Abstract
Identification and differentiation of Candida spp. yeasts, especially clinically relevant isolates, is of high importance with respect to their origin, pathogenic potential, colonization pattern, and resistance to antimycotics. Currently, numerous typing methods with varying or unknown discriminatory power are used. This study evaluated the utility of five methods—biotyping using the API system, ITS1 and ITS4 sequence analysis, ITS region polymorphism, multiplex PCR of ITS1, ITS3, and ITS4 regions, and karyotyping—for typing 42 strains differing in origin (24 clinical and 18 food-borne). The highest discriminatory power was obtained for ITS sequencing and karyotyping, both yielding a discrimination index of 1.000. The discrimination indices for other methods ranged from 0.957 for genotyping based on ITS region polymorphism to 0.997 for multiplex PCR-genotyping. Although biotyping showed relatively high discriminatory potential, its use led to misclassification of 64.3% of isolates compared to ITS sequencing. These findings emphasize the importance of applying a typing method with a discrimination index of 1.000 to ensure accurate interpretation of strain-relatedness and origin. Methods with lower indices may reflect methodological limitations rather than actual genetic relatedness. Determining the discrimination index is therefore essential when selecting appropriate tools for yeast typing, particularly in clinical and epidemiological contexts.
1. Introduction
Candida yeasts constitute a heterogeneous group of microorganisms capable of colonizing various ecological niches. Due to their clinical importance, research has primarily focused on pathogenic species, with C. albicans considered the most important representative [1,2,3]. In addition to C. albicans, five non-albicans Candida (NAC) species are considered the most clinically relevant, namely C. glabrata (current name Nakaseomyces glabratus), C. tropicalis, C. parapsilosis, C. krusei (current name Issatchenkia orientalis), and the emerging global public health threat C. auris [1,2,3,4,5,6]. Invasive Candida infections are associated with high morbidity and mortality, as well as increasing antifungal resistance. Invasive candidiasis affects approximately 250,000 to 700,000 individuals worldwide each year, with a mortality rate ranging from 40% to 55% [2,5,6].
It is noteworthy that Candida species are predominantly considered opportunistic pathogens, with the majority of infections having an endogenous origin. The pathogenesis of candidiasis is closely related to a number of Candida virulence factors, including secretion of hydrolytic enzymes, expression of adhesins and invasins on the cell wall surface, pleomorphism, phenotypic switching, and biofilm formation [2,3]. Furthermore, metabolic plasticity, efficient nutrient acquisition, and a remarkable ability to adapt to environmental stressors contribute significantly to their pathogenic potential.
Different Candida species appear to have evolved distinct pathogenic strategies. For instance, C. albicans is particularly adept at filamentation and biofilm formation, which contribute to tissue invasion and fungal persistence [3]. In contrast, non-albicans species such as C. glabrata, C. parapsilosis, and C. auris exhibit limited filamentation. C. tropicalis, on the other hand, demonstrates a higher capacity for biofilm formation and produces more potent candidalysin of cytolytic activity compared to C. albicans [3]. Moreover, C. auris is more resistant to phagocytosis by neutrophils than C. albicans, likely due to the protective properties of its outer cell wall mannan layer [3,7]. Unlike other Candida species, C. glabrata lacks secreted aspartyl proteases (SAPs) but expresses cell wall-associated aspartic proteases known as yapsins [8]. In addition to these unique virulence traits, antifungal susceptibility remains a key therapeutic concern, with the limited availability of effective treatment options posing a significant challenge in the case of multidrug-resistant species such as C. auris and C. glabrata [7,8].
Many Candida species, including C. kefyr (current name Kluyveromyces marxianus), C. boidinii, C. lipolytica, C. shehatae, C. pseudotropicalis, C. famata, C. guilliermondii (current name Meyerozyma guilliermondii), and C. inconspicua, are recognized as part of natural food microbiota, are used in food production, or have been isolated from food products [9,10]. The number of yeasts in these products can reach levels as high as 106–108 CFU/mL or CFU/g [11]. Recent metagenomic data from 2500 food samples demonstrated that Candida yeasts occur at low frequency in food environments, with C. parapsilosis detected in 2.7% of samples, C. sake in 2.5%, and both Pichia kudriavzevii (current name Issatchenkia orientalis, formerly Candida krusei) and Diutina catenulata (formerly C. catenulata) in 1.3% [12]. In general, yeasts isolated from food that are not part of its native microbiota are considered food spoilage microorganisms.
Candida spp. are capable of colonizing the human gastrointestinal tract, contributing to the development of diarrhea and other gastrointestinal symptoms in at-risk individuals. In such cases, fecal samples may contain yeast counts exceeding 106 CFU/g [13]. Talwar et al. [14] identified C. albicans as the primary etiologic agent of gastroenteritis, although other species, including C. tropicalis, C. kefyr, C. krusei, C. parapsilosis, C. lusitaniae (current name Clavispora lusitaniae), and C. guilliermondii, have also been implicated. In addition, yeasts present in ingested foods may exacerbate Crohn’s disease and induce intestinal inflammatory responses [15]. Nevertheless, gastrointestinal infections caused by food-derived yeast are extremely rare, and only sporadic cases of yeast-induced gastroenteritis have been reported [11].
We have previously shown that food-borne NAC strains may exhibit relevant similarity to clinical C. albicans, classifying them within the group of risk of potential pathogens [16]. However, the preliminary and key criterion remains the inability of some food-derived Candida spp. strains to grow at human body temperature. To date, no clear evidence has been found to support the transmission of yeasts from food sources to humans. However, the presence of some food-derived Saccharomyces cerevisiae isolates in the human gut indicates the potential for yeast transmission through food consumption [12].
Currently, Candida spp. infections represent a significant public health problem, highlighting the importance of accurate identification and typing of yeasts in the diagnosis of such infections, as well as in the choice of appropriate antifungal therapy. It also enables the identification of the source of infection and facilitates tracking the development of antifungal drug resistance in epidemiological studies. To identify and distinguish isolates of Candida spp. from different sources, numerous molecular typing methods have been proposed, the most recently and commonly used being duplex PCR, restriction fragment length polymorphisms (RFLP), multilocus sequence typing (MLST), randomly amplified polymorphic DNA (RAPD), and microsatellites [17,18,19,20,21]. The utility of these methods has been well documented in previous publications, although literature data concern their application to determine genetic relatedness primarily among clinical Candida strains.
The aim of the study was to evaluate the usefulness of various typing methods in the identification and differentiation of Candida yeasts isolated from diverse environments, including both clinical and food-related sources. Given their different origins, these strains are likely to vary in virulence and, consequently, in the potential risk they pose to human health [11,16,22]. This study specifically aimed to assess the discriminatory power of selected bio- and genotyping methods that could enhance the accurate differentiation of Candida strains from diverse origins. To our best knowledge, most studies to date have focused either on clinical or environmental strains and have not covered such a diverse group of yeasts.
