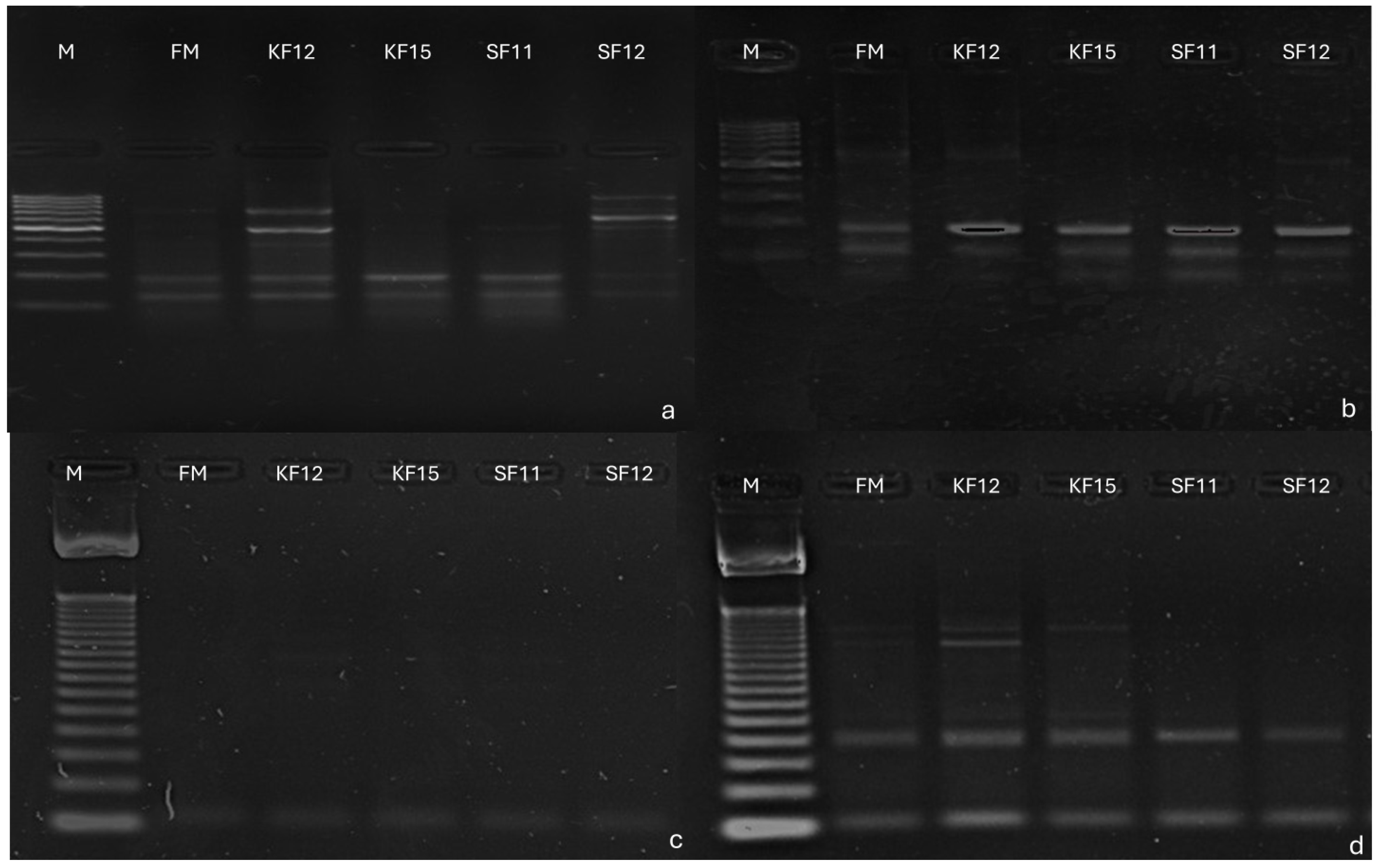
3.3. Identification of SilicoDArT Markers Linked to Candidate Genes Determining Fusarium Resistance in Maize
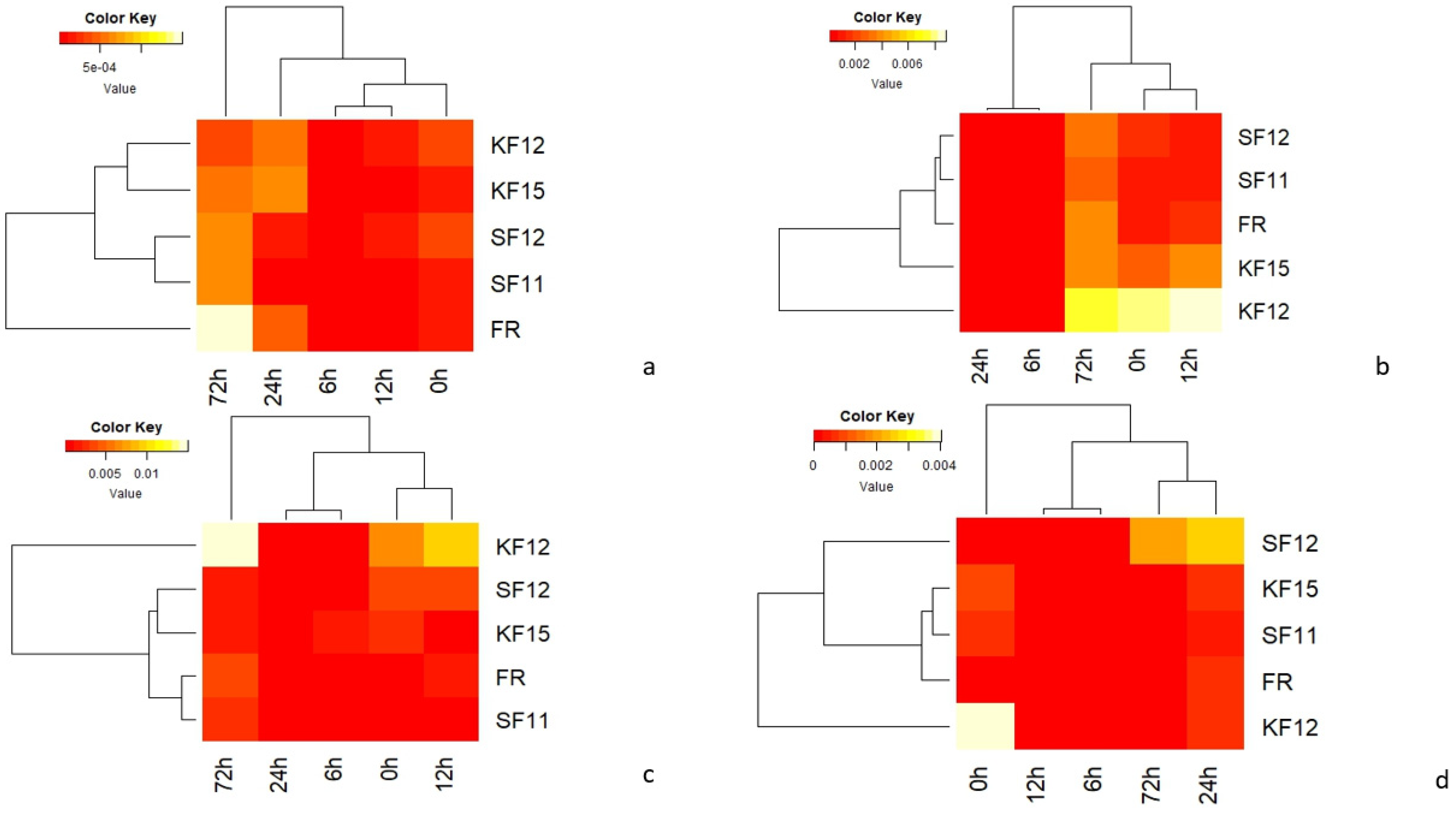
3.4. Transcriptomic Data Analysis
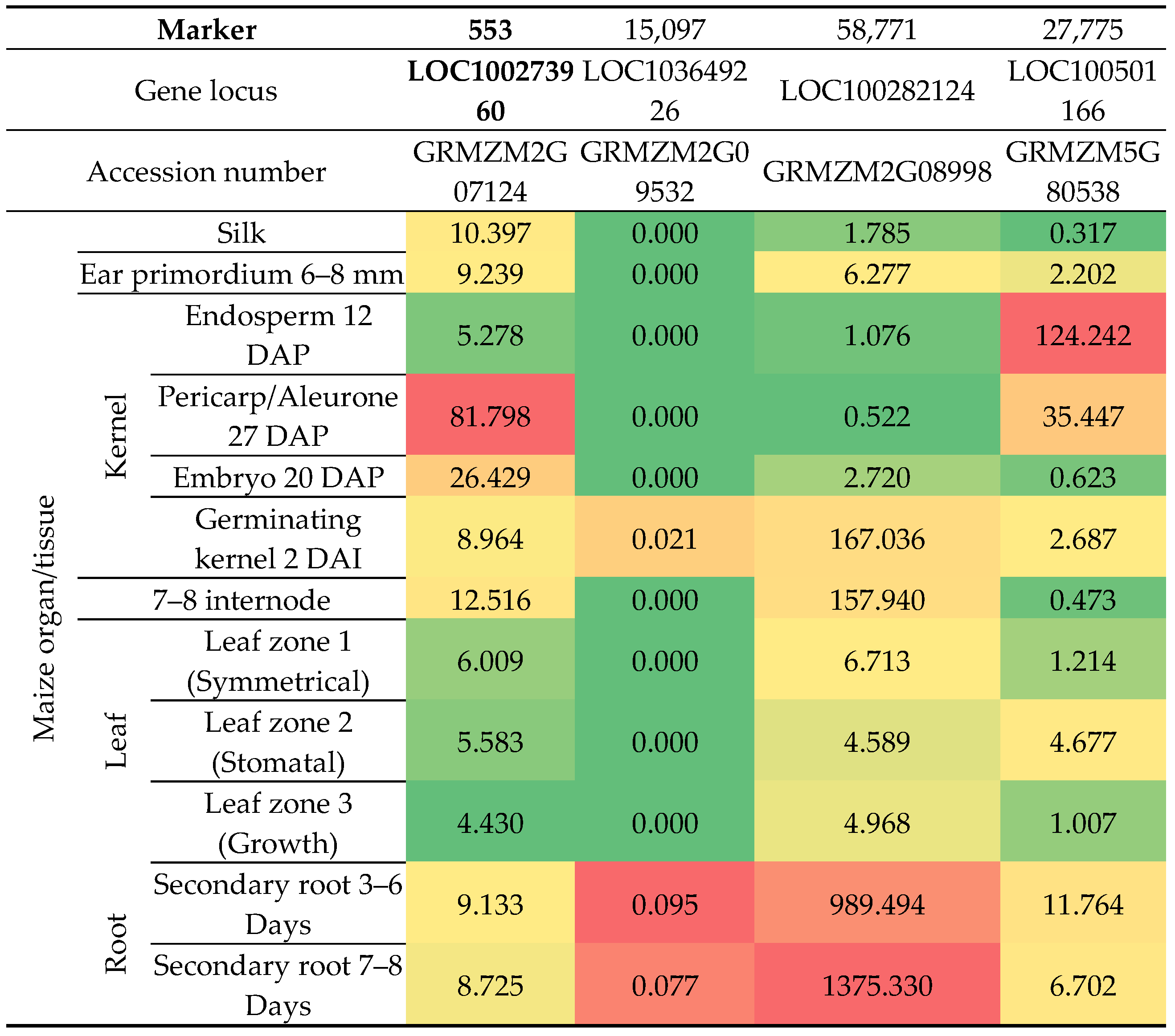
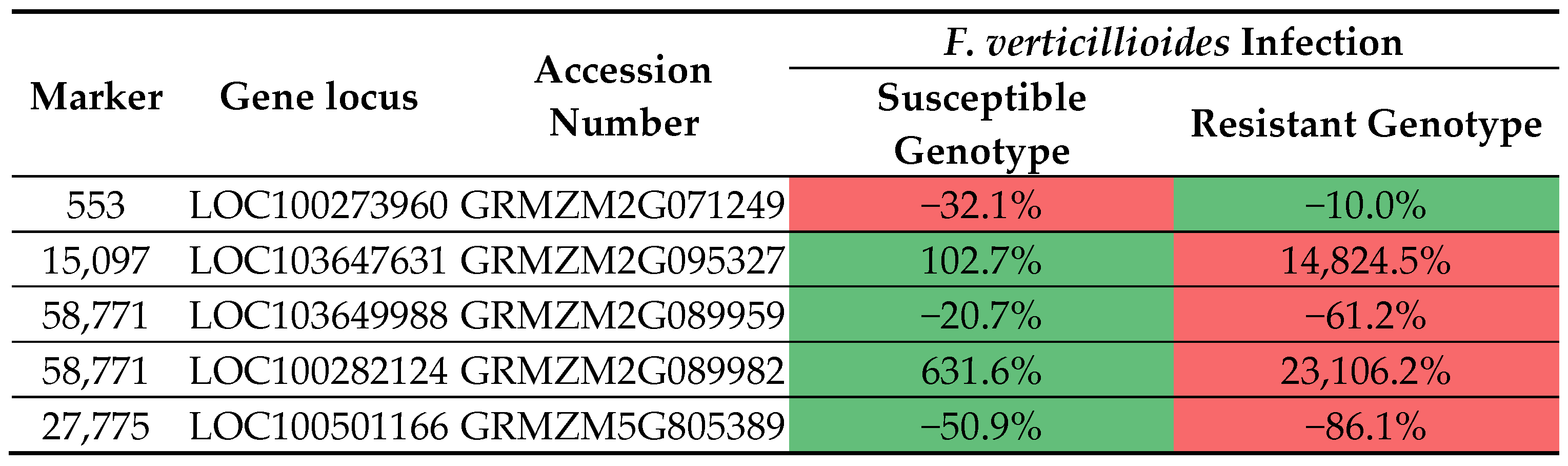
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Awika, J. Major cereal grains production and use around the world. In Advances in Cereal Science: Implications to Food Processing and Health Promotion; Awika, J.M., Piironen, V., Bean, S., Eds.; American Chemical Society: Atlantic City, NJ, USA; Washington, DC, USA, 2011; pp. 1–13. [Google Scholar] [CrossRef]
- Kennett, D.J.; Prufer, K.M.; Culleton, B.J.; George, R.J.; Robinson, M.; Trask, W.R.; Buckley, G.M.; Moes, E.; Kate, E.J.; Harper, T.K.; et al. Early isotopic evidence for maize as a staple grain in the Americas. Sci. Adv. 2020, 6, eaba3245. [Google Scholar] [CrossRef]
- García-Lara, S.; Serna-Saldivar, S.O. Corn History and Culture. In Corn, 3rd ed.; Serna-Saldivar, S.O., Ed.; AACC International Press: Oxford, UK, 2019; pp. 1–18. [Google Scholar] [CrossRef]
- FAOSTAT. Statistical Database of the Food and Agriculture of the United Nations; FAO: Rome, Italy, 2021; Available online: http://www.fao.org (accessed on 10 April 2021).
