The Importance of Networking: Plant Polycomb Repressive Complex 2 and Its Interactors
Abstract
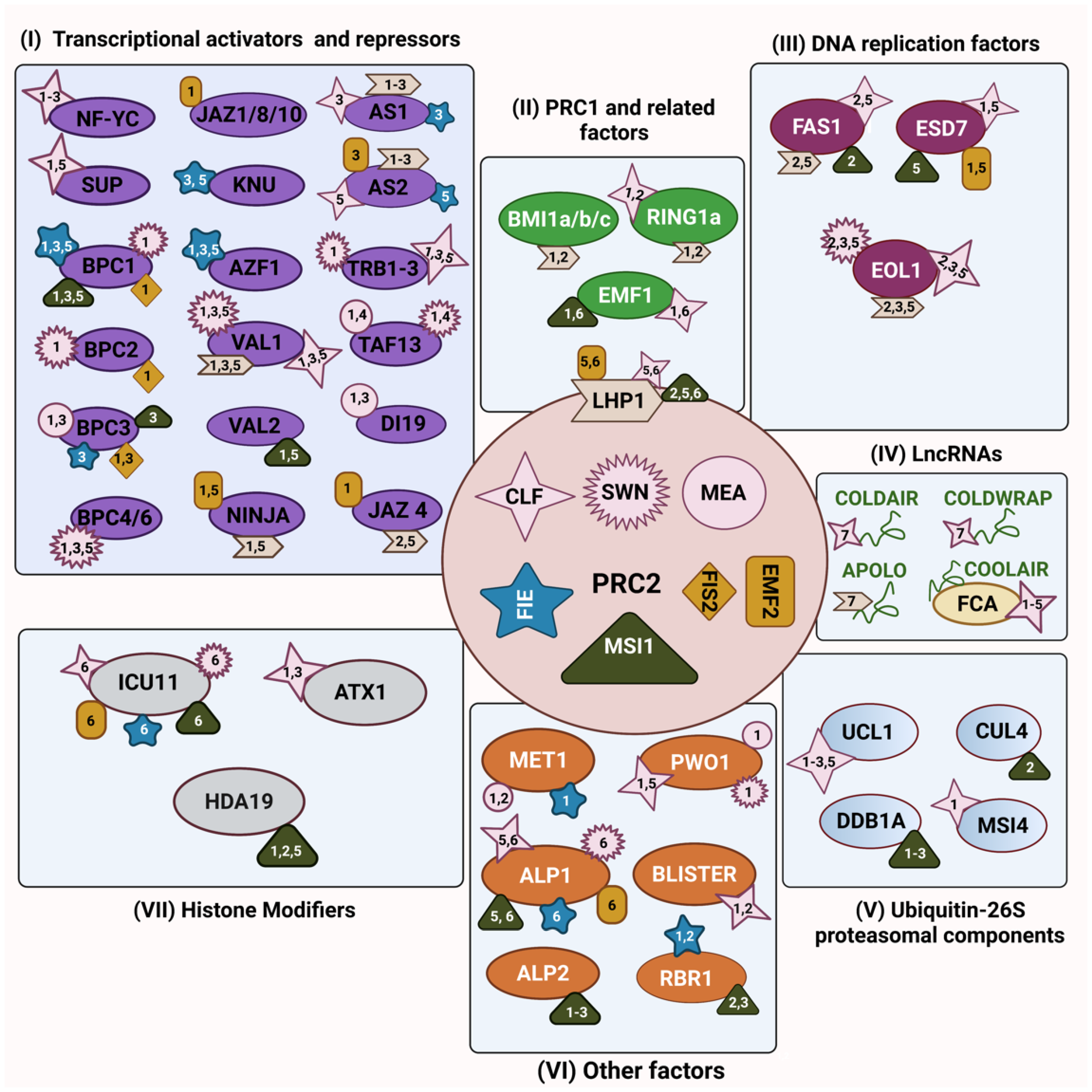
1. Background
2. PRC2’s Interaction with Transcriptional Activators and Repressors
3. Interaction between PRC2 and PRC1 Components
4. PRC2’s Interaction with Ubiquitin-26S Proteasomal Components
5. PRC2’s Interaction with DNA Replication Components
5.1. FASCIATA 1
5.2. ENHANCER OF LHP1 (EOL1)
5.3. DNA Polymerases
6. PRC2’s Interaction with Histone Modifiers
6.1. INCURVATA 11 (ICU11)
6.2. ARABIDOPSIS HOMOLOG OF TRITHORAX 1 (ATX1)
6.3. HISTONE DEACETYLASES (HDAC)
7. Other PRC2 Interactors
7.1. RETINOBLASTOMA RELATED 1 (RBR1)
7.2. DNA METHYLTRANSFERASE 1 (MET1)
7.3. PWWP-DOMAIN INTERACTOR OF POLYCOMBS 1 (PWO1)
7.4. ANTAGONIST OF LIKE HETEROCHROMATIN PROTEIN 1 and 2 (ALP1 and 2)
7.5. BLISTER
8. PRC2’s Interaction with Long Non-Coding RNAs
9. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef]
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb Repressive Complex 2 in Eukaryotes—An Evolutionary Perspective. Epigenomes 2022, 6, 3. [Google Scholar] [CrossRef]
- Baile, F.; Gómez-Zambrano, A.; Calonje, M. Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 2021, 100267. [Google Scholar] [CrossRef] [PubMed]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef]
- Mozgova, I.; Köhler, C.; Hennig, L. Keeping the gate closed: Functions of the polycomb repressive complex PRC2 in development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef]
- Deevy, O.; Bracken, A.P. PRC2 functions in development and congenital disorders. Development 2019, 146, dev181354. [Google Scholar] [CrossRef]
- Deng, W.; Buzas, D.M.; Ying, H.; Robertson, M.; Taylor, J.; Peacock, W.J.; Dennis, E.S.; Helliwell, C. Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genom. 2013, 14, 593. [Google Scholar] [CrossRef]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef]
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic Regulation of H3K27 Trimethylation during Arabidopsis Differentiation. PLoS Genet. 2011, 7, e1002040. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Chen, C.; Thapa, R.K.; Bian, S.; Nguyen, V.; Yu, K.; Yuan, Z.-C.; Liu, J.; Kohalmi, S.E.; Li, C.; et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 2019, 3, e00100. [Google Scholar] [CrossRef] [PubMed]