2. Materials and Methods
2.1. Yeast Strains
The study was conducted on 23 clinical isolates (provided by the Department of Laboratory Diagnostics of the Polish Mother’s Memorial Hospital Research Institute in Lodz, Poland) and 1 reference Candida albicans strain ATCC 10231, as well as a group of 18 food-borne strains, including 5 strains from the Culture Collection of Microorganisms LOCK 105 (Table 1). Both the food-derived and clinical Candida strains were obtained from diverse sample types and were selected randomly, reflecting the heterogeneity of the source materials used in this study.

Table 1.
Yeast strains used in the study.
2.2. Yeast Biotyping
In yeast biotyping, the identification system API 20 C AUX (bioMérieux S.A., Marcy-l’Etoile, France), based on the biochemical assimilation of 19 carbohydrates, was used, according to the manufacturer’s instructions. The API test readings and interpretations were performed independently by two researchers to minimize the potential for bias resulting from subjective assessment. Strain identification was performed on the basis of the obtained numerical profiles, using ApiwebTM software v.1.3.1 (bioMérieux S.A., Marcy-l’Etoile, France).
2.3. Sequence Analysis of ITS Regions
Genomic DNA was extracted employing the Genomic Mini AX Yeast (A&A Biotechnology, Gdansk, Poland) in accordance with the manufacturer’s protocol. Spectrophotometric evaluation of DNA was conducted both qualitatively and quantitatively (Implen). DNA at a minimum concentration of 10 ng was used as a template for the PCR reaction. The required DNA purity was determined based on the ratio of absorbance (nm): 260/280 and 260/230, and their values in the ranges of 1.8–2.0 and 2.0–2.2, respectively, were considered acceptable. PCR master mix amplifying the ITS regions contained 12.0 μL of REDTaq Ready Mix polymerase (Sigma-Aldrich, St. Louis, MO, USA), 0.2 μL of ITS1 and ITS4 primers (Genomed Inc., Warsaw, Poland) [23,24] (Table 2), and 10–20 ng of DNA as template (1.0 μL). The PCR was run using the following thermal cycling program: initial denaturation at 94 °C for 2 min, 34 cycles including denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, elongation at 72 °C for 2 min, and a final extension step at 72 °C for 2 min. The amplicons were separated on 1.0% (w/v) agarose gel in 0.5× TBE buffer (Sigma-Aldrich, St. Louis, MO, USA) and then purified and sequenced by the Sanger method. The strains were identified by analyzing the sequences (approximately 500 bp in size) using the BLAST+ 2.16.0 (available at https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 April 2025)) and then compared with the sequences deposited in the GenBank database (NCBI).

Table 2.
Sequences of primers used in this study.
2.4. Multiplex PCR Analysis
Multiplex PCR was performed using three universal primers, ITS1, ITS3, and ITS4 (Table 2), targeting the 18S, 5.8S, and 28S rDNA conserved regions [23,24]. To each PCR reaction, 12.0 μL DreamTaq™ Green DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 0.2 μL of each primer, and 1.0 μL of DNA template were added and made up to a total volume of 25.0 μL with PCR-grade water (Sigma-Aldrich, St. Louis, MO, USA). The multiplex PCR reaction was performed using a thermal cycle as described above, with the primer annealing temperature set at 53 °C. Obtained amplicons were separated on 1.0% (w/v) agarose gel in 0.5× TBE buffer (Sigma-Aldrich, St. Louis, MO, USA), and the PerfectTM 100 bp DNA Ladder was used as a size standard (EURx, Gdańsk, Poland).
2.5. Yeast Karyotyping
Isolation of yeast chromosomal DNA was performed using the CHEF Yeast Genomic DNA Plug Kit (Bio-Rad, Hercules, CA, USA) exactly according to the manufacturer’s instructions. Chromosomes were separated by pulsed-field gel electrophoresis in 0.8% agarose gel (Pulsed-Field Certified Agarose; Bio-Rad, Hercules, CA, USA) by means of CHEF-DR II apparatus (Bio-Rad, Hercules, CA, USA). Electrophoresis was carried out in 0.5× TBE buffer (Sigma-Aldrich, St. Louis, MO, USA) cooled to 10 °C, using the following conditions in two blocks, i.e., block 1: voltage 4.5 V/cm, pulse duration 120 s, separation time 24 h; block 2: voltage 4.5 V/cm, linearly increasing pulse duration from 240 to 360 s, separation time 24 h. After separation, the gels were stained in ethidium bromide solution (50 μg/mL) at room temperature for 15 min [25]. The gels were washed in distilled water at room temperature for 30 min and then photographed. To estimate the molecular weight of chromosomes Saccharomyces cerevisiae YNN295 and Schizosaccharomyces pombe 972 h DNA size standards (Bio-Rad, Hercules, CA, USA) were used.
2.6. Statistical Analysis
2.6.1. Agglomeration Analysis
To determine the similarity of the studied yeast features, the UPGMA cluster analysis (unweighted pair-group method using arithmetic averages) based on the maximum composite likelihood method was used. The similarity analysis of the electrophoretic profiles of ITS regions was performed using GelJ v.2.3 software [26]. Likewise, the karyotype similarity analysis was also conducted employing this software. The dendrogram of the similarity of ITS regions sequences was generated using MEGA 12 program [27]. The procedure for creating dendrogram included 42 nucleotide sequences, and the bootstrap consensus tree was inferred from 500 replicates. The optimal tree with the sum of 5.488 branch length was shown. The pairwise-deletion option was applied to all ambiguous positions for each sequence pair, resulting in a final data set of 693 positions.
2.6.2. Discrimination Index
An index of discrimination (D), expressing the ability of a typing method to distinguish between different strains, was calculated according to the Simpson index
where D is the discrimination index, N is the number of tested strains, s is the number of types described, and nj is the number of strains belonging to the j type [28]. A D value of 1.0 indicates maximal discriminatory power of the method, indicating its ability to uniquely distinguish each individual strain.
3. Results
In this study, 42 Candida strains were characterized using both biotyping and four genotyping methods. Biotyping was performed based on yeast assimilation capabilities using API tests. The genotyping methods included (1) ITS1 and ITS4 sequence analysis, (2) size polymorphism analysis of the ITS1 and ITS4 regions, (3) multiplex PCR targeting the ITS1, ITS3, and ITS4 regions, and (4) karyotyping. In addition to strain typing, biotyping and ITS sequence-based genotyping were also used to identify the tested strains.