- Dragomir, V.; Ioan Sebastian, B.; Alina, B.; Victor, P.; Tanasă, L.; Horhocea, D. An overview of global maize market compared to Romanian production. Rom. Agric. Res. 2022, 39, 535–544. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Maragos, C.M.; Proctor, R.H. Maize ear rot and moniliformin contamination by cryptic species of Fusarium subglutinans. J. Agric. Food Chem. 2006, 54, 7383–7390. [Google Scholar] [CrossRef] [PubMed]
- Pfordt, A.; Ramos Romero, L.; Schiwek, S.; Karlovsky, P.; von Tiedemann, A. Impact of Environmental Conditions and Agronomic Practices on the Prevalence of Fusarium Species Associated with Ear-and Stalk Rot in Maize. Pathogens 2020, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.; Yu, J.H. Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef]
- Bryła, M.; Pierzgalski, A.; Zapaśnik, A.; Uwineza, P.A.; Ksieniewicz-Woźniak, E.; Modrzewska, M.; Waśkiewicz, A. Recent Research on Fusarium Mycotoxins in Maize—A Review. Foods 2022, 11, 3465. [Google Scholar] [CrossRef]
- Chukwudi, U.P.; Kutu, F.R.; Mavengahama, S. Mycotoxins in maize and implications on food security: A Review. Agric. Rev. 2021, 140, 42–49. [Google Scholar] [CrossRef]
- Bouwer, G.A. Framework for effective Bt maize IRM Programs: Incorporation of lessons learned from Busseola fusca resistance development. Front. Bioeng. Biotechnol. 2020, 8, 717. [Google Scholar] [CrossRef]
- Zila, C.T.; Ogut, F.; Romay, M.C.; Gardner, C.A.; Buckler, E.S.; Holland, J.B. Genome-wide association study of Fusarium ear rot disease in the U.S.A. maize inbred line collection. BMC Plant Biol. 2014, 14, 372. [Google Scholar] [CrossRef]
- Chen, J.; Shrestha, R.; Ding, J.; Zheng, H.; Mu, C.; Wu, J.; Mahuku, G. Genome-wide association study and QTL mapping reveal genomic loci associated with Fusarium ear rot resistance in tropical maize germplasm. G3-Genes Genomes Genet. 2016, 6, 3803–3815. [Google Scholar] [CrossRef]
- Ayesiga, S.B.; Rubaihayo, P.; Oloka, B.M.; Dramadri, I.O.; Sserumaga, J.P. Genome-wide association study and pathway analysis to decipher loci associated with Fusarium ear rot resistance in tropical maize germplasm. Genet. Resour. Crop Evol. 2024, 71, 2435–2448. [Google Scholar] [CrossRef]
- Martin, M.; Schipprack, W.; Miedaner, T.; Dhillon, B.S.; Kessel, B.; Ouzunova, M.; Melchinger, A. Variation and covariation for Gibberella ear rot resistance and agronomic traits in testcrosses of doubled haploid maize lines. Euphytica 2012, 185, 441–451. [Google Scholar] [CrossRef]
- Butrón, A.; Reid, L.M.; Santiago, R.; Cao, A.; Malvar, R.A. Inheritance of maize resistance to Gibberella and Fusarium ear rots and kernel contamination with deoxynivalenol and fumonisins. Plant Pathol. 2015, 64, 1053–1060. [Google Scholar] [CrossRef]
- Bocianowski, J. Using NGS Technology and Association Mapping to Identify Candidate Genes Associated with Fusarium Stalk Rot Resistance. Genes 2024, 15, 106. [Google Scholar] [CrossRef]
- Champeil, A.; Doré, T.; Fourbet, J.F. Fusarium head blight: Epidemiological origin of the effects of cultural practices on head blight attacks and the production of mycotoxins by Fusarium in wheat grains. Plant Sci. 2004, 166, 1389–1415. [Google Scholar] [CrossRef]
- Kathiresan, S.; Voight, B.F.; Purcell, S.; Musunuru, K.; Ardissino, D.; Mannucci, P.M.; Anand, S.; Engert, J.C.; Samani, N.J.; Schunkert, H.; et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009, 41, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Huang, Y.S.; Vilhjalmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Hackett, C.A.; McLean, K.; Bryan, G.J. Linkage analysis and QTL mapping using SNP dosage data in a tetraploid potato mapping population. PLoS ONE 2013, 8, e63939. [Google Scholar] [CrossRef] [PubMed]
- Lanubile, A.; Ferrarini, A.; Maschietto, V.; Delledonne, M.; Marocco, A.; Bellin, D. Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genomics 2014, 15, 710. [Google Scholar] [CrossRef]
- Diversity Arrays Technology. Available online: https://www.diversityarrays.com/ (accessed on 12 May 2024).
- Tomkowiak, A.; Nowak, B.; Sobiech, A.; Bocianowski, J.; Wolko, Ł.; Spychała, J. The Use of DArTseq Technology to Identify New SNP and SilicoDArT Markers Related to the Yield-Related Traits Components in Maize. Genes 2022, 13, 848. [Google Scholar] [CrossRef]
- Sobiech, A.; Tomkowiak, A.; Bocianowski, J.; Szymańska, G.; Nowak, B.; Lenort, M. Identification and Analysis of Candidate Genes Associated with Maize Fusarium Cob Resistance Using Next-Generation Sequencing Technology. Int. J. Mol. Sci. 2023, 24, 16712. [Google Scholar] [CrossRef]
- Sobiech, A.; Tomkowiak, A.; Nowak, B.; Bocianowski, J.; Wolko, Ł.; Spychała, J. Associative and Physical Mapping of Markers Related to Fusarium in Maize Resistance, Obtained by Next-Generation Sequencing (NGS). Int. J. Mol. Sci. 2022, 23, 6105. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Wang, M.; Zhang, S.; Du, Y.; Cong, J.; Yan, H.; Guo, H.; Xu, B.; Zhou, Z. GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection. Int. J. Mol. Sci. 2023, 24, 10343. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Shen, S.; Wang, D.; Zhang, F.; Zhang, C.; Wang, Z.; Zhou, Q.; Wang, R.; Tao, H.; He, F.; et al. Function of hydroxycinnamoyl transferases for the biosynthesis of phenolamides in rice resistance to Magnaporthe oryzae. J. Genet. Genomics 2022, 49, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Roumani, M.; Le Bot, J.; Boisbrun, M.; Magot, F.; Péré, A.; Robin, C.; Hilliou, F.; Larbat, R. Transcriptomics and Metabolomics Analyses Reveal High Induction of the Phenolamide Pathway in Tomato Plants Attacked by the Leafminer Tuta absoluta. Metabolites 2022, 12, 484. [Google Scholar] [CrossRef]
