Abstract
Varroa destructor is a major health problem for honey bees (Apis mellifera). Selective breeding of Varroa-resistant bees is a suitable long-term solution to Varroa parasitism. After three generations of selecting honey bees for lower (resistant) and higher (susceptible) V. destructor population growth (LVG and HVG, respectively), LVG bees showed increased behavioral, cellular, and humoral immunity against Varroa. To further analyze resistance, the transcriptomes of both bee genotypes were examined, revealing that parasitized LVG bees had fewer differentially expressed genes (DEGs) than parasitized HVG bees, indicating a reduced impact by Varroa with greater resistance. Annotations of the altered DEGs showed that both genotypes were affected with an increased demand for energy, protein, and repair during parasitism. However, there were also DEGs in LVG bees, possibly related to resistance, such as up-regulation of odorant binding protein genes and down-regulation of the corazonin receptor gene, whereas DEGs in the HVG bees may be more related to stress, such as up-regulation of ATP synthase and down-regulation of the transcription factor dorsal. Overall, this work shows that selection for LVG and HVG bees resulted in genotypes with widespread differences in gene expression during Varroa parasitism, which may be related to resistance and susceptibility.
1. Introduction
Western honey bees (Apis mellifera) experience increased mortality due to the parasitic mite Varroa destructor, resulting in major losses each year [1]. For example, a review of colony losses in Europe showed that most colonies died after three years if only typical colony management was used without Varroa control treatment [2]. Varroa punctures the bee cuticle and consumes fat body tissue and hemolymph from both brood and adults [3,4], as well as injecting a variety of proteins in its saliva that can damage hemocytes [5]. It can also inject or stimulate latent viruses like deformed wing virus (DWV) [6,7]. Both Varroa parasitism and DWV infections decrease the immune response and lifespan of bees [1].
Parasitism by Varroa can also alter the transcriptome of the honey bee. For example, parasitized adult bees had 99 up-regulated differentially expressed genes (DEGs) mostly related to immune and sphingolipid mechanisms, and 79 down-regulated DEGs mostly related to oxidative stress and olfactory recognition, resulting in the conclusion that immunity was induced but detection of the parasite and stress responses were suppressed [8]. Honey bees exposed to Varroa showed up-regulation of 126 DEGs and down-regulation of 46 DEGs including those related to stress, immunity, and neurological system function [9]. Another example is 43 down-regulated DEGs and 57 up-regulated DEGs found in the brains of Varroa-parasitized bees, some of which were related to neurodegenerative disorders and viral myocarditis, indicating that the DEGs could help to explain neurological dysfunction and viral damage in parasitized bees [10]. Thus, transcriptomic studies have been very useful to better understand the interactions of bees with Varroa, demonstrating a diversity of functions that are affected during parasitism.
Control of Varroa primarily depends upon the use of synthetic acaricides, but Varroa can develop resistance against them [11]. Thus, alternative control methods are needed. One alternative to control Varroa is to breed honey bees for resistance against the mite, such as by selecting for low Varroa population growth (LVG) [12,13] or selecting for specific resistant mechanisms against the mite like increased hygienic behavior [14] or grooming behavior [15,16,17]. Changes in gene expression have been associated with selection for resistance to the mite, such as Varroa-parasitized bees selected for high Varroa-sensitive hygiene (VSH) versus low VSH; up-regulated DEGS were associated with neural connections and brood care behavior, whereas down-regulated DEGs were related to visual and olfactory aspects of behavior resulting in the conclusion that some aspects of neural activity and behavior were increased while others were decreased [18]. Parasitized bees selected for lower Varroa infestation levels, compared to non-selected bees, showed up-regulation of a transcriptional activator gene and down-regulation of an embryonic central nervous system development gene [19]. Morfin et al. [20] found that bees selected for LVG had up-regulated DEGs related to odorant binding proteins, neuron regulation, and ecdysone synthesis compared to those bees selected for high Varroa population growth (HVG) when undergoing grooming behavior, indicating that selection for LVG affected parasite detection and neural functions associated with increased hygienic behavior.
The goal of this work was to examine the transcriptomes of bees described by De la Mora et al. [13], who reported that three generations of selecting colonies for LVG and HVG resulted in adult LVG honey bees with 72% less mite infestation and 46% greater longevity compared to adult HVG bees. The LVG bees also differed in many aspects associated with Varroa resistance, including 14–61% more grooming behavior (proportion of groomer bees and mutilated mites), 35% more hygienic behavior (percentage of cleaned cells), 55–80% more hemocytes, and 62–82% higher gene expression for hymenoptaecin and defensin 2 [21]. This indicated widespread differences following selection, and this study sought to further examine the differences between the genotypes by comparing the transcriptomes of LVG and HVG bees, with and without Varroa parasitism, to identify gene expression differences that may be associated with resistance or susceptibility.
2. Materials and Methods
2.1. Samples of Bees Selected for LVG and HVG That Were Treated with V. destructor
Honey bee colonies were previously selected for three generations for lower (resistant) and higher (susceptible) rates of Varroa population growth as per De la Mora et al. [13]. Briefly, Varroa populations were determined by assessing the number of mites that fell onto sticky boards per day in spring and summer. Then, the rate of Varroa population growth was calculated by estimating the proportional difference between the Varroa counts in late summer (August) and those in spring (May). Colonies with the lowest Varroa population growth were classified as LVG, whereas colonies with the highest Varroa population growth were classified as HVG. Three colonies per genotype were randomly chosen from the third generation of selection for LVG and HVG [13]. Combs from each colony were incubated (Lab-Line Instruments, Inc., Dubuque, IA, USA) at 35 °C and 60% RH overnight in screened emerging cages (50.3 × 7.3 × 25.2 cm), and newly emerged bees were collected [10]. Varroa from highly infested colonies that were unrelated to the project were collected from adult bees using CO2 as per Dietemann et al. [22]. To expose bees to Varroa, ten newly emerged bees from each colony were placed in a wooden three-hole cage (75 × 25 × 16 mm), and to ensure that individual bees were challenged with Varroa parasitism, one mite was placed on each bee using a fine paintbrush. There were five cages per treatment with four treatments: LVG bees without Varroa (LVG-C), LVG bees with Varroa (LVG-V), HVG bees without Varroa (HVG-C), and HVG bees with Varroa (HVG-V). To allow Varroa to sufficiently parasitize the bees, the cages were incubated for 8 days at 32–35 °C and 60% RH with queen candy (sugar syrup mixed with icing sugar) and water provided. Dead bees were removed daily. Live bees were collected and stored at −80 °C.
2.2. RNA Extraction, Sequencing, and Library Preparation
To extract RNA from entire bees, 15 frozen bees per sample were macerated with 5 mL of One Step RNA Reagent (BioBasic, Markham, ON, Canada) as per Morfin et al. [20] with modifications as per De la Mora et al. [13], following the manufacturer’s instructions. Fifteen bees per biological replicate were used as this was found to be the fewest bees to provide low variability in gene expression analysis among worker bees from the same hive, which are half-sisters with potentially different drones as fathers [23]. The macerate was transferred, incubated at 20–22 °C for 5 min, and 300 μL of chloroform (MilliporeSigma, Burlington, MA, USA) added. After vortexing (Fisher Scientific, Waltham, MA, USA) for 15 s at 7000 rpm, the sample was incubated for 2–3 min at 20–22 °C, and then centrifuged (Symphony 417R, VWR, Mississauga, ON, Canada) at 12,000× g for 15 min at 4 °C. The aqueous phase was transferred, and then 500 μL 99% isopropanol was added followed by incubation for 10 min at 20–22 °C. After centrifugation at 4 °C for 10 min at 12,000× g, the RNA pellet was washed three times with 70% ethanol and dried for 10 min at 20–22 °C. The pellet was dissolved in 30 μL of Invitrogen UltraPure H2O (Fisher Scientific, Waltham, MA, USA) and sent to Génome Québec Innovation Centre, Montreal, QC, Canada, for high-throughput sequencing using an Illumina NovaSeq (25 M reads per lane) (Illumina, San Diego, CA, USA) with 100 bp paired-end reads. The library type was polyA-enriched RNA.