3.1. Biotyping and Yeasts Identification by API System
As a result of biotyping, 30 various assimilation profiles were obtained, with 4 profiles shared by more than one strain (Table A1). The first group of yeasts with identical biochemical profiles included five clinical strains: cl/MP/04, cl/MP/07, cl/MP/12, cl/MP/2K, and cl/OZ/g2 (Table 3). The next group consisted of six clinical isolates: cl/MP/02, cl/MP/05, cl/MP/09, cl/MP/4K, cl/MP/3M, and cl/MP/4M. Strains from both of these groups were identified as C. albicans. Another assimilation profile was obtained for three food-derived strains: fo/BM/02, fo/MP/02, and LOCK 0009, which were classified as C. krusei/C. inconspicua (Table 3). The last cluster consisted of two food-borne collection strains: LOCK 0004 and LOCK 0006, identified as C. lusitaniae. The other 26 strains were characterized by unique assimilation profiles. Despite differences in the API profiles, 21 clinical isolates and the reference strain ATCC 10231 were identified as C. albicans. Only two clinical isolates were classified as non-albicans Candida species, namely cl/KL/01 as C. glabrata and cl/KL/02 as C. lusitaniae (Table 3). Greater species diversity was obtained among food-derived strains. In this group, a total of nine yeast species were identified, i.e., C. lusitaniae (four isolates), C. krusei/C. inconspicua (four), C. boidinii (three), C. famata (two), C. parapsilosis (one), C. colliculosa (one), C. tropicalis (one), C. rugosa (one), and C. pelliculosa (one). Notably, in the Apiweb database, strains are reported as C. krusei/C. inconspicua, indicating taxonomic ambiguity even at high probability levels.

Table 3.
Identification of clinical and food-borne yeasts. If different, current species names according to www.indexfungorum.org (accessed on 4 June 2025) are given in parentheses. Strains that were differently classified into species based on assimilation profiles (API 20 C AUX) and ITS region sequences are in bold.
The discrimination index for yeast biotyping based on their assimilation capabilities by means of the API system was equal to 0.966.
3.2. Genotyping and Yeast Identification Based on ITS Region Sequences
Partially different results of yeast identification were obtained based on the sequences of the ITS1 and ITS4 regions (Table 3). Reclassification concerned 14 out of 24 clinical isolates and 13 out of 18 food-borne yeasts, representing slightly over 58% and 72% of the strains tested, respectively. This underscores the limited reliability of identification based on API tests alone and highlights the importance of molecular methods in yeast classification.
Moreover, based on API system results, all the tested yeasts were classified within the genus Candida, and nearly 89% of clinical isolates were identified as C. albicans. In contrast, among the clinical strains, ITS region sequencing revealed two Candida species, C. albicans (nine strains) and C. boidinii (five), whereas the most frequently represented species was Clavispora lusitaniae (10 isolates). These findings indicate a possible overidentification of C. albicans when using the API system.
Furthermore, isolates classified within the same species differed in their origin. C. lusitaniae strains were isolated from feces (five isolates), vagina (two), throat (one), blood (one), and urinary tract (one). The sources of C. albicans isolates included feces (six), throat (one), stomach (one), and, in the case of the reference strain ATCC 10231, lungs. Clinical C. boidinii strains were obtained from feces (three), stomach (one), and vagina (one).
Among the strains isolated from food, four species belonging to the genus Candida were identified (C. albicans, C. lusitaniae, C. boidinii, and C. tropicalis), along with four yeast species currently taxonomically reassigned from the Candida genus (Pichia membranifaciens, Pichia fermentans, Wickerhamomyces anomalus, and Meyerozyma guilliermondii). It is worth emphasizing that strain fo/79/01 isolated from fruit yogurt and fo/BM/01 originating from pickled cucumber were classified as C. albicans (Table 3).
Differentiation of yeasts based on the ITS region sequence was characterized by high discriminatory power, as no identical sequences were obtained in the group of the tested strains. The discrimination index for this method reached the highest possible value, amounting to 1.000.
The ITS sequence similarity dendrogram grouped the yeasts into four larger clusters (Figure 1). Cluster I consisted of 7 C. boidinii strains, including all three food-derived isolates and four out of five clinical strains classified as this species (cl/MP/12, cl/MP/3M, cl/OZ/k1, and cl/MP/2K), along with single representatives of M. guilliermondii (LOCK 0007) and W. anomalus (LOCK 0004). Cluster II included two P. membranifaciens (fo/MP/02 and fo/BG/05) and two P. fermentans isolates (fo/BM/02 and LOCK 0009), all originating from food. Cluster III comprised 10 C. albicans strains, representing all clinical isolates of this species included in the study, as well as 1 of the 2 C. albicans strains derived from food (fo/BM/01). This cluster is particularly notable for grouping all clinical C. albicans strains together, suggesting a high degree of genetic similarity among them. The presence of a single food-derived isolate within this cluster indicates potential overlap between clinical and food-associated C. albicans isolates. The second food-borne C. albicans strain fo/79/01 showed higher similarity to C. lusitaniae fo/LI/02 (Figure 1).
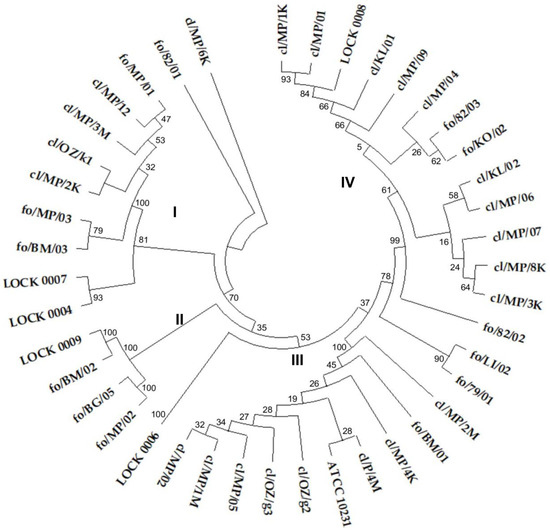
Figure 1.
UPGMA cluster analysis tree based on a similarity matrix obtained from ITS1 and ITS4 region sequences of food-borne and clinical yeast isolates. A bootstrap consensus tree was inferred from 500 replicates, and the optimal tree with the sum of 5.488 branch length was shown. The percentage of replicate trees in which the related taxa were clustered together in the bootstrap test is shown next to the branches.
Cluster IV contained 13 strains of C. lusitaniae, including all 9 clinical isolates of this species and 3 out of 6 isolated from food (fo/82/03, fo/KO/02, and LOCK 0008). C. lusitaniae strain fo/82/01 showed lower similarity to other isolates of this species. On the other hand, the lowest similarity of the ITS region sequence in the tested group of yeasts was exhibited by C. boidinii cl/MP/6K (Figure 1). The clustering of yeasts in accordance with their ITS-based species identification reflected a clear congruence between molecular taxonomy and the observed cluster structure. However, it is worth noting the high similarity of clinical and food-borne strains and their frequent common grouping in the same cluster.