- Fang, H.; Shen, S.; Wang, D.; Zhang, F.; Zhang, C.; Wang, Z.; Ning, Y. A monocot-specific hydroxycinnamoylputrescine gene cluster contributes to immunity and cell death in rice. Sci. Bull. 2021, 66, 2381–2393. [Google Scholar] [CrossRef]
- Onkokesung, N.; Gaquerel, E.; Kotkar, H.; Kaur, H.; Baldwin, I.T.; Galis, I. MYB8 Controls Inducible Phenolamide Levels by Activating Three Novel Hydroxycinnamoyl-Coenzyme A:Polyamine Transferases in Nicotiana attenuata. Plant Physiol. 2012, 158, 389–407. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, B.C. Pentatricopeptide repeat proteins in plants: Cellular functions, action mechanisms and potential applications. Plant Commun. 2024, 6, 101203. [Google Scholar] [CrossRef]
- Dong, X.; Gao, Y.; Chen, W.; Wang, W.; Gong, L.; Liu, X.; Luo, J. Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol. Plant 2015, 8, 111–121. [Google Scholar] [CrossRef]
- Tanabe, K.; Hojo, Y.; Shinya, T.; Galis, I. Molecular evidence for biochemical diversification of phenolamide biosynthesis in rice plants. J. Integr. Plant Biol. 2016, 58, 903–913. [Google Scholar] [CrossRef]
- Dietz, K.J. Thiol-based peroxidases and ascorbate peroxidases: Why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 2016, 39, 20. [Google Scholar] [CrossRef]
- Johansson, T.; Welinder, K.G.; Nyman, P.O. Isozymes of lignin peroxidase and manganese (II) peroxidase from the white-rot basidiomycete Trametes versicolor: II. Partial sequences, peptide maps, and amino acid and carbohydrate compositions. Arch. Biochem. Biophys. 1993, 300, 57–62. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef]
- Barceló, A.R.; Pomar, F. Oxidation of cinnamyl alcohols and aldehydes by a basic peroxidase from lignifying Zinnia elegans hypocotyls. Phytochemistry 2001, 57, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Schultz, J.C. Differential activity of peroxidase isozymes in response to wounding, gypsy moth, and plant hormones in northern red oak (Quercus rubra L.). J. Chem. Ecol. 2004, 30, 1363–1379. [Google Scholar] [CrossRef] [PubMed]
- Gazaryan, I.G.; Lagrimini, L.M.; Ashby, G.A.; Thorneley, R.N. Mechanism of indole-3-acetic acid oxidation by plant peroxidases: Anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases. Biochem. J. 1996, 313, 841–847. [Google Scholar] [CrossRef]
- Liszkay, A.; van der Zalm, E.; Schopfer, P. Production of reactive oxygen intermediates (O2−, H2O2, and OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004, 136, 3114–3123. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Minibayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar] [CrossRef]
- Schopfer, P.; Liszkay, A.; Bechtold, M.; Frahry, G.; Wagner, A. Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta 2002, 214, 821–828. [Google Scholar] [CrossRef]
- Delannoy, E.; Jalloul, A.; Assigbetsé, K.; Marmey, P.; Geiger, J.P.; Lherminier, J.; Nicole, M. Activity of class III peroxidases in the defense of cotton to bacterial blight. Mol. Plant-Microbe Interact. 2003, 16, 1030–1038. [Google Scholar] [CrossRef]
- Liszkay, A.; Kenk, B.; Schopfer, P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 2003, 217, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Laloi, C.; Apel, K.; Danon, A. Reactive oxygen signalling: The latest news. Curr. Opin. Plant Biol. 2004, 7, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Booth, C. Methods in Microbiology; Academic Press: New York, NY, USA, 1971. [Google Scholar] [CrossRef]
- Nelson, P.E.; Toussoun, T.A.; Cook, R.J. Fusarium: Diseases, Biology and Taxonomy; Pennsylvania State University Press: University Park, PA, USA, 1981. [Google Scholar]
- Kwaśna, H.; Chełkowski, J. Ecology and taxonomy of Fusarium species in Poland. Mycotoxin Res. 1991, 7, 58–63. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press, Inc.: San Diego, CA, USA, 1990. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- NCBI BLAST. Available online: http://blast.ncbi.nlm.nih.gov/ (accessed on 9 September 2024).
- Lin, Y.; Zhang, C.; Lan, H.; Gao, S.; Liu, H.; Liu, J.; Cao, M.; Pan, G.; Rong, T.; Zhang, S. Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PLoS ONE 2014, 9, e95445. [Google Scholar] [CrossRef]
- Srdić, J.; Nikolić, A.; Pajić, Z.; Mladenović Drinić, S.; Filipović, M. Genetic similarity of sweet corn inbred lines in correlation with heterosis. Maydica 2011, 56, 251–256. [Google Scholar]
- Spychała, J.; Tomkowiak, A.; Noweiska, A.; Bobrowska, R.; Bocianowski, J.; Książkiewicz, M.; Sobiech, A.; Kwiatek, M. Expression profiling of the slow rusting resistance genes Lr34/Yr18 and Lr67/Yr46 in common wheat (Triticum aestivum L.) and associated miRNAs patterns. Genes 2023, 14, 1376. [Google Scholar] [CrossRef]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 2 August 2024).