2.3. Bioinformatic Analyses
Bioinformatic analyses were performed at the Bioinformatics Core Facility, Montreal Clinical Research Institute, Montreal, QC, Canada. The quality of the raw reads was assessed with FASTQC v0.11.8 [24], and the results summarized using MultiQC, v1.14 [24]. Raw counts were calculated with FeatureCounts v1.6.0, based on the honey bee reference genome (Amel_HAv3.1) [25]. Then, trimming was performed with TRIMMOMATIC v0.36 [26]. The reads were aligned to the Apis mellifera reference genome (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_003254395.2/; accessed on 23 March 2024) with STAR v2.7.6a [27] using default parameters, which allows up to 30% mismatches and tolerates gaps in read sequences. Aligned RNA-seq normalized fragment counts were assembled into transcripts, and their abundance in fragments per kilobase of exon per million fragments mapped (FPKM) was determined through Cufflinks [28]. Reads per kilobase of transcript per million mapped reads (RPKM) were also calculated with the percent relative error (PRE) of the RPKM values determined for each sample. Differential Expression Analysis (DEA) was performed to determine differentially expressed genes (DEGs) with the DESeq2 R package [29] and edge R Bioconductor package v3.19 [30] based on the raw read counts. DEG heatmaps were drawn based on the z-score of the normalized count. Functional enrichment analysis of DEGs based on gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/, accessed on 24 May 2024) pathway enrichment were performed with the gprofiler2 R package v0.2.3 (accessed on 23 March 2024) [31].
Venn diagrams were created for the DEGs using the Bioinformatics and Evolutionary Genomics website (http://bioinformatics.psb.ugent.be/webtools/Venn/, accessed on 24 May 2024) [32]. GO and KEGG pathway enrichment analyses were performed by using the g:profiler search for biological process terms, considering a depth of two hierarchical levels [31]. ShinyGO v0.741 search (https://bioinformatics.sdstate.edu/go74/; accessed on 24 May 2024) [33] was used for GO and KEGG analysis. GO analysis results (biological process, cellular component, and molecular function) were related to physiological function using the Mouse Genome Database (MGD) (https://www.informatics.jax.org/vocab/gene_ontology; accessed on 24 May 2024) [34].
2.4. Statistical Analyses
Transcript expression levels and tests for significant differences (p < 0.05) were calculated with Cuffdiff [28]. The log2 fold change (log2FC) estimated the fold change between the treatments, based on the distribution of the reads. Lists of up- and down-regulated DEGs were determined using thresholds for the adjusted p-value (padj) and log2FC. For graphical representations, the first threshold yielding less than 1000 DEGs in each comparison was used to generate both heatmaps and volcano plots. Statistical analyses were performed with the R 4.1.1. software (R Development Core Team, Auckland, The Netherlands) [35]. Principal component analysis (PCA) was generated with FactorMineR and was based on the regularized log-transformed normalized counts. The Shapiro–Wilk test was used to analyze normality for the number of reads, number of aligned reads, percentage of aligned reads, and percentage of GC content. The Student t-test was used for pairwise comparisons between genotypes and between treatments within genotypes. The chi-square test was used to make pairwise comparisons of DEGs between treatments within genotypes (HVG-V versus HVG-C and LVG-V versus LVG-C) and between genotypes regardless of treatment (LVG versus HVG).
3. Results
3.1. Treatments and Reads
A total of 939,413,112 reads were obtained with an average of 78.3 million reads per sample (Table S1). The average percent alignment to the reference genome of A. mellifera was 59%, with 78.1% (±5.50) for HVG-C and HVG-V bees, which was significantly higher than that of LVG-C and LVG-V bees at 39.2% (±2.70) (t = 6.344, p < 0.05, df = 11). The average GC contents of the reads were 42.5% (±0.62) for HVG-C and HVG-V, and 42.3% (±0.54) for LVG-C and LVG-V bees, which were not significantly different from each other (t = −0.102, p > 0.05). After mapping the reads to the A. mellifera reference genome (Amel_HAv3.1), the gene counts between samples clustered into four groups based on principal component analysis (PCA), corresponding to the genotype and treatment, except for LVG-V replicate 2, which differed from all other samples (Figure 1).
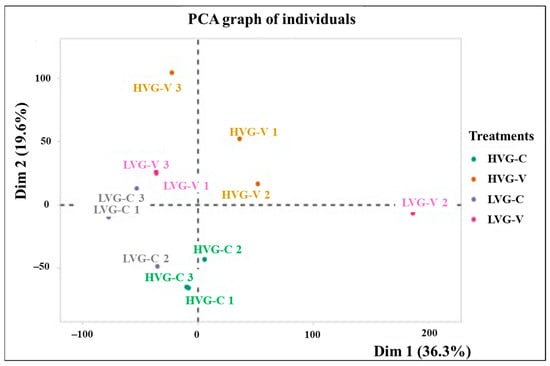
Figure 1.
Principal component analysis (PCA) of the transcriptome of colonies 1, 2 and 3 for each treatment (LVG-C, HVG-C, LVG-V, and HVG-V). The proportion of variance is represented with the numbers in parentheses. Dim 1 and Dim 2 represent the top two dimensions of the genes among treatments (36.3% and 19.6%, respectively).
3.2. DEGs from Treatment Comparisons
There was a total of 1332 DEGs based on a significant change of log2 fold or more among the treatment/genotype comparisons (Table S2). There were significantly more up-regulated DEGs per sample for the HVG than the LVG genotype (χ2 = 4.758, p < 0.05) and significantly more down-regulated DEGs per sample for the HVG than the LVG genotype (χ2 = 1.571, p < 0.05), indicating that the total effect on the transcriptome due to Varroa parasitism was greater with the HVG than the LVG genotype. Volcano plots showed an approximately even distribution of the fold changes for up- and down-regulated DEGs in each comparison (Figure S1).
The fewest total DEGs were found for the comparison between the non-parasitized bees of the two genotypes with no up-regulated genes and one down-regulated gene for LVG-C versus HVG-C (Table S2; Figure 2). That DEG was not shared with any other comparison. The next lowest was found for the comparison between the parasitized bees of the two genotypes with 10 and 16 up-regulated and down-regulated DEGs, respectively, for LVG-V versus HVG-V. Those up-regulated DEGs were unique to that comparison, while 15 of the down-regulated DEGs were unique and 1 was shared with the HVG-V versus HVG-C comparison.
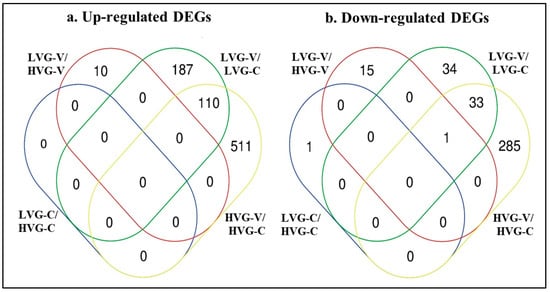
Figure 2.