- Huan, Q.; Mao, Z.; Chong, K.; Zhang, J. Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytol. 2018, 219, 1373–1387. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Poza-Viejo, L.; Martín-Uriz, P.S.; Lara-Astiaso, D.; Wilkinson, M.D.; Crevillén, P. Genome-wide analysis of the H3K27me3 epigenome and transcriptome in Brassica rapa. GigaScience 2019, 8, 1–13. [Google Scholar] [CrossRef]
- He, G.; Zhu, X.; Elling, A.A.; Chen, L.; Wang, X.; Guo, L.; Liang, M.; He, H.; Zhang, H.; Chen, F.; et al. Global Epigenetic and Transcriptional Trends among Two Rice Subspecies and Their Reciprocal Hybrids. Plant Cell 2010, 22, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Makarevitch, I.; Eichten, S.; Briskine, R.; Waters, A.J.; Danilevskaya, O.N.; Meeley, R.B.; Myers, C.L.; Vaughn, M.; Springer, N.M. Genomic Distribution of Maize Facultative Heterochromatin Marked by Trimethylation of H3K27. Plant Cell 2013, 25, 780–793. [Google Scholar] [CrossRef]
- De Lucia, F. Epigenetic Control by Plant Polycomb Proteins: New Perspectives and Emerging Roles in Stress Response; Woodhead Publishing Limited: Sawston, UK, 2013; ISBN 9781907568299. [Google Scholar]
- Shen, Q.; Lin, Y.; Li, Y.; Wang, G. Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 2021, 10, 1165. [Google Scholar] [CrossRef]
- Mozgová, I.; Muñoz-Viana, R.; Hennig, L. PRC2 Represses Hormone-Induced Somatic Embryogenesis in Vegetative Tissue of Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006562. [Google Scholar] [CrossRef]
- Chen, T.; Dent, S.Y.R. Chromatin modifiers: Regulators of cellular differentiation Taiping. Nat Rev Genet. 2014, 15, 93–106. [Google Scholar] [CrossRef]
- Yang, Z.; Qian, S.; Scheid, R.N.; Lu, L.; Chen, X.; Liu, R.; Du, X.; Lv, X.; Boersma, M.D.; Scalf, M.; et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat. Genet. 2018, 50, 1247–1253. [Google Scholar] [CrossRef]
- Li, Z.; Fu, X.; Wang, Y.; Liu, R.; He, Y. Polycomb-mediated gene silencing by the BAH–EMF1 complex in plants. Nat. Genet. 2018, 50, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Nashun, B.; Hill, P.W.S.; Hajkova, P. Reprogramming of cell fate: Epigenetic memory and the erasure of memories past. EMBO J. 2015, 34, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendía, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.; Tomlins, S.; Mehra, R.; Laxman, B.; Cao, X.; Kleer, C.G.; et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar] [CrossRef]