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Briggs, S.P. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef]
- Bocianowski, J.; Jakubowska, M.; Zawada, D.; Dobosz, R. The Effect of Acaricide Control of the Two-Spotted Spider Mite Tetranychus urticae Koch on the Cultivation of Sugar Beet (Beta vulgaris L.) and on the Size and Quality of the Yield. Appl. Sci. 2022, 12, 12139. [Google Scholar] [CrossRef]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Acad. Sci. USA 1936, 12, 49–55. [Google Scholar]
- Bocianowski, J.; Majchrzak, L. Analysis of effects of cover crop and tillage method combinations on the phenotypic traits of spring wheat (Triticum aestivum L.) using multivariate methods. Appl. Ecol. Environ. Res. 2019, 17, 15267–15276. [Google Scholar] [CrossRef]
- Warzecha, T.; Bathelt, R.; Skrzypek, E.; Warchoł, M.; Bocianowski, J.; Sutkowska, A. Studies of Oat-Maize Hybrids Tolerance to Soil Drought Stress. Agriculture 2023, 13, 243. [Google Scholar] [CrossRef]
- VSN International Genstat for Windows, 23rd ed.; VSN International: Hemel Hempstead, UK, 2023; Available online: https://vsni.co.uk/software/genstat/ (accessed on 1 May 2023).
- Lanubile, A.; Bernardi, J.; Marocco, A.; Logrieco, A.; Paciolla, C. Differential activation of defense genes and enzymes in maize genotypes with contrasting levels of resistance to Fusarium verticillioides. Environ. Exp. Bot. 2012, 78, 39–46. [Google Scholar] [CrossRef]
- Tomkowiak, A. Identification of SNP and SilicoDArT Markers and Characterization of Their Linked Candidate Genes Associated with Maize Smut Resistance. Int. J. Mol. Sci. 2024, 25, 11358. [Google Scholar] [CrossRef]
- Nelson, M.T. Interactions of divalent cations with single calcium channels from rat brain synaptosomes. J. Gen. Physiol. 1986, 87, 201–222. [Google Scholar] [CrossRef]
- Escrivá, L.; Font, G.; Manyes, L. In vivo toxicity studies of fusarium mycotoxins in the last decade: A review. Food Chem. Toxicol. 2015, 78, 185–206. [Google Scholar] [CrossRef]
- Ling, H. Sequence analysis of GDSL lipase gene family in Arabidopsis thaliana. Pak. J. Biol. Sci. 2008, 11, 763–767. [Google Scholar] [CrossRef]
- Volokita, M.; Rosilio-Brami, T.; Rivkin, N.; Zik, M. Combining comparative sequence and genomic data to ascertain phylogenetic relationships 1 and explore the evolution of the large GDSL- lipase family in land-plants. Mol. Biol. Evol. 2011, 28, 551–565. [Google Scholar] [CrossRef]
- Chepyshko, H.; Lai, C.P.; Huang, L.M.; Liu, J.H.; Shaw, J.F. Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: New insights from bioinformatics analysis. BMC Genomics 2012, 13, 309. [Google Scholar] [CrossRef]
- Hong, J.K.; Choi, H.W.; Hwang, I.S.; Kim, D.S.; Kim, N.H.; Choi, D.S.; Kim, Y.J.; Hwang, B.K. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 2008, 227, 539–558. [Google Scholar] [CrossRef]
- Su, H.G.; Zhang, X.H.; Wang, T.T.; Wei, W.L.; Wang, Y.X.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; et al. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef]
- Shen, G.; Sun, W.; Chen, Z.; Shi, L.; Hong, J.; Shi, J. Plant GDSL Esterases/Lipases: Evolutionary, Physiological and Molecular Functions in Plant Development. Plants 2022, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Wang, F.; Shen, H.; Xie, S.; Chen, X.; Xie, Q.; Li, R.; Cao, A.; Li, H. Identification, evolution, and expression of GDSL-type Esterase/Lipase (GELP) gene family in three cotton species: A bioinformatic analysis. BMC Genomics 2023, 24, 795. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Dong, Z.; Tian, Y.; Xie, K.; Wu, S.; Zhu, T.; Wan, X. ZmMs30 Encoding a Novel GDSL Lipase is Essential for Male Fertility and Valuable for Hybrid Breeding in Maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef]
- Walters, D. Resistance to plant pathogens: Possible roles for free polyamines and polyamine catabolism. New Phytol. 2003, 159, 109–115. [Google Scholar] [CrossRef]
- Wang, L.; Qian, J.; Li, M.; Zheng, H.; Yang, X.; Zheng, M.; Hsu, Y.F. Arabidopsis PDE1 confers phosphate-deficiency tolerance in primary root growth. Plant Cell Rep. 2024, 43, 8. [Google Scholar] [CrossRef]