Venn diagrams showing number of DEGs only found or shared with treatment comparisons of LVG-C versus HVG-C, LVG-V versus HVG-V, LVG-V versus LVG-C, and HVG-V versus HVG-C, based on Differential Expression Analysis (DEA). (a) Up-regulated DEGs. (b) Down-regulated DEGs. The ovals marked with blue, red, green and yellow lines contain the number of DEGs in the LVG-C versus HVG-C, LVG-V versus HVG-V, LVG-V versus LVG-C, and HVG-V versus HVG-C comparisons, respectively.
In contrast, the most DEGs were found with the comparison of Varroa-parasitized to non-parasitized HVG bees (HVG-V versus HVG-C) at 621 and 319 up-regulated and down-regulated DEGs, respectively (Table S2; Figure 2). Among those up-regulated DEGs, 511 were unique to the comparison, while 110 were shared with the Varroa-parasitized to non-parasitized LVG bees (LVG-V versus LVG-C) comparison. For those down-regulated DEGs, 285 were unique to the comparison, while 33 were shared with the LVG-V versus LVG-C comparison, and 1 was shared with the non-parasitized LVG bees and non-parasitized HVG bees (LVG-C versus HVG-C) comparison. The number of DEGs comparing Varroa-parasitized to non-parasitized LVG bees (LVG-V versus LVG-C) was less with 297 and 68 up-regulated and down-regulated DEGs, respectively. Among those up-regulated DEGs, 187 were unique to the comparison, while 110 were shared with the Varroa-parasitized to non-parasitized HVG bees (HVG-V versus HVG-C) comparison. For those down-regulated DEGs, 34 were unique to the comparison, while 33 were shared with the Varroa-parasitized to non-parasitized HVG bees (HVG-V versus HVG-C) comparison and 1 shared with the non-parasitized LVG bees and non-parasitized HVG bees (LVG-C versus HVG-C) comparison. The differences between those comparisons were all significant (χ2 = 43.71, p < 0.05). Thus, the effect on the number of DEGs was more due to Varroa parasitism than genotype, and although some of the effects on gene expression due to Varroa parasitism were shared between genotypes, a large amount was unique to each genotype.
3.3. Up-Regulated DEGs
Among the annotations of the 511 up-regulated DEGs only observed in the comparison of Varroa-treated versus control bees of the HVG genotype (HVG-V versus HVG-C), the most common coded for ribosomal proteins (16 DEGs), ATP synthase (7 DEGs), histone (7 DEGs), and vesicle transport (4 DEGs) (Table S3). For those DEGs with GO biological processes, all were related to nitrogen metabolism, and the GO cellular components were mostly related to intracellular organelles, organelles, and membranes (Figure 3a). The GO molecular functions were mostly related to structural activity and the ribosome, and the KEGG pathways were all related to the ribosome and vesicle transport.
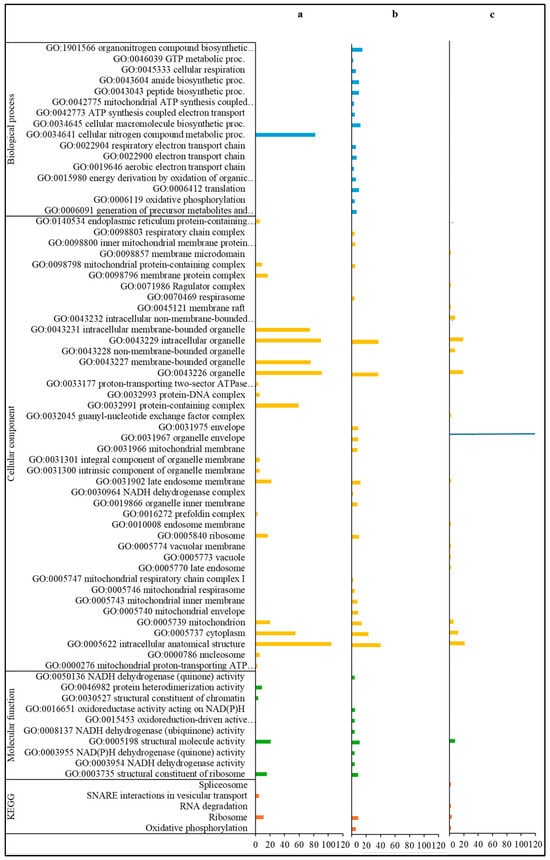
Figure 3.
Gene ontology classification and KEGG pathways of up-regulated DEGs. DEGs only detected in the HVG-V versus HVG-C comparison (p < 0.05) (a), DEGs only found in the LVG-V versus LVG-C comparison (p < 0.05) (b), and DEGs shared between LVG-V versus LVG-C and HVG-V versus HVG-C comparisons (p < 0.05) (c). The y-axis indicates the GO (biological process, cellular component, and molecular function) and KEGG classification. The x-axis indicates the number of DEGs matching the classification. Bars in blue, yellow, green and orange colors indicate the number of DEGs for GO biological processes, GO cellular components, GO molecular functions, and KEGG pathways, respectively.
For the annotations of the 187 up-regulated DEGs only found in the comparison of parasitized versus non-parasitized LVG bees (LVG-V versus LVG-C), the most common were ribosomal proteins (15 DEGs), NADH dehydrogenase (7 DEGs), mitochondrion-related (7 DEGs), cytochrome (5 DEGs), and spliceosomal RNA (5 DEGs) (Table S4). For GO biological processes, most were related to the generation of energy and metabolites, biosynthesis of macromolecules and organonitrogen compounds, and nitrogen compound metabolism (Figure 3b). The GO cellular components were mostly related to intracellular structures and organelles, particularly the mitochondrion, and the GO molecular functions were mostly related to oxidoreductase activity. The KEGG pathways were mostly related to intracellular structures and the generation of energy and metabolites.
Among the 110 up-regulated DEGs shared between comparisons of the two genotypes for Varroa versus control bees (LVG-V versus LVG-C shared with HVG-V versus HVG-C), the most common annotations were 4 DEGs for ribosomal proteins, 4 for mitochondrion related, 3 for histone, and 3 for snRNA (Table S5). None of the DEGs were characterized by a GO biological process, but for those linked to a GO cellular component, most were related to organelles and intracellular structures (Figure 3c). The most common GO molecular function was related to the activity of structural molecules. The KEGG pathways were widely distributed without any one being more dominant.
Among the 10 up-regulated DEGs only detected in the comparison of Varroa-treated bees between the two genotypes (LVG-V versus HVG-V), two had closely related annotations, both of which were odorant binding proteins (Table S6). Based on the GO biological process, three DEGs were related to the regulation of molecular function, organic substance metabolism, localization, and response to other organisms (Figure S2). The GO cellular component of one DEG was related to the extracellular region. No DEGs were associated with a GO molecular function. The KEGG pathway of one DEG was related to organic substance metabolism and intracellular structure.
3.4. Down-Regulated DEGs
For the annotations of the 285 down-regulated DEGs only found in the comparison of Varroa to control in HVG bees (HVG-V versus HVG-C), the most common were related to serine/threonine kinase (7 DEGs), homeobox protein (6 DEGs), odorant binding and receptor (5 DEGs), acetyl-coenzyme A (4 DEGs), facilitating trehalose transporter (4 DEGs), and zinc finger protein and transporter (4 DEGs) (Table S7). For DEGs with biological GO processes, all were related to localization, organic acid, carbohydrate and small molecule metabolism, transmembrane transport, and oxidoreductase activity (Figure 4a and Figure 5a). There were no DEGs with a GO cellular component, but the DEGs with a GO molecular function were all related to transporter/transmembrane transporter activity, and small molecule, DNA, and carbohydrate binding (Figure 5a). The DEGs with KEGG pathways were all related to precursor and carbon metabolism, energy, and amino acid synthesis (Figure 5a).