Since it seems that more reliable results are obtained using the ITS region sequencing method, the species identification of the tested strains obtained by means of this method was further used.
3.3. Yeasts Genotyping Based on ITS Region Polymorphism
As a result of electrophoretic separation of PCR products of the ITS region, 20 different electrophoretic profiles were obtained. Identical band sizes were observed in several groups of strains (Figure 2). Identical profiles exhibited three clinical C. albicans strains (cl/OZ/g2, cl/MP/4M, and cl/MP/2M) and food-derived C. tropicalis LOCK 0006, all showing the band size of 457 bp (cluster I). Cluster II included 3 C. boidinii isolates, i.e., two clinical isolates (cl/OZ/k1 and cl/MP/3M) and food-borne strain fo/BM/03, characterized by a band of 526 bp. A different electrophoretic profile with a band size of 463 bp was obtained for three clinical C. albicans strains in cluster III (cl/OZ/g3, cl/MP/1M, and cl/MP/4K). Cluster IV included three clinical C. lusitaniae isolates (cl/MP/8K, cl/KL/01, and cl/MP/3K), food-derived C. albicans fo/79/01, and food-borne C. lusitaniae fo/82/01, all of which exhibited a band size of 369 bp. The next cluster consisted of four C. lusitaniae strains, including two food-borne strains (fo/82/02 and fo/82/03) and two clinical isolates (cl/MP/07 and cl/MP/06), with an electrophoretic profile characterized by a 384 bp band. Another profile, with a single band of 377 bp, was observed for three clinical C. lusitaniae isolates in cluster VI (cl/KL/02, cl/MP/04, and cl/MP/01).
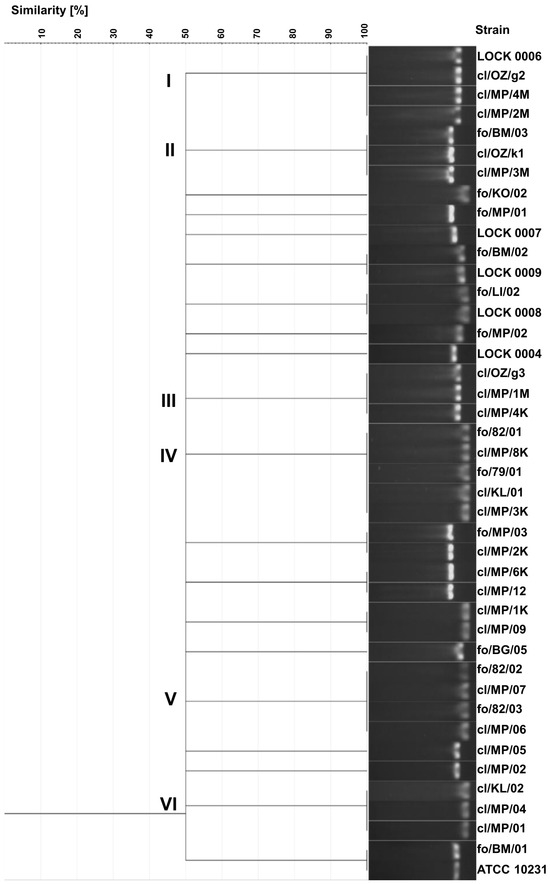
Figure 2.
UPGMA dendrogram illustrating the similarity among food-borne and clinical yeast strains based on PCR-amplified ITS region profiles visualized by gel electrophoresis banding patterns.
In addition, identical electrophoretic profiles were obtained for the following pairs of strains: P. fermentans fo/BM/02 and LOCK 0009, C. lusitaniae fo/LI/02 and LOCK 0008, C. boidinii fo/MP/03 and cl/MP/2K, C. boidinii cl/MP/6K and cl/MP/12, C. lusitaniae cl/MP/1K and cl/MP/09, and C. albicans fo/BM/01 and ATCC 10231. Only eight strains (C. lusitaniae fo/KO/02, C. boidinii fo/MP/01, W. anomalus LOCK 0007, P. membranifaciens fo/MP/02 and fo/BG/05, M. guilliermondii LOCK 0004, C. albicans cl/MP/05, and cl/MP/02) exhibited unique electrophoretic profiles of ITS region PCR products (Figure 2).
A total of 20 distinct electrophoretic profiles were obtained using this method, with yeasts of the same species generally clustering together. However, four electrophoretic profiles were indistinguishable among isolates from different species, indicating a limitation in the method’s taxonomic utility. Furthermore, clinical and food-derived strains were frequently grouped within the same cluster. The discrimination index of the yeast genotyping method based on ITS region polymorphism, relying on electrophoretic separation of the PCR product, was the lowest among all methods used, amounting to 0.957. This reduced discriminatory power may be attributable to limited resolution in band size differentiation and low strain heterogeneity. Electrophoretic profiles were scored with software support (GelJ), which was used for both band calling and cluster analysis.
3.4. Genotyping Based on Multiplex PCR
As a result of the multiplex PCR analysis, 39 different electrophoretic profiles were obtained, demonstrating greater diversity than that observed in the ITS region polymorphisms analysis. The higher discriminatory power of this method resulted from the increased complexity of band patterns generated in multiplex PCR, which captures polymorphisms across multiple loci, compared to the single band produced by ITS region-based genotyping.
Identical electrophoretic profiles were noted for only three pairs of strains, namely C. albicans cl/OZ/g2 and cl/MP/1M, C. lusitaniae LOCK 0008 and cl/KL/02, and C. lusitaniae fo/LI/02 and cl/KL/01 (Figure 3). Yeast strains were grouped into six internally diversified clusters, within which the similarity of electrophoretic profiles ranged from 50% to 100%. Cluster I included six strains belonging exclusively to one species, C. albicans, including five clinical isolates (cl/MP/1M, cl/OZ/g2/, cl/MP/4M, cl/OZ/g3, and cl/MP/2M) and food-borne fo/BM/01. Cluster II consisted of six clinical C. lusitaniae isolates (cl/MP/04, cl/MP/07, cl/MP/09, cl/MP/3K, cl/MP/8K, and cl/MP/06) and two food-derived P. fermentans strains (LOCK 0009 and fo/BM/02). Cluster III comprised both strains of P. membranifaciens originating from food (fo/BG/05 and fo/MP/02), as well as two clinical isolates of C. lusitaniae (cl/MP/1K and cl/MP/01). Cluster IV grouped together three clinical C. boidinii isolates (cl/MP/12, cl/MP/2K, and cl/MP/6K) and five C. lusitaniae, i.e., two clinical (cl/KL/01 and cl/KL/02) and three originating from food (fo/LI/02, LOCK 0008, and fo/82/01). Cluster V was the most diverse and contained three C. boidinii strains of various origins (cl/MP/3M, cl/OZ/k1, and fo/MP/01), clinical C. albicans cl/MP/02, as well as food-derived W. anomalus LOCK 0007 and M. guilliermondii LOCK 0004. Cluster VI was dominated by food-derived yeasts, namely C. lusitaniae fo/KO/02, fo/82/02, and fo/82/03, C. albicans fo/79/01, as well as C. boidinii fo/BM/03 and fo/MP/03. This group also included two clinical isolates of C. albicans (cl/MP/05 and cl/MP/4K) and the collection strain ATCC 10231 (Figure 3).