- Wojtasik, W.; Kulma, A.; Namysl, K.; Preisner, M.; Szopa, J. Polyamine metabolism in flax in response to treatment with pathogenic and non-pathogenic Fusarium strains. Front. Plant Sci. 2015, 6, 291. [Google Scholar] [CrossRef]
- Jimenez-Bremont, J.F.; Marina, M.; Guerrero-Gonzalez, M.D.; Rossi, F.R.; Sanchez-Rangel, D.; Rodriguez-Kessler, M.; Ruiz, O.; Garriz, A. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 2014, 5, 95. [Google Scholar] [CrossRef]
- Gauthier, L.; Atanasova-Penichon, V.; Chéreau, S.; Richard-Forget, F. Metabolomics to Decipher the Chemical Defense of Cereals against Fusarium graminearum and Deoxynivalenol Accumulation. Int. J. Mol. Sci. 2015, 16, 24839–24872. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Gall, H.L.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Ruszczyńska, M.; Sytykiewicz, H. New Insights into Involvement of Low Molecular Weight Proteins in Complex Defense Mechanisms in Higher Plants. Int. J. Mol. Sci. 2024, 25, 8531. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar] [CrossRef] [PubMed]
- Murillo, I.; Cavallarin, L.; San Segundo, B. The maize pathogenesis-related PRms protein localizes to plasmodesmata in maize radicles. Plant Cell 1997, 9, 145–156. [Google Scholar] [CrossRef]
- Lambarey, H.; Moola, N.; Veenstra, A.; Murray, S.; Rafudeen, M.S. Transcriptomic Analysis of a Susceptible African Maize Line to Fusarium verticillioides Infection. Plants 2020, 9, 1112. [Google Scholar] [CrossRef]


| Marker | Primer Sequences | Annealing Temperature (°C) | Product Size (bp) | Candidate Genes | Reference |
|---|---|---|---|---|---|
| 553 | F: TTGTCGACGTACACGACCG | 60.0 | 116 | GDSL esterase/lipase gene (LOC100273960) | [26] |
| R: TTCGGGTGCGTGAAAAGCTA | |||||
| 15,097 | F: GGCTCACCTTCCCGTTCTAC | 59.0 | 107 | Hydroxycinnamyltransferase gene (LOC103649226) | [26] |
| R: GTACGAAGGCACCAGGAACA | |||||
| 58,771 | F: TGCTAGCACAAGTGCATTTCAA | 58.0 | 103 | Peroxidase 72 gene (LOC100282124) | [26] |
| R: TGAAGGTGTTGCAAGCGAAT | |||||
| 27,775 | F: ACCAGAAGAACATTCTGCAG | 57.5 | 167 | Gene encoding for an uncharacterized protein (LOC100501166) | [25] |
| R: GAACGAGCTCACTCAGAAGC |
| Gene Abbreviation | Gene Name | The Accession Number | Gene Symbol | Primer Sequence | Product Size on mRNA(bp) | Length Including Intron | Amplification Efficiency (%E) | R2 | Tm (°C) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | |||||||||
| F1 | GDSL esterase/lipase | NM_001148347.1 | LOC100273960 | AAAGCGTCAAGCGAAGCCTA | GAACACCATGAAGCTGCGAG | 98 | 224 | 123.9 | 1 | 60 |
| F2 | Putrescine hydroxy cinnamyl transferase | XM_008673542.3 | LOC103649226 | GTGGAGGTGGTGCAGGTG | CGAGGAAGTCGCTGGTGG | 109 | - | 120.8 | 0.998 | 60 |
| F3 | Uncharacterized gene | NM_001195961.1 | LOC100501166 | GTGCGCTTGTCGTACTTCTTG | ACTACATAGGTAGGAGGAGCTCTA | 105 | 207 | 93.8 | 1 | 60 |
| F4 | Peroxidase 72 gene | NM_001155037.2 | LOC100282124 | ATTGTGCAGTCCATTGTGGC | GTTGTCCAACAGCACCGAAG | 123 | 755 | 103.8 | 0.999 | 60 |
| CYT | Cyclophilin | M55021/ | LOC MZECYP | CTACCTCACGGCATCTGCTATGT | AACACGAATCAAGCAGAG | 139 | - | 85.7 | 1 | 60 |
| Zm00001eb312970 (umc2057) | ||||||||||
| β-TUB | β-tubulin | NP_001112117 | LOC1920235 | CTGAGTGGTGGTCTTAGT | GTCACACACACTCGACTTCACG | 100 | - | 111.5 | 1 | 60 |
| Source of Variation | d.f. | F1 | F2 | F3 | F4 | CYT | TUB | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m.s. | F | m.s. | F | m.s. | F | m.s. | F | m.s. | F | m.s. | F | ||
| Line | 4 | 0.0000011 | 18.18 *** | 0.000053 | 16.85 *** | 0.000048 | 9.03 ** | 0.000004 | 3.44 | 12.566 | 8.62 ** | 12.911 | 9.32 ** |
| Residual | 10 | 0.0000001 | 0.36 | 0.000003 | 0.97 | 0.000005 | 1.44 | 0.000001 | 0.85 | 1.457 | 0.5 | 1.385 | 0.79 |
| Term | 4 | 0.0000001 | 0.77 | 0.000039 | 12.03 *** | 0.000077 | 20.73 *** | 0.000003 | 1.83 | 18.904 | 6.43 ** | 20.51 | 11.69 *** |
| Term × Line | 16 | 0.0000002 | 0.99 | 0.000007 | 2.16 | 0.000021 | 5.6 *** | 0.000003 | 1.8 | 5.156 | 1.75 | 2.037 | 1.16 |
| Residual | 40 | 0.0000002 | 0.000003 | 0.000004 | 0.000001 | 2.94 | 1.755 | ||||||