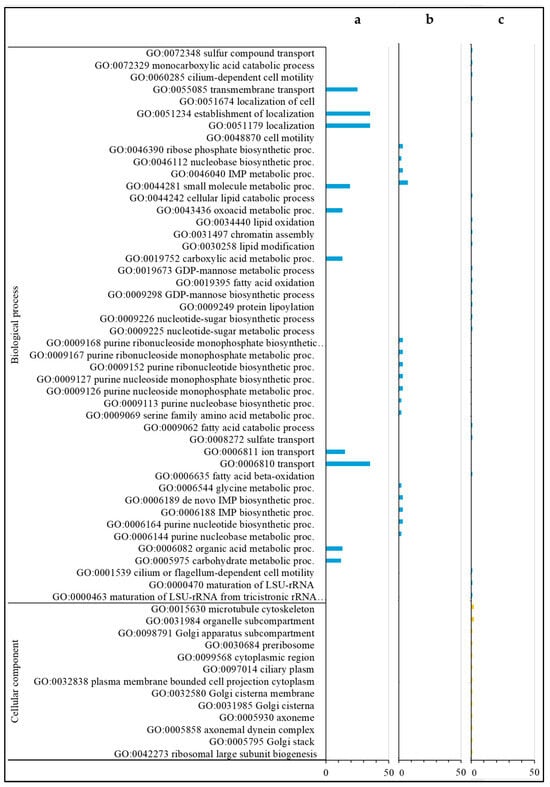
Figure 4.
Gene ontology (biological processes and cellular components) classification of down-regulated DEGs. DEGs only found in the HVG-V versus HVG-C comparison (p < 0.05) (a), DEGs only found in the LVG-V versus LVG-C comparison (p < 0.05) (b), and DEGs shared between LVG-V versus LVG-C and HVG-V versus HVG-C comparisons (p < 0.05) (c). The y-axis indicates the GO (biological process, cellular component, and molecular function) and KEGG results. The x-axis indicates the number of genes matching the classification. Bars in blue, yellow, green and orange colors indicate the number of DEGs for GO biological processes, GO cellular components, GO molecular functions, and KEGG pathways, respectively.
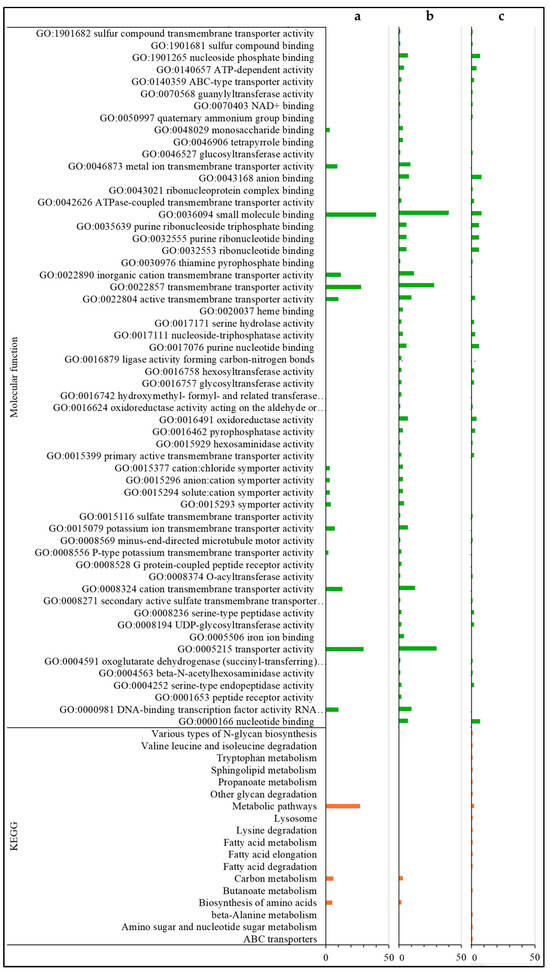
Figure 5.
Gene ontology (molecular function) classification and KEGG pathways of down-regulated DEGs. DEGs only found in the HVG-V versus HVG-C comparison (p < 0.05) (a), DEGs only found in the LVG-V versus LVG-C comparison (p < 0.05) (b), and DEGs shared between LVG-V versus LVG-C and HVG-V versus HVG-C comparisons (p < 0.05) (c). The y-axis indicates the GO (biological process, cellular component, and molecular function) and KEGG results. The x-axis indicates the number of genes matching the classification. Bars in blue, yellow, green and orange colors indicate the number of DEGs for GO biological processes, GO cellular components, GO molecular functions, and KEGG pathways, respectively.
The most common annotations of the 34 down-regulated DEGs only detected in the comparison of Varroa versus control in LVG bees (LVG-V versus LVG-C) were related to the ATP binding cassette (2 DEGs) (Table S8). For the DEGs with a GO biological process, most were related to the metabolism of carbohydrates, cellular lipid, protein, lipid/fatty acid metabolism, and cellular mobility (Figure 4b). The GO cellular components were mostly related to the Golgi apparatus and membrane organelles (Figure 4b), and the GO molecular functions were mostly related to nucleotide binding, ion binding, hydrolase activity, ATP-dependent activity, and transferase activity (Figure 5b). KEGG pathway classifications were mostly related to amino acid, fatty acid, and lipid metabolism (Figure 5b).
For the annotations of the 33 down-regulated DEGs shared between the comparison of Varroa versus control of both LVG and HVG bees (LVG-V versus LVG-C shared with HVG-V versus HVG-C), the most common were major royal jelly protein (4 DEGs) and receptor related (3 DEGs) (Table S9). For GO biological processes, almost all were related to nucleobase compound metabolism, oxidoreductase activity, and small molecule metabolism for energy, protein metabolism, and oxidoreductase activity (Figure 4c). The DEGs with GO cellular components were mostly related to the Golgi and cytoskeleton (Figure 4c), and GO molecular functions related to oxidoreductase activity, ion and cyclic organic compound binding, and peptide receptors (Figure 5c). KEGG pathway classifications were mostly for amino acid biosynthesis and carbon metabolism (Figure 5c).
There were no annotations of the 15 down-regulated DEGs only found in the comparison of Varroa treated bees between genotypes (LVG-V versus HVG-V) (Table S10). Only one DEG could be characterized by GO and KEGG with the extracellular region as a GO cellular component, and carbohydrate derivative binding as a GO molecular function. The RNA catabolic process was the KEGG pathway.
There were also no annotations for the one down-regulated DEG only detected in the Varroa non-treated bees between genotypes (LVG-V versus HVG-V) (Table S11). It could not be characterized by GO or KEGG analysis.
The annotation for the one down-regulated DEG shared between the three-way comparison of Varroa between genotypes, Varroa versus control of both LVG and HVG bees (LVG-V versus HVG-V shared with LVG-V versus LVG-C and shared with HVG-V versus HVG-C) denoted major royal jelly protein (Table S12). The GO molecular function was related to DNA binding, and the GO cellular component was extracellular. The KEGG pathway was RNA binding.
4. Discussion
The percent alignment to the reference genome of A. mellifera was much less for LVG than HVG bees, indicating that selection for LVG bees resulted in more sequence variants from the reference genome than HVG bees. Such differences also implied that gene expression would differ between LVG and HVG bees. The largest number of DEGs was found between non-parasitized and parasitized bees in each genotype, suggesting that changes in gene expression are mainly triggered by V. destructor. Shared DEGs between the Varroa-triggered DEGs in LVG and HVG bees indicate that there were some gene expression changes during parasitism that were not greatly affected by selection. However, more DEGs triggered by Varroa were unique to each genotype, indicating that not only the number of DEGs but their functions were affected by selection.