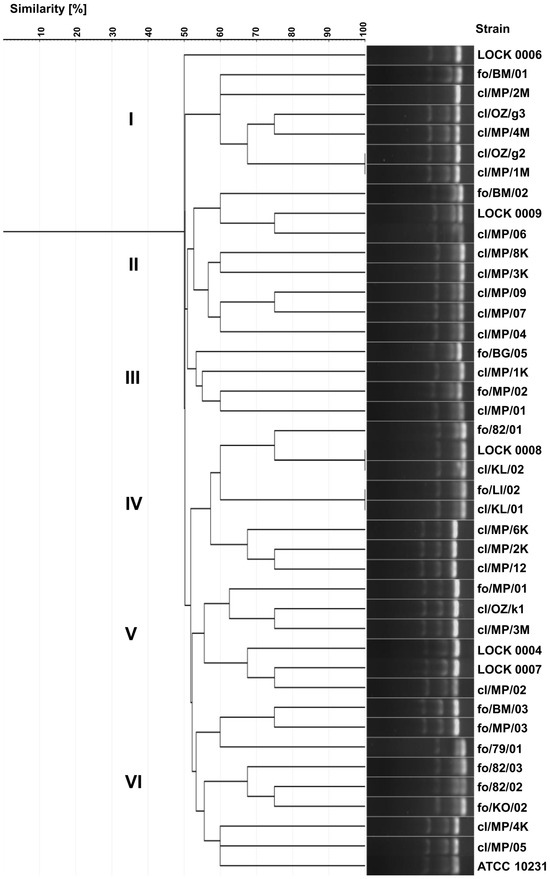
Figure 3.
UPGMA dendrogram illustrating the similarity of food-borne and clinical yeast profiles generated by multiplex PCR targeting the 18S, 5.8S, and 28S rDNA regions and visualized as electrophoretic banding patterns.
In multiplex PCR-based genotyping, as previously observed, both clinical and food-borne strains were grouped into the same clusters. Strains representing different species were also grouped together, which may reflect strain convergence. Notably, identical electrophoretic profiles were obtained only for yeasts classified within the same species. The discrimination index of the yeast genotyping method based on electrophoretic separation of multiplex PCR products was 0.997, and it was higher than the indices of both biotyping and ITS region-based genotyping.
3.5. Yeasts Karyotyping
Karyotyping revealed 42 different electrophoretic profiles of yeast chromosomal DNA (Figure 4), indicating that each of the strains tested exhibited a unique karyotype. The highest profile similarity observed between any two strains was ~90% and was observed in three pairs of yeasts, i.e., two food-derived C. boidinii strains (fo/MP/03 and fo/MP/01), two clinical C. lusitaniae isolates (cl/MP/07 and cl/MP/06), and two C. lusitaniae strains originating from food (fo/82/03 and fo/82/02).

Figure 4.
UPGMA dendrogram showing the similarity of electrophoretic karyotypes of food-borne and clinical yeast strains.
On the dendrogram, 75% of clinical strains were grouped into three clusters—cluster I, III, and VI—composed only of clinical isolates. However, in terms of species composition, all three clusters were internally diverse. Cluster I comprised two C. albicans isolates (cl/MP/1M and cl/OZ/g2) and C. boidinii cl/MP/3M. Cluster III included 6 out of 10 C. lusitaniae strains (cl/MP/09, cl/MP/07, cl/MP/06, cl/MP/04, cl/MP/8K, and cl/KL/02), along with two C. albicans isolates (cl/MP/05 and cl/MP/4M). Cluster VI encompassed three C. boidinii strains (cl/MP/2K, cl/MP/6K, and cl/OZ/k1), three C. albicans isolates (cl/MP/02, cl/OZ/g3, and cl/MP/2M), and C. lusitaniae cl/KL/01 (Figure 4). In cluster II, apart from C. albicans cl/MP/4K, food-derived strains were grouped, namely C. albicans fo/79/01, as well as all food-borne strains identified as C. boidinii (fo/BM/03, fo/MP/01, and fo/MP/03), P. fermentans (fo/BM/02 and LOCK 0009), P. membranifaciens (fo/BG/05 and fo/MP/02), C. tropicalis (LOCK 0006), M. guilliermondii (LOCK 0004), and W. anomalus (LOCK 0007). Cluster IV was the most diverse, containing both clinical and food-borne strains of C. lusitaniae (cl/MP/01, cl/MP/1K, fo/LI/02, fo/82/03, and fo/82/02), C. albicans (fo/BM/01 and ATCC 10231), and C. boidinii cl/MP/12. In contrast, cluster V included only C. lusitaniae strains, but differing in origin, i.e., three food-derived yeasts (LOCK 0008, fo/KO/02, and fo/82/01) and one clinical isolate cl/MP/3K.
Karyotyping, similarly to genotyping based on ITS sequences, proved to be a highly effective method for typing yeast, with the discrimination index of 1.000. Among the applied methods, karyotyping appeared to allow the most structured and coherent grouping of yeasts according to their origin.
4. Discussion
The evaluation of the effectiveness of yeast typing methods is typically based on several parameters, i.e., typeability, reproducibility, and discriminatory power [28]. Among these characteristics, in this study we focused on the discriminatory power of typing methods, which is defined as the ability to distinguish between unrelated strains [28]. From both clinical and epidemiological points of view, but also from a scientific perspective, differentiation of Candida spp. strains in terms of their origin is crucial, particularly in relation to sources of infection, yeast’s pathogenic potential, colonization patterns, resistance to antimycotics, as well as strain microevolution within species [4,6,18,19,29].
High discriminatory power does not always imply clinical relevance or reproducibility. The usefulness of numerous methods in this regard has been well described in previous publications [17,18,19,20,21,29]. Nevertheless, comparing the discriminatory power of different typing methods is difficult since the discrimination index was rarely reported. Available data indicate that the discrimination index for the microsatellites method may be 0.85–0.91 [18]; for DNA typing, 0.868 [28]; for RAPD fingerprinting, 0.984; and for karyotyping, only 0.630 [30]. It is assumed that for typing results to be interpreted with confidence, the discrimination index should be greater than 0.90 [28].