| Line | Term | F1 | F2 | F3 | F4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T/0 h | K-S Test | Levene’s | t-Student | T/0 h | K-S Test | Levene’s | t-Student | T/0 h | K-S Test | Levene’s | t-Student | T/0 h | K-S Test | Levene’s | t-Student | ||
| FR | 00 h | 1 | 0.916 | 1.000 | 0.770 | 1.000 | 0.928 | 1.000 | 0.696 | ||||||||
| FR | 06 h | 2.916 | 0.026 | 0.963 | 10.608 | 0.024 | 0.249 | 10.667 | 0.053 | 0.252 | 17.565 | 0.039 | 0.756 | ||||
| FR | 12 h | 0.930 | 0.292 | 0.914 | 3.608 | 0.289 | 0.872 | 5.246 | 0.034 | 0.986 | 3.522 | 0.227 | 0.774 | ||||
| FR | 24 h | 1.168 | 0.236 | 0.226 | 1.351 | 0.287 | 0.001 *** | 1.070 | 0.082 | 0.020 * | 3.348 | 0.110 | 0.063 | ||||
| FR | 72 h | 2.608 | 0.192 | 0.303 | 2.284 | 0.105 | 0.561 | 5.807 | 0.059 | 0.199 | 0.304 | 0.037 | 0.932 | ||||
| KF12 | 00 h | 1.000 | 0.987 | 1.000 | 0.898 | 1.000 | 0.892 | 1.000 | 0.702 | ||||||||
| KF12 | 06 h | 0.252 | 0.167 | 0.454 | 2.188 | 0.103 | 0.176 | 0.734 | 0.038 | 0.187 | 12.000 | 0.093 | 0.964 | ||||
| KF12 | 12 h | 0.682 | 0.252 | 0.215 | 1.625 | 0.714 | 0.513 | 2.625 | 0.232 | 0.807 | 0.000 | 0.016 * | 1.000 | ||||
| KF12 | 24 h | 0.467 | 0.393 | 0.091 | 0.688 | 0.047 | 0.020 * | 0.719 | 0.029 | 0.814 | 1.000 | 0.148 | 0.460 | ||||
| KF12 | 72 h | 0.813 | 0.587 | 0.650 | 0.938 | 0.534 | 0.887 | 0.594 | 0.058 | 0.730 | 23.000 | 0.018 | 0.144 | ||||
| KF15 | 00 h | 1.000 | 0.968 | 1.000 | 0.853 | 1.000 | 0.860 | 1.000 | 0.891 | ||||||||
| KF15 | 06 h | 1.583 | 0.046 | 0.673 | 4.961 | 0.065 | 0.094 | 6.142 | 0.095 | 0.450 | 17.000 | 0.057 | 0.650 | ||||
| KF15 | 12 h | 0.777 | 0.442 | 0.589 | 2.169 | 0.036 | 0.534 | 0.206 | 0.111 | 0.563 | 4.000 | 0.078 | 0.472 | ||||
| KF15 | 24 h | 0.709 | 0.066 | 0.290 | 0.590 | 0.090 | <0.001 *** | 0.394 | 0.397 | <0.001 *** | 6.000 | 0.022 | 0.053 | ||||
| KF15 | 72 h | 1.524 | 0.040 | 0.344 | 0.680 | 0.035 | 0.625 | 2.277 | 0.022 | 0.234 | 0.000 | 0.158 | 0.891 | ||||
| SF11 | 00 h | 1.000 | 0.949 | 1.000 | 0.954 | 1.000 | 0.939 | 1.000 | 0.933 | ||||||||
| SF11 | 06 h | 1.211 | 0.082 | 0.590 | 2.400 | 0.187 | 0.389 | 5.733 | 0.030 | 0.657 | 0.825 | 0.974 | 0.992 | ||||
| SF11 | 12 h | 1.304 | 0.249 | 0.200 | 1.667 | 0.181 | 0.907 | 4.000 | 0.029 | 0.907 | 0.988 | 0.032 | 0.515 | ||||
| SF11 | 24 h | 0.249 | 0.350 | 0.709 | 1.133 | 0.267 | 0.088 | 1.333 | 0.332 | 0.162 | 0.450 | 0.158 | 0.810 | ||||
| SF11 | 72 h | 0.310 | 0.153 | 0.236 | 0.600 | 0.819 | 0.581 | 5.200 | 0.154 | 0.208 | 3.338 | 0.819 | 0.016 * | ||||
| SF12 | 00 h | 1.000 | 0.874 | 1.000 | 0.969 | 1.000 | 0.796 | 1.000 | 0.693 | ||||||||
| SF12 | 06 h | 0.230 | 0.067 | 0.182 | 1.916 | 0.128 | 0.942 | 4.074 | 0.104 | 0.369 | 6.667 | 0.022 | 0.995 | ||||
| SF12 | 12 h | 0.409 | 0.501 | 0.196 | 1.050 | 0.105 | 0.758 | 0.360 | 0.225 | 0.745 | 0.667 | 0.636 | 0.997 | ||||
| SF12 | 24 h | 0.430 | 0.264 | 0.091 | 0.780 | 0.179 | 0.213 | 0.771 | 0.249 | 0.001 *** | 1.000 | 0.795 | 0.863 | ||||
| SF12 | 72 h | 0.434 | 0.761 | 0.199 | 0.908 | 0.219 | 0.899 | 0.371 | 0.169 | 0.376 | 70.667 | 0.034 | 0.054 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobiech, A.; Tomkowiak, A.; Jamruszka, T.; Kosiada, T.; Spychała, J.; Lenort, M.; Bocianowski, J. Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.). Pathogens 2025, 14, 779. https://doi.org/10.3390/pathogens14080779
Sobiech A, Tomkowiak A, Jamruszka T, Kosiada T, Spychała J, Lenort M, Bocianowski J. Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.). Pathogens. 2025; 14(8):779. https://doi.org/10.3390/pathogens14080779
Chicago/Turabian StyleSobiech, Aleksandra, Agnieszka Tomkowiak, Tomasz Jamruszka, Tomasz Kosiada, Julia Spychała, Maciej Lenort, and Jan Bocianowski. 2025. "Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.)" Pathogens 14, no. 8: 779. https://doi.org/10.3390/pathogens14080779
APA StyleSobiech, A., Tomkowiak, A., Jamruszka, T., Kosiada, T., Spychała, J., Lenort, M., & Bocianowski, J. (2025). Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.). Pathogens, 14(8), 779. https://doi.org/10.3390/pathogens14080779