There were approximately 2.5-times more DEGs due to Varroa parasitism in HVG than LVG bees, even though the samples had the same number of bees and similar biomasses. This indicates that Varroa has a broader impact on the susceptible genotype. This may be due to greater damage and stress in the susceptible than the resistant genotype or perhaps the induction of more, but possibly less effective, resistance mechanisms in the susceptible genotype. However, another possibility is that there were fewer DEGs with the resistant genotype because selection for LVG may have reduced gene expression noise (random variation in gene expression among cells, even in the same environment) [36]. Increased expression noise can reduce the fitness of a cell by at least 25%, and this reduction cannot be substantially attenuated by gene overexpression. Also, expression noise may result in organisms being both less adapted and less adaptable [37]. Thus, in addition for selecting for Varroa resistance, selection of LVG bees may have resulted in less expression noise and therefore better adaptability to Varroa parasitism.
Up-regulated DEGs in Varroa-parasitized LVG bees in comparison to Varroa-parasitized HVG bees were those genes that may be related to resistance as they were significantly more expressed in the parasitized resistant genotype than the susceptible genotype (up-regulated DEGs only observed in the LVG-V to HVG-V comparison; Table S6). They included two odorant binding protein genes that may be related to the greater ability of LVG bees to detect odors. Volatile odors from Varroa-infested pupae can be learned and discriminated from non-infested pupae [38], and higher expression of odorant binding proteins was associated with bees triggering more rapid self-grooming instances [20]. Thus, higher expression of odorant binding proteins in LVG-V than HVG-V bees could indicate enhanced hygienic and grooming behaviors, as reported in bees with enhanced hygienic behavior [38]. KEGG analysis of the DEGs indicated some were related to lysozyme and glycosaminoglycan degradation. Greater lysozyme could increase Varroa resistance as insect lysozymes have antibacterial and antifungal activity [39]. Higher expression of the lysozyme genes was observed in Varroa-parasitized worker bees, which could have occurred as part of the response of the innate immune system [40]. Greater glycosaminoglycan degradation in LVG bees could increase resistance as glycosaminoglycans can promote pathogenesis by facilitating pathogen attachment, invasion, or evasion of host defense mechanisms [41]. As Varroa can suppress bee immunity [1], more glycosaminoglycan degradation in the LVG genotype could restrain opportunistic pathogens, such as DWV, which increases in Varroa-parasitized bees [6,7]; this may help explain why DWV infection levels were reduced in parasitized LVG bees compared to parasitized HVG bees [13,21].
Among the up-regulated DEGs due to the effects of Varroa in comparison to the control bees that were only found with LVG bees, some could be related to greater Varroa resistance mechanisms (up-regulated DEGs only observed in the LVG-V to LVG-C comparison; Table S4). The relatively large number of ribosomal genes among these DEGs indicates greater protein production; this is consistent with up-regulated DEGs for the spliceosome, which removes introns from a transcribed pre-mRNA [42]. More protein production is also consistent with GO analysis showing biosynthesis of macromolecules and organonitrogen compounds, and nitrogen compound metabolism. There was also up-regulation of DEGs for mitochondria indicating greater energy production, including NADH dehydrogenase, the first enzyme in the respiratory chain. This is consistent with GO and KEGG analyses showing up-regulated DEGs related to oxidoreductase activity and energy generation. Perhaps more protein- and energy-requiring processes in LVG bees during Varroa parasitism enable the bees to compensate for Varroa damage. NADH levels decrease in Varroa-parasitized bees due to lower energy production as the bees become weaker [43]. More protein- and energy-requiring processes may also provide energy and compounds needed for resistance mechanisms. Another more up-regulated DEG unique to parasitized LVG bees was related to cytochrome-related function, indicating that LVG bees may perform more detoxification than HVG bees during parasitism.
The majority of up-regulated DEGs were related to the effects of Varroa in comparison to control bees that were only found with HVG bees (up-regulated DEGs only observed in the HVG-V to HVG-C comparison; Table S3). Many were possibly related to greater stress responses or less successful resistance in the susceptible genotype. Like the LVG genotype, there were many DEGs for protein production, such as ribosomal proteins, which is consistent with GO analysis detecting DEGs related to nitrogen metabolism. Also, similar to parasitized LVG bees, there were up-regulated DEGs in HVG bees for energy, such as ATP synthase. In addition to possibly less successful resistance and greater stress, perhaps these changes are part of the compensation for greater nutrient loss due to Varroa feeding of the susceptible genotype. Up-regulation of DEGs for SNARE interactions could be related to the susceptibility of HVG bees as higher expression of SNARE genes is associated with a potentially higher risk of pathogen exposure, such as bacterial infection of Drosophila melanogaster [44].
Up-regulated DEGs triggered by Varroa in both LVG and HVG bees are likely due to the effects of Varroa that were not significantly altered from the parental material during the three rounds of selection (up-regulated DEGs shared between LVG-V to LVG-C comparison and HVG-V to HVG-C comparison; Table S5). Like the up-regulated DEGs due to Varroa observed only in each genotype, these DEGs included those for protein production such as ribosomal proteins, spliceosomal RNA, and energy generation, such as mitochondrion-related genes. This indicates that some increased protein and energy production occurs with parasitism similarly in both genotypes. However, unlike the DEGs triggered by Varroa in only the LVG or HVG bees, many of the GO terms for these DEGs shared between the genotypes were related to membranes, indicating that membrane damage may be occurring during Varroa feeding, regardless of resistance. One reason for bee membrane damage could be the activity of proteins in Varroa saliva that can lyse hemocytes, presumably due to membrane disruption [5], as well as lysis of fat body cells during their digestion by Varroa [3]. One up-regulated DEG in both genotypes that may have a direct relationship with susceptibility to Varroa is for guanyl-nucleotide exchange factor, which could result in increased exchange of GDP to GTP and activate signaling GTPases. Dysregulation of Rho GTPase function corresponds to certain pathologies, such as cancer progression and mental disabilities in humans [45]. Also, Rab GTPases are key regulators of vesicular transport that can be altered by infectious diseases as part of host defenses or as a way for pathogens to avoid host defenses [46]. Thus, changes in GTPase in both genotypes during parasitism could contribute to reduced functioning of key elements of bee metabolism and allowing Varroa to avoid certain defense responses [46].
Down-regulated DEGs in Varroa-parasitized LVG in comparison to Varroa-parasitized HVG bees were those genes that may be less suppressed by Varroa in the parasitized resistant genotype than the parasitized susceptible genotype (down-regulated DEGs only observed in the LVG-V to HVG-V comparison; Table S10). They had diverse functions with many annotated as uncharacterized protein genes, indicating that there may be resistance mechanisms not yet discovered. Only extracellular chitin binding and cell wall integrity were identified by GO and KEGG analyses. Varroa punctures the exoskeleton of bees, but insect cuticles have regeneration and repair mechanisms [47]. Chitin binding proteins are involved in cuticle formation [48] and are highly expressed in the integument (single layer of epidermal cells) of honey bees during their development [49]. Therefore, one might expect the up-regulation of such genes to increase damage repair during parasitism. Also, invertebrate chitin-binding proteins can have antimicrobial activity [50] and, thus, might also be expected to be increased with resistance. As those genes were suppressed instead, it is possible that the response of the more resistant bees to Varroa parasitism requires less repair to the cuticle or less antimicrobial activity.