In this study, discrimination indices were determined for five selected typing methods, namely biotyping based on yeast assimilation profiles using the popular and commonly used API system, genotyping based on ITS region polymorphism and ITS sequencing, multiplex PCR of ITS regions, and karyotyping. The highest discriminatory capacity (D = 1.000), enabling differentiation of all Candida spp. strains tested, was achieved through karyotyping and ITS sequence analysis. The multiplex PCR method demonstrated very high discriminatory ability (D = 0.997), while biotyping showed lower discriminatory power (D = 0.966). ITS region-based genotyping was the least discriminatory among the methods evaluated, with a discrimination index of 0.957. However, despite the differences in the index values, all tested typing methods fulfilled the criterion for results to be interpreted with confidence [28]. On the other hand, a high index does not guarantee correct species identification, as demonstrated by the API 20C AUX system, which exhibited a misclassification rate of 64.3%. It should also be noted that the discrimination index value strongly depends on the number and diversity of strains analyzed.
Among the methods used in this study, karyotyping is also applicable for yeast identification. However, this method is challenging for species identification due to yeast chromosomal polymorphism, genomic instability, intraspecies variation, aneuploidy, and the occurrence of co-migrating bands [31]. Its primary utility lies in strain-level differentiation [31], which was confirmed by our results, as each isolate exhibited a distinct karyotype even among conspecific strains. Therefore, the utility of karyotyping is primarily in distinguishing strains rather than in species identification.
According to ITS sequence-based identification, a significantly higher species diversity was observed among both food-derived and clinical yeasts compared to biotyping results. Among the clinical strains, in addition to C. albicans and C. lusitaniae, isolates belonging to C. boidinii were also identified, with C. lusitaniae being the predominant species. This finding is partly consistent with previous reports on NAC species associated with human infections [6,17,32]. However, C. boidinii is rarely mentioned in this context, although clinical strains of this species have been reported [33]. Similarly, among food-borne yeasts, the identification of two strains as C. albicans is surprising, as this species has not been previously associated with food sources. Other strains identified among food-derived yeasts, i.e., W. anomalus and C. lusitaniae, are also considered opportunistic pathogens that may cause infections, especially in people at risk [17,32,34].
These findings are consistent with previous studies suggesting the inability to distinguish between clinical and environmental strains. In line with this, no genetic distinction was found between 20 clinical C. krusei isolates and 12 environmental P. kudriavzevii isolates, indicating that these yeasts belong to the same species [35]. High genetic congruence was also observed for yeasts originating from different environments in various regions of Mumbai (soil adjacent to urinals, sewage water, beach water, and hospital soil), and among the identified species, C. albicans, C. tropicalis, and C. krusei were found in descending order of abundance [19]. Moreover, environmental and food-borne strains may exhibit similar levels of drug resistance to clinically relevant isolates [19,35,36]. Douglass et al. [35] further hypothesize that food-borne or environmental exposure may contribute to colonization or infection in vulnerable individuals.
These findings are consistent with the One Health concept, which assumes the potential transmission of microorganisms between animals, humans, and ecosystems, with a particular focus on emerging and endemic zoonoses [37]. Antimicrobial resistance also remains a critical concern, as resistance can arise in humans, animals, or the environment and may spread between these reservoirs. Candida strains that were thermotolerant to human body temperature, pathogenic, and resistant to at least one antifungal drug were detected on plastic pollutants in aquatic ecosystems [38]. These plastic pollutants may serve as important vehicles for the dissemination of human pathogens and the potential exchange of antimicrobial resistance genes across various ecosystems, including terrestrial environments [39,40]. The significance of the natural environment in the epidemiology of infectious diseases—as a source of pathogens—was documented for C. auris, a recently emerged human pathogenic yeast. Environmental reservoirs of antibiotic-resistant C. auris include terrestrial, freshwater, and marine ecosystems, with colonized humans or animals or contaminated clinical waste, likely serving as sources of environmental contamination [41]. Other pathogenic Candida species—including C. albicans, C. glabrata, C. dubliniensis, C. krusei, and C. parapsilosis—were detected at various stages of wastewater treatment [42], further reinforcing the One Health paradigm.
The phenomenon of microorganism transmission between different environments may partly explain the high similarity of strains of diverse origins observed in this study, as well as the clustering of food-borne and clinical isolates within the same groups.
5. Conclusions
This study contributes to a better understanding of the discriminatory capacity and limitations of various typing methods used for yeast identification and strain differentiation. The highest discriminatory power (D = 1.000) for distinguishing Candida spp. strains from diverse sources was observed with genotyping based on ITS region sequencing and karyotyping. Identification using ITS sequences proved significantly more reliable than that based on assimilation profiles using the API system. Our findings highlight the need for a cautious interpretation of the discrimination index, particularly when D < 1.000 may reflect methodological limitations rather than true strain identity. The fact that all strains were differentiated by two out of five tested methods suggests distinct strain origins and highlights the importance of applying at least one typing method with a D = 1.000 in such studies. This approach minimizes the risk of misinterpreting strain-relatedness and origin. In this context, our findings may provide practical recommendations for researchers and clinicians when selecting appropriate tools for taxonomic and epidemiological investigations.
On the other hand, the results of this study should be regarded as preliminary, emphasizing the relevance of the discriminatory index. Very high D values may sometimes lead to an overestimation of diversity due to a method’s sensitivity to minor, potentially irrelevant variations. Moreover, this study focused primarily on discriminatory power, without including detailed phylogenetic or clinical outcome analyses. Future research should involve broader comparative approaches, including, for example, whole-genome sequencing of yeast strains. Integrating complementary techniques—such as combining molecular and phenotypic methods—may further improve the accuracy and reliability of yeast identification and strain typing.
Our primary objective was to draw attention to the potential for misinterpretation that may arise from methodological limitations. Moreover, these findings support the growing recognition of genetic-relatedness among Candida strains originating from varied environments, which has implications for infection control, outbreak investigation, and microbial ecology within the One Health framework.
Author Contributions
Conceptualization, K.R. and A.O.; methodology, K.R. and A.O.; investigation, K.R., A.O. and D.S.; data curation, K.R., A.O. and D.S.; writing—original draft preparation, review and editing, K.R. and A.O.; visualization, K.R. and A.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All nucleotide sequences of the ITS regions analyzed during the current study are available in the GeneBank NCBI database under the accession numbers PV670444 to PV670466, PV785352, and PV686762 to PV686779.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
API profiles of clinical and food-borne yeast strains.
Table A1.