Among the down-regulated DEGs due to the effects of Varroa in comparison to the control bees that were found only with LVG bees, some could be related to the lower impact of Varroa parasitism in the resistant genotype (down-regulated DEGs only observed in the LVG-V to LVG-C comparison; Table S8). GO and KEGG analyses showed that many were related to the metabolism of carbohydrates, proteins, lipids and fatty acids, such as hydrolase, and ATP-dependent and transferase activities. This indicates a suppression of general metabolism by Varroa in the resistant genotype, perhaps as there is a shift to resistance mechanisms. One down-regulated DEG coded for chitooligosaccharidolytic β-N-acetylglucosamindase involved in chitin degradation [51]. While it is important in molting, less activity may allow for greater retention of the cuticle when damaged by Varroa. Down-regulation of a corazonin receptor DEG could result in less detection of the neuropeptide hormone corazonin in the hemolymph, which is involved in insect development [52]. The corazonin receptor is expressed in salivary glands and adipocytes of the liver-like fat body, and reduced expression of its receptor increases fruit fly resistance to starvation, desiccation, and oxidative stress and affects gene expression in the fat body [53]. In honey bees, corazonin gene expression is increased with dietary stress [54]. Considering that Varroa removes nutrients and water from the bee when feeding, lower expression of the corazonin receptor gene in parasitized LVG bees could be an indicator that they are experiencing less nutrient stress than in parasitized HVG bees.
The majority of down-regulated DEGs were related to the effects of Varroa in comparison to controls that were only observed with HVG bees, and many were possibly related to higher stress impact by Varroa in the susceptible genotype (down-regulated DEGs only observed in the HVG-V to HVG-C comparison; Table S7). Many had GO and KEGG classifications related to carbon metabolism, indicating reduced production of various carbohydrates and intermediates in carbon metabolism, as well as amino acids. This may also be linked to down-regulated DEGs for various transporter and membrane proteins. Also, these DEGs included annotations with many regulatory elements, such as the transcription factor dorsal [55], transcription initiation factor IIB that helps to recruit RNA polymerase II into the initiation complex [56], upstream activation factor subunit spp27 [57], and poly(U)-specific endoribonuclease, which is a promoter of translation of mRNA [58]. Down-regulation of such regulatory genes could result in broad scale changes in HVG bees during parasitism, perhaps reflecting a strong suppression of many honey bee cell functions during Varroa parasitism of the susceptible genotype.
Down-regulated DEGs triggered by Varroa in both the LVG and HVG genotypes reflect similar down-regulated functions due to parasitism that were not differentially selected during selection (down-regulated DEGs shared between LVG-V to HVG-V comparison, LVG-V to LVG-C comparison, and HVG-V to HVG-C comparison; Table S9). Among these, there were DEGs for four types of major royal jelly protein. Major royal jelly protein is the main component of brood food for developing larvae [59], and its functions include neural activity related to honey bee development and division of labor [60], as well as affecting antioxidant regulation in the bodies of worker bees [61]. Thus, lower expression of these genes would result in a variety of negative impacts, such as poorly developed larvae, reduced brain function, and higher oxidative stress, which may have occurred both in parasitized LVG and HVG bees. There were also many down-regulated DEGs related to energy, protein metabolism, and nucleobase metabolism, indicating reduced production of key metabolites in parasitized bees of both genotypes.
This study has revealed that changes in gene expression occur differently following parasitism of LVG and HVG bees. Thus, selection has clearly altered the response of the bees. Many of the DEGs identified in this study may contribute directly to resistance to Varroa or indirectly to resistance to the stresses created during parasitism. Other genes may contribute to the susceptibility of bees to the mite. However, this does not establish a functional role for these genes in the response of either LVG or HVG bees to Varroa. Future research can examine the function of these genes in the response of LVG and HVG bees to Varroa, such as by disrupting their function using CRISPR/Cas9 mediated knockout [62].
5. Conclusions
Overall, this study demonstrated that a wide range of changes occurred in gene expression with Varroa parasitism of both resistant and susceptible bees, but based on the number of DEGs, the impacts were less broad in LVG than in HVG bees. Perhaps, one element of Varroa resistance is the ability of LVG bees to better limit the stresses caused by parasitism, viral replication, and immunosuppression. There are many DEGs triggered by Varroa in both genotypes, likely due to stresses caused by the parasite regardless of resistance. However, there were more DEGs up- or down-regulated only with either the LVG or HVG genotype, showing that selection affected bee gene expression. Some of these DEGs, such as those with higher expression in LVG for odorant binding protein, were also observed in the brains of non-parasitized LVG bees undergoing grooming behavior [20], possibly contributing to behavioral resistance to Varroa. Others DEGs like those for glucuronosyltransferase activity and cytochrome P450 that were up-regulated in LVG bees were also reported by Le Conte et al. [18] and may possibly contribute to humoral resistance, detoxification, and stress resistance to Varroa. The transcriptome of LVG bees provides new insights into the diversity of mechanisms by which these bees respond differently than HVG bees to V. destructor. These transcriptomic insights could be used as markers in future selective breeding strategies aimed at enhancing honey bee resistance to Varroa.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pathogens14111077/s1, Figure S1: Volcano plots of DEGs of pairwise comparisons. (a) LVG-C versus HVG-C. (b) LVG-V versus HVG-V. (c) LVG-V versus LVG-C. (d) HVG-V versus HVG-C. The horizontal line indicates the log2FC, and vertical line indicates the -log10 adjusted p-value. The green dots indicate DEGs with log2FC (p > 0.05), and the red dots indicate DEGs with log2FC (p < 0.05); Figure S2: Gene ontology classification and KEGG pathways of up-regulated DEGs unique to the LVG-V versus HVG-V comparison (p < 0.05). The y-axis indicates the GO (biological process, cellular component, and molecular function) and KEGG classification. The x-axis indicates the number of DEGs matching the classification; Table S1: Number of reads, aligned reads to Apis mellifera reference genome (Amel_HAv3.1), and GC content of the reads. Treatments were HVG control bees without Varroa (HVG-C), LVG control bees without Varroa (LVG-C), HVG bees with Varroa (HVG-V), and LVG bees with Varroa (LVG-V). Each treatment had three replicates; Table S2: Pairwise comparisons of DEGs between treatments based on log2 fold-change (p < 0.05); Table S3: Characteristics of up-regulated DEGs unique to HVG-V versus HVG-C comparison (p < 0.05, log2FC); Table S4: Characteristics of up-regulated DEGs unique to the LVG-V versus LVG-C comparison (p < 0.05, log2FC; Table S5: Characteristics of up-regulated DEGs shared between LVG-V versus LVG-C and HVG-V versus HVG-C comparisons (p < 0.05, log2FC); Table S6: Characteristics of up-regulated DEGs unique to the LVG-V versus HVG-V comparison (p < 0.05, log2FC); Table S7: Characteristics of down-regulated DEGs unique from HVG-V versus HVG-C comparison (p < 0.05, log2FC); Table S8: Characteristics of down-regulated unique DEGs from LVG-V versus LVG-C comparison (p < 0.05, log2FC); Table S9; Characteristics of down-regulated DEGs shared between LVG-V versus LVG-C and HVG-V versus HVG-C comparisons (p < 0.05, log2FC); Table S10: Characteristics of down-regulated DEGs unique to the LVG-V versus HVG-V comparison (p < 0.05, log2FC); Table S11: Characteristics of the down-regulated DEGs unique to the LVG-C versus LVG-C comparison (p < 0.05, log2FC); Table S12: Characteristics of down-regulated DEGs shared between LVG-V versus HVG-V, LVG-V versus LVG-C, and HVG-V versus HVG-C comparisons (p < 0.05, log2FC).
Author Contributions
Conceptualization, E.G.-N., P.H.G. and A.D.l.M.; methodology, A.D.l.M., P.H.G. and E.G.-N.; formal analyses, T.P. and A.D.l.M.; resources, E.G.-N.; investigation, A.D.l.M. and P.H.G.; data curation, T.P.; writing, A.D.l.M., E.G.-N., P.H.G. and T.P.; supervision, E.G.-N. and P.H.G.; funding acquisition, E.G.-N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially funded by the Ontario Ministry of Agriculture, Food and Rural Affairs, Grant No. ND2017-3142. It was also partially funded by a Canadian Bee Research Grant from the Canadian Honey Council.