API profiles of clinical and food-borne yeast strains.
| Substrate 1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cl/MP/02 | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/4K | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/1M | + | + | + | − | + | + | + | + | + | + | + | + | − | − | + | + | + | − | − |
| cl/MP/2M | + | − | + | − | − | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/4M | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/OZ/g2 | + | − | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/OZ/g3 | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | − | − | − |
| cl/MP/05 | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| ATCC 10231 | + | − | + | − | + | + | + | + | + | + | + | + | − | − | + | + | + | − | − |
| cl/MP/01 | + | + | + | + | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/1K | + | + | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/3K | + | − | + | − | − | + | + | + | + | + | + | + | − | − | + | + | + | − | − |
| cl/MP/8K | + | − | + | − | − | + | + | + | + | + | − | + | − | − | + | + | + | − | − |
| cl/MP/04 | + | − | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/06 | + | − | + | − | − | + | + | + | − | + | − | + | − | − | + | + | + | − | − |
| cl/MP/07 | + | − | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/09 | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/KL/02 | + | + | + | − | + | + | + | + | − | + | − | − | + | − | + | + | + | + | − |
| cl/KL/01 | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − |
| cl/MP/12 | + | − | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/3M | + | − | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/OZ/k1 | + | + | + | − | + | − | + | + | − | + | + | + | − | − | + | + | − | − | − |
| cl/MP/2K | + | − | + | − | + | − | + | + | − | + | + | + | − | − | + | + | + | − | − |
| cl/MP/6K | + | + | + | − | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
| fo/82/01 | + | + | + | − | + | − | − | − | − | + | + | + | + | − | + | + | + | + | − |
| fo/82/02 | + | + | + | − | − | + | + | + | − | + | + | + | + | + | + | + | + | + | − |
| fo/82/03 | + | + | + | + | − | + | − | + | − | + | − | + | − | − | + | + | + | + | − |
| fo/KO/02 | + | + | + | − | − | − | + | − | − | + | − | − | − | − | − | + | + | − | + |
| fo/LI/02 | + | + | + | − | − | + | + | + | − | + | + | + | + | + | + | + | − | + | + |
| LOCK 0008 | + | − | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| fo/79/01 | + | + | + | − | − | − | − | + | − | + | − | + | + | − | + | + | − | + | − |
| fo/BM/01 | + | − | + | − | + | + | − | + | − | + | − | + | − | − | + | − | + | − | − |
| fo/MP/01 | + | + | − | − | + | + | + | − | − | + | − | + | − | − | − | + | − | − | − |
| fo/MP/03 | + | + | − | − | + | + | + | − | − | + | − | + | − | − | − | − | − | − | − |
| fo/BM/03 | + | + | − | − | + | + | + | − | − | + | − | + | + | − | − | − | − | − | + |
| LOCK 0006 | + | + | + | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − |
| fo/MP/02 | + | + | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| fo/BG/05 | + | − | − | − | + | − | − | + | − | + | − | − | − | − | − | − | − | − | − |
| fo/BM/02 | + | + | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| LOCK 0009 | + | + | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| LOCK 0004 | + | + | + | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − |
| LOCK 0007 | + | + | − | − | − | − | − | − | − | − | + | − | − | − | + | + | − | + | − |
1 Substrates: 1—D-glucose, 2—glycerol, 3—calcium 2-keto-gluconate, 4—L-arabinose, 5—D-xylose, 6—adonitol, 7—xylitol, 8—D-galactose, 9—inositol, 10—D-sorbitol, 11—methyl-αD-glucopyranoside, 12—N-acetyl-glucosamine, 13—D-cellobiose, 14—D-lactose, 15—D-maltose, 16—D-saccharose, 17—D-trehalose, 18—D-melezitose, 19—D-raffinose.
References
- Lopes, J.P.; Lionakis, M.S. Pathogenesis and virulence of Candida albicans. Virulence 2021, 13, 89–121. [Google Scholar] [CrossRef] [PubMed]
- Parambath, S.; Dao, A.; Kim, H.Y.; Zawahir, S.; Izquierdo, A.A.; Tacconelli, E.; Govender, N.; Oladele, R.; Colombo, A.; Sorrell, T.; et al. Candida albicans—A systematic review to inform the World Health Organization Fungal Priority Pathogens List. Med. Mycol. 2024, 62, myae045. [Google Scholar] [CrossRef] [PubMed]
- Gaffar, N.R.; Valand, N.; Venkatraman Girija, U. Candidiasis: Insights into virulence factors, complement evasion and antifungal drug resistance. Microorganisms 2025, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Rhomberg, P.R.; Castanheira, M. CD101, a long-acting echinocandin, and comparator antifungal agents tested against a global collection of invasive fungal isolates in the SENTRY 2015 Antifungal Surveillance Program. Int. J. Antimicrob. Agents 2017, 50, 352–358. [Google Scholar] [CrossRef]
- Soriano, A.; Honore, P.M.; Puerta-Alcalde, P.; Garcia-Vidal, C.; Pagotto, A.; Gonçalves-Bradley, D.C.; Verweij, P.E. Invasive candidiasis: Current clinical challenges and unmet needs in adult populations. J. Antimicrob. Chemother. 2023, 78, 1569–1585. [Google Scholar] [CrossRef]
- Pinho, S.; Miranda, I.M.; Costa-de-Oliveira, S. Global epidemiology of invasive infections by uncommon Candida species: A systematic review. J. Fungi 2024, 10, 558. [Google Scholar] [CrossRef]
- Watkins, R.R.; Gowen, R.; Lionakis, M.S.; Ghannoum, M. Update on the pathogenesis, virulence, and treatment of Candida auris. Pathog. Immun. 2022, 7, 46–65. [Google Scholar] [CrossRef]
- Frías-De-León, M.G.; Hernández-Castro, R.; Conde-Cuevas, E.; García-Coronel, I.H.; Vázquez-Aceituno, V.A.; Soriano-Ursúa, M.A.; Farfán-García, E.D.; Ocharán-Hernández, E.; Rodríguez-Cerdeira, C.; Arenas, R.; et al. Candida glabrata antifungal resistance and virulence factors, a perfect pathogenic combination. Pharmaceutics 2021, 13, 1529. [Google Scholar] [CrossRef]
- Kieliszek, M.; Kot, A.M.; Bzducha-Wróbel, A.; Błażejak, S.; Gientka, I.; Kurcz, A. Biotechnological use of Candida yeasts in the food industry: A review. Fungal Biol. Rev. 2017, 31, 185–198. [Google Scholar] [CrossRef]
- Johnson, E.A.; Echavarri-Erasun, C. Yeast biotechnology. In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: London, UK, 2011; pp. 22–44. [Google Scholar]
- Fleet, G.H.; Balia, R. The public health and probiotic significance of yeasts in foods and beverages. In Yeasts in Food and Beverages, 1st ed.; Querol, A., Fleet, G.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 381–398. [Google Scholar]