Institutional Review Board Statement
Research with honey bees in Canada does not requires ethics committee approval, because insects are not considered animals for research purposes and therefore do not require approval from ethics committees.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data from this study will be provided by the corresponding author upon reasonable request. The data are not publicly available due to future publications and collaborations related to the breeding lines.
Acknowledgments
We thank the staff and volunteers from the Honey Bee Research Center, University of Guelph, Technology Transfer Program, Ontario Beekeepers’ Association, and Ontario Bee Breeders’ Association.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ATP | Adenosine triphosphate |
| bp | Base pairs |
| CA | California |
| CO2 | Carbon dioxide |
| DEA | Differential expression analysis |
| DEGs | Differentially expressed genes |
| Dim | Dimension |
| DNA | Deoxyribonucleic acid |
| DWV | Deformed wing virus |
| FPKM | Kilobase of exon per million fragments mapped |
| GC | Guanine and cytosine content |
| GO | Gene ontology |
| GTP | Guanosine triphosphate |
| GTPase | Guanosine triphosphatases |
| H2O | Water |
| HVG | Higher rates of Varroa population growth |
| HVG-C | HVG bees without Varroa |
| HVG-V | HVG bees with Varroa |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LVG | Lower rates of Varroa population growth |
| LVG-C | LVG bees without Varroa |
| LVG-V | LVG bees with Varroa |
| Log | Logarithm |
| log2FC | Logarithm 2 fold change |
| logFC | Logarithm fold change |
| μL | Microliter |
| M | Millions |
| MA | Massachusetts |
| Min | Minutes |
| mm | Millimeter |
| NADH | Nicotinamide adenine dinucleotide (NAD) + hydrogen (H) |
| ON | Ontario |
| p | p-value |
| padj | Adjusted p-value |
| PCA | Principal component analysis |
| PRE | Percent relative error |
| QC | Québec |
| RPKM | Kilobase of transcript per million mapped reads |
| RH | Relative humidity |
| RNA | Ribonucleic acid |
| RNA-seq | RNA sequencing |
| rpm | Revolutions per minute |
| s | Seconds |
| srnRNA | Small nuclear RNA |
| USA | United States of America |
| V. destructor | Varroa destructor |
| VSH | Varroa sensitive hygiene |
| ×g | G-force |
| χ2 | Chi-square test |
References
- Morfin, N.; Goodwin, P.H.; Guzman-Novoa, E. Varroa destructor and its impacts on honey bee biology. Front. Bee Sci. 2023, 1, 1272937. [Google Scholar] [CrossRef]
- Brook, G. Varroa treatment-free colony losses in the European honey bee (Apis mellifera): A review of published literature. J. Apic. Res. 2025, 1–10. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef]
- Han, B.; Wu, J.; Wei, Q.; Liu, F.; Cui, L.; Rueppell, O.; Xu, S. Life-history stage determines the diet of ectoparasitic mites on their honey bee hosts. Nat. Commun. 2024, 15, 725. [Google Scholar] [CrossRef]
- Richards, E.H.; Jones, B.; Bowman, A. Salivary secretions from the honeybee mite, Varroa destructor: Effects on insect haemocytes and preliminary biochemical characterization. Parasitology 2024, 138, 602–608. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, 1004230. [Google Scholar] [CrossRef]
- Shen, M.; Yang, X.; Cox-Foster, D.; Cui, L. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 2005, 342, 141–149. [Google Scholar] [CrossRef]
- Kunc, M.; Dobeš, P.; Ward, R.; Lee, S.; Čegan, R.; Dostálková, S.; Holušová, K.; Hurychová, J.; Eliáš, S.; Pinďáková, E.; et al. Omics-based analysis of honey bee (Apis mellifera) response to Varroa sp. Parasitisation and associated factors reveals changes impairing winter bee generation. Insect Biochem. Mol. Biol. 2023, 152, 103877. [Google Scholar] [CrossRef]
- Zanni, V.; Galbraith, D.A.; Annoscia, D.; Grozinger, C.M.; Nazzi, F. Transcriptional signatures of parasitism and markers of colony decline in Varroa-infested honey bees (Apis mellifera). Insect Biochem. Mol. Biol. 2017, 87, 1–13. [Google Scholar] [CrossRef]
- Morfin, N.; Goodwin, P.H.; Hunt, G.J.; Guzman-Novoa, E. Effects of sublethal doses of clothianidin and/or V. destructor on honey bee (Apis mellifera) self-grooming behavior and associated gene expression. Sci. Rep. 2019, 9, 5196. [Google Scholar] [CrossRef]
- Wallner, K. Varroacides and their residues in bee products. Apidologie 1999, 30, 235–248. [Google Scholar] [CrossRef]
- Rinderer, T.E.; Harris, J.W.; Hunt, G.J.; De Guzman, L.I. Breeding for resistance to Varroa destructor in North America. Apidologie 2010, 41, 409–424. [Google Scholar] [CrossRef]
- De La Mora, A.; Goodwin, P.H.; Emsen, B.; Kelly, P.G.; Petukhova, T.; Guzman-Novoa, E. Selection of honey bee (Apis mellifera) genotypes for three generations of low and high population growth of the mite Varroa destructor. Animals 2024, 14, 3537. [Google Scholar] [CrossRef]
- Spivak, M.; Reuter, G.S.; Lee, K.; Ranum, B. Hygienic Stock of Bees is in Good Hands! Am. Bee J. 2009, 149, 965–967. Available online: https://www.researchgate.net/publication/279896898_The_Future_of_the_MN_Hygienic_Stock_of_Bees_is_in_Good_Hands (accessed on 10 October 2025).
- Guzman-Novoa, E.; Correa Benítez, A. Selection of honey bees (Apis mellifera L.) for resistance to the mite Varroa jacobsoni O. Vet. Mex. 1996, 27, 149–158. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19980200441 (accessed on 10 October 2025).
- Boecking, O.; Spivak, M. Behavioral defenses of honey bees against Varroa jacobsoni Oud. Apidologie 1999, 30, 141–158. [Google Scholar] [CrossRef]
- Guzman-Novoa, E.; Emsen, B.; Unger, P.; Espinosa-Montaño, L.G.; Petukhova, T. Genotypic variability and relationships between mite infestation levels, mite damage, grooming intensity, and removal of Varroa destructor mites in selected strains of worker honey bees (Apis mellifera L.). J. Invertebr. Pathol. 2012, 110, 314–320. [Google Scholar] [CrossRef]
- Le Conte, Y.; Alaux, C.; Martin, J.-F.; Harbo, J.R.; Harris, J.W.; Dantec, C.; Séverac, D.; Cros-Arteil, S.; Navajas, M. Social immunity in honeybees (Apis mellifera): Transcriptome analysis of Varroa-hygienic behaviour: Genomics of social immunity. Insect Mol. Biol. 2011, 20, 399–408. [Google Scholar] [CrossRef]
- Hakanoğlu, H. Identification of Potential Transcriptomic Biomarkers for Varroa Resistance in Honey Bees (Apis mellifera anatoliaca). Ph.D. Thesis, Sabancı University, Istanbul, Turkey, 2020. [Google Scholar]
- Morfin, N.; Harpur, B.A.; De La Mora, A.; Guzman-Novoa, E. Breeding honey bees (Apis mellifera L.) for low and high Varroa destructor population growth: Gene expression of bees performing grooming behavior. Front. Insect Sci. 2020, 3, 951447. [Google Scholar] [CrossRef]
- De La Mora, A.; Goodwin, P.H.; Morfin, N.; Petukhova, T.; Guzman-Novoa, E. Diversity of potential resistance mechanisms in honey bees (Apis mellifera) selected for low population growth of the parasitic mite, Varroa destructor. Insects 2025, 16, 385. [Google Scholar] [CrossRef]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D.L.; Locke, B.; Delaplane, K.S.; Wauquiez, Q.; Tannahill, C.; Frey, E.; Ziegelmann, B.; et al. Standard methods for varroa research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Morfin, N. (University of Manitoba, Winnipeg, MB, Canada). Personal communication, 2025.