- Carlino, N.; Blanco-Míguez, A.; Punčochář, M.; Mengoni, C.; Pinto, F.; Tatti, A.; Manghi, P.; Armanini, F.; Avagliano, M.; Barcenilla, C.; et al. Unexplored microbial diversity from 2,500 food metagenomes and links with the human microbiome. Cell 2024, 187, 5775–5795.e15. [Google Scholar] [CrossRef]
- Levine, J.; Dykoski, R.K.; Janoff, E.N. Candida-associated diarrhea: A syndrome in search of credibility. Clin. Infect. Dis. 1995, 21, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Talwar, P.; Chakrabarti, A.; Chalwa, A.; Mehta, S.; Walza, B.N.S.; Lumar, L.; Chung, K.S. Fungal diarrhoea: Association of different fungi and seasonal variation and their incidence. Mycopathologia 1990, 110, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Barclay, G.R.; McKenzie, H.; Pennington, J.; Parratt, D.; Pennington, C.R. The effect of dietary yeast on the activity of stable chronic Crohn’s disease. Scand. J. Gastroenterol. 1992, 27, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Rajkowska, K.; Kunicka-Styczyńska, A. Typing and virulence factors of food-borne Candida spp. isolates. Int. J. Food Microbiol. 2018, 279, 57–63. [Google Scholar] [CrossRef]
- Arafa, S.H.; Elbanna, K.; Osman, G.E.H.; Abulreesh, H.H. Candida diagnostic techniques: A review. J. Umm Al-Qura Univ. Appll. Sci. 2023, 9, 360–377. [Google Scholar] [CrossRef]
- Bonfim-Mendonça, P.S.; Fiorini, A.; Shinobu-Mesquita, C.S.; Baeza, L.C.; Fernandez, M.A.; Svidzinski, T.I.E. Molecular typing of Candida albicans isolates from hospitalized patients. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 385–391. [Google Scholar] [CrossRef]
- Jha, V.; Giri, R.; Koli, J.; Poojari, D.; Dhamapurkar, V.; Jhangiani, A.; Nikumb, D.; Rumani, S.; Markam, M.; Sahu, A.; et al. RAPD typing, antibiotic resistance profiling and genetic diversity of Candida isolates. Microbiol. Infect. Dis. 2022, 6, 1–8. [Google Scholar] [CrossRef]
- Bautista-Muñoz, C.; Boldo, X.M.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Identification of Candida spp. by randomly amplified polymorphic DNA analysis and differentiation between Candida albicans and Candida dubliniensis by direct PCR methods. J. Clin. Microbiol. 2003, 41, 414–420. [Google Scholar] [CrossRef]
- Małek, M.; Paluchowska, P.; Bogusz, B.; Budak, A. Molecular characterization of Candida isolates from intensive care unit patients, Krakow, Poland. Rev. Iberoam. Micol. 2017, 34, 10–16. [Google Scholar] [CrossRef]
- Makled, A.F.; Ali, S.A.M.; Labeeb, A.Z.; Salman, S.S.; Shebl, D.Z.M.; Hegazy, S.G.; Sabal, M.S. Characterization of Candida species isolated from clinical specimens: Insights into virulence traits, antifungal resistance and molecular profiles. BMC Microbiol. 2024, 24, 388. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Fujita, S.I.; Senda, Y.; Nakaguchi, S.; Hashimoto, T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 2001, 39, 3617–3622. [Google Scholar] [CrossRef] [PubMed]
- Kunicka-Styczyńska, A.; Rajkowska, K. Phenotypic and genotypic diversity of wine yeasts used for acidic musts. World J. Microbiol. Biotechnol. 2012, 28, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V. GelJ—A tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. Molecular evolutionary genetics analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]
- Bartie, K.L.; Williams, D.W.; Wilson, M.J.; Potts, A.J.; Lewis, M.A. PCR fingerprinting of Candida albicans associated with chronic hyperplastic candidosis and other oral conditions. J. Clin. Microbiol. 2001, 39, 4066–4075. [Google Scholar] [CrossRef]
- Giammanco, G.M.; Lopes, M.M.; Coimbra, R.S.; Pignato, S.; Grimont, P.A.; Grimont, F.; Freitas, G.; Giammanco, G. Value of morphotyping for the characterization of Candida albicans clinical isolates. Mem. Inst. Oswaldo Cruz. 2005, 100, 483–490. [Google Scholar] [CrossRef]
- Gouliamova, D.; Dimitrov, R.; Petrova, P.; Stoyancheva, G.; Petrov, K. Genomic approaches to yeast taxonomy. Biotechnol. Biotechnol. Equip. 2009, 23 (Suppl. S1), 519–523. [Google Scholar] [CrossRef]
- Mendoza-Reyes, D.F.; Gómez-Gaviria, M.; Mora-Montes, H.M. Candida lusitaniae: Biology, pathogenicity, virulence factors, diagnosis, and treatment. Infect. Drug Resist. 2022, 15, 5121–5135. [Google Scholar] [CrossRef]
- Linton, C.J.; Borman, A.M.; Cheung, G.; Holmes, A.D.; Szekely, A.; Palmer, M.D.; Bridge, P.D.; Campbell, C.K.; Johnson, E.M. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom mycology reference laboratory. J. Clin. Microbiol. 2007, 45, 1152–1158. [Google Scholar] [CrossRef]
- Ioannou, P.; Baliou, S.; Kofteridis, D.P. Fungemia by Wickerhamomyces anomalus—A narrative review. Pathogens 2024, 13, 269. [Google Scholar] [CrossRef] [PubMed]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.R.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef] [PubMed]
- Maroszyńska, M.; Kunicka-Styczyńska, A.; Rajkowska, K.; Maroszyńska, I. Antibiotics sensitivity of Candida clinical and food-borne isolates. Acta Biochim. Pol. 2013, 60, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.S.; Jeggo, M. The One Health approach-why is it so important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Metcalf, R.; Akinbobola, A.; Woodford, L.; Quilliam, R.S. Thermotolerance, virulence, and drug resistance of human pathogenic Candida species colonising plastic pollution in aquatic ecosystems. Environ. Sci. Pollut. Res. 2025. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Tyler, A.C.; Hoffman, M.J.; Savka, M.A.; Hudson, A.O. Is plastic pollution in aquatic and terrestrial environments a driver for the transmission of pathogens and the evolution of antibiotic resistance? Environ. Sci. Technol. 2019, 53, 1744–1745. [Google Scholar] [CrossRef]
- Gkoutselis, G.; Rohrbach, S.; Harjes, J.; Obst, M.; Brachmann, A.; Horn, M.A.; Rambold, G. Microplastics accumulate fungal pathogens in terrestrial ecosystems. Sci. Rep. 2021, 11, 13214. [Google Scholar] [CrossRef]
- Akinbobola, A.B.; Kean, R.; Hanifi, S.M.A.; Quilliam, R.S. Environmental reservoirs of the drug-resistant pathogenic yeast Candida auris. PLoS Pathog. 2023, 19, e1011268. [Google Scholar] [CrossRef]
- Biedunkiewicz, A.; Ozimek, T. Qualitative and quantitative changes of potentially pathogenic fungi in a hydrophyte wastewater treatment plant. Pol. J. Environ. Stud. 2009, 18, 161–166. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).