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Roberts, A.; Pimentel, H.; Trapnell, C.; Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011, 27, 2325–2329. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. (Online Ed.) 2010, 11, R106. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler—A web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T.; De Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Blake, J.A.; Baldarelli, R.; Kadin, J.A.; Richardson, J.E.; Smith, C.L.; Bult, C.J.; Anagnostopoulos, A.V.; Beal, J.S.; Bello, S.M.; Blodgett, O.; et al. Mouse Genome Database (MGD): Knowledgebase for mouse–human comparative biology. Nucleic Acids Res. 2021, 49, D981–D987. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: R-Project, 2019 [Computer Software]. Available online: https://www.R-project.org/ (accessed on 23 March 2024).
- Raser, J.M.; O’Shea, E.K. Noise in gene expression: Origins, consequences, and control. Science 2005, 309, 2010–2013. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc. Natl. Acad. Sci. USA 2011, 108, E67–E76. [Google Scholar] [CrossRef] [PubMed]
- Chakroborty, N.K.; Bienefeld, K.; Menzel, R. Odor learning and odor discrimination of bees selected for enhanced hygienic behavior. Apidologie 2015, 46, 499–514. [Google Scholar] [CrossRef]
- Fiołka, M.J.; Ptaszyńska, A.A.; Czarniawski, W. Antibacterial and antifungal lysozyme-type activity in Cameraria ohridella pupae. J. Invertebr. Pathol. 2005, 90, 1–9. [Google Scholar] [CrossRef]
- Zaobidna, E.A.; Żółtowska, K.; Łopieńska-Biernat, E. Expression and activity of lysozyme in Apis mellifera carnica brood infested with Varroa destructor. J. Apic. Sci. 2017, 61, 253. [Google Scholar] [CrossRef]
- Jinno, A.; Park, P.W. Role of Glycosaminoglycans in infectious disease. Glycosaminoglycans 2015, 1229, 567–585. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; van Wijnen, A.J.; Azizi, P.; Abiri, R.; Ashkani, S.; Taheri, S. Towards understanding pre-mRNA splicing mechanisms and the role of SR proteins. Gene 2016, 587, 107–119. [Google Scholar] [CrossRef]
- Kunat-Budzyńska, M.; Staniszewska, P.; Olszewski, K.; Cytryńska, M.; Strachecka, A. The efficiency of the Krebs cycle and the respiratory chain in physiologically and prematurely aging bees (Apis mellifera). Int. J. Mol. Sci. 2025, 26, 7294. [Google Scholar] [CrossRef]
- Margiotta, A. Role of SNAREs in neurodegenerative diseases. Cells 2021, 10, 991. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. The role of Rho GTPases in disease development. Gene 2002, 286, 155–174. [Google Scholar] [CrossRef]
- Seabra, M.C.; Mules, E.H.; Hume, A.N. Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 2002, 8, 23–30. [Google Scholar] [CrossRef]
- Parle, E.; Dirks, J.-H.; Taylor, D. Damage, repair and regeneration in insect cuticle: The story so far, and possibilities for the future. Arthropod Struct. Dev. 2017, 46, 49–55. [Google Scholar] [CrossRef]
- Willis, J.H. Structural cuticular proteins from arthropods: Annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010, 40, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Micas, A.F.D.; Ferreira, G.A.; Laure, H.J.; Rosa, J.C.; Bitondi, M.M.G. Proteins of the integumentary system of the honeybee, Apis mellifera. Arch. Insect Biochem. Physiol. 2016, 93, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, S.I.; Nagayama, R.; Hirata, M.; Shigenaga, T.; Agarwala, K.L.; Saito, T.; Cho, J.; Nakajima, H.; Takagi, T.; Iwanaga, S. Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J. Biochem. 1996, 120, 1253–1260. [Google Scholar] [CrossRef]
- Zhang, B.; Li, C.; Luan, Y.; Lu, Y.; Hu, H.; Liu, Y.; Lu, K.; Zhang, G.; Dai, F.; Tong, X. The role of chitooligosaccharidolytic β-N-acetylglucosamindase in the molting and wing development of the silkworm Bombyx mori. Int. J. Mol. Sci. 2022, 23, 3850. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, W.J.; Park, S.-J. Analyzing gut microbial community in Varroa destructor-infested western honeybee (Apis mellifera). J. Microbiol. Biotechnol. 2023, 33, 1495–1505. [Google Scholar] [CrossRef]
- Kubrak, O.I.; Lushchak, O.V.; Zandawala, M.; Nässel, D.R. Systemic corazonin signalling modulates stress responses and metabolism in Drosophila. Open Biol. 2016, 6, 160152. [Google Scholar] [CrossRef]
- Obernesser, B.; Snyder, L.; Hoffman, A.; Bennett, M.M.; Corby-Harris, V. Corazonin responds to nutrient stress and increases flight activity in Apis mellifera workers. J. Apic. Res. 2025, 1–8. [Google Scholar] [CrossRef]
- Fernandez-Ayala, D.J.; Chen, S.; Kemppainen, E.; O’Dell, K.M.; Jacobs, H.T. Gene expression in a Drosophila model of mitochondrial disease. PLoS ONE 2010, 5, e8549. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef]
- Ptushkina, M.; Poolman, T.; Iqbal, M.; Ashe, M.; Petersen, J.; Woodburn, J.; Rattray, M.; Whetton, A.; Ray, D. A non-transcriptional role for the glucocorticoid receptor in mediating the cell stress response. Sci. Rep. 2017, 7, 12101–12112. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Fu, C.Y.; Lin, C.Y.; Hu, J.R.; Huang, T.Y.; Lo, K.Y.; Tsai, H.Y.; Sheu, J.C.; Tsai, H.J. Poly (U)-specific endoribonuclease ENDOU promotes translation of human CHOP mRNA by releasing uORF element-mediated inhibition. EMBO J. 2021, 40, e104123. [Google Scholar] [CrossRef] [PubMed]
- Drapeau, M.D.; Albert, S.; Kucharski, R.; Prusko, C.; Maleszka, R. Evolution of the yellow/major royal jelly protein family and the emergence of social behavior in honey bees. Genome Res. 2006, 16, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Feng, M.; Ma, C.; Rueppell, O.; Li, J. Major royal jelly proteins influence the neurobiological regulation of the division of labor among honey bee workers. Int. J. Biol. Macromol. 2023, 225, 848–860. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, B.Y.; Deng, Y.; Park, H.G.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Antioxidant capacity of major royal jelly proteins of honeybee (Apis mellifera) royal jelly. J. Asia-Pac. Entomol. 2020, 23, 445–448. [Google Scholar] [CrossRef]
- Nie, H.Y.; Liang, L.Q.; Li, Q.F.; Li, Z.H.Q.; Zhu, Y.N.; Guo, Y.K.; Zheng, Q.L.; Lin, Y.; Yang, D.L.; Li, Z.G.; et al. CRISPR/Cas9 mediated knockout of Amyellow-y gene results in melanization defect of the cuticle in adult Apis mellifera. J. Insect Physiol. 2021, 132, 104264. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).