- Casanova, M.; Preissner, T.; Cerase, A.; Poot, R.; Yamada, D.; Li, X.; Appanah, R.; Bezstarosti, K.; Demmers, J.; Koseki, H.; et al. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development 2011, 138, 1471–1482. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Huo, Y.; Yan, Z.; Zhang, B.; Wang, X. Recruitment of Polycomb Repressive Complex 2 is Essential to Suppress the Target Chromatin in Arabidopsis. Crit. Rev. Plant Sci. 2016, 35, 131–145. [Google Scholar] [CrossRef]
- Hepworth, J.; Dean, C. Flowering Locus C’s Lessons: Conserved Chromatin Switches Underpinning Developmental Timing and Adaptation. Plant Physiol. 2015, 168, 1237–1245. [Google Scholar] [CrossRef]
- Xu, S.; Chong, K. Remembering winter through vernalisation. Nat. Plants 2018, 4, 997–1009. [Google Scholar] [CrossRef]
- Sharma, N.; Geuten, K.; Giri, B.S.; Varma, A. The molecular mechanism of vernalization in Arabidopsis and cereals: Role of Flowering Locus C and its homologs. Physiol. Plant. 2020, 170, 373–383. [Google Scholar] [CrossRef]
- Qüesta, J.I.; Song, J.; Geraldo, N.; An, H.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Luo, X.; Li, Z.; Yang, W.; Wang, Y.; Liu, R.; Du, J.; He, W.Y.X.L.Z.L.Y.W.R.L.J.D.Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016, 48, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Bieluszewski, T.; Xiao, J.; Yang, Y.; Wagner, D. PRC2 activity, recruitment, and silencing: A comparative perspective. Trends Plant Sci. 2021, 26, 1186–1198. [Google Scholar] [CrossRef]
- Bauer, M.; Trupke, J.; Ringrose, L. The quest for mammalian Polycomb response elements: Are we there yet? Chromosoma 2015, 125, 471–496. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Krause, K.; Yang, T.; Dongus, J.A.; Zhang, Y.; Turck, F. Telobox motifs recruit CLF/SWN–PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 2018, 50, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.D.; Brown, J.L.; Kassis, J.A. Characterization of the Polycomb Group Response Elements of the Drosophila melanogaster invected Locus. Mol. Cell. Biol. 2010, 30, 820–828. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Kahn, T.G.; Nix, D.A.; Li, X.-Y.; Bourgon, R.; Biggin, M.; Pirrotta, V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006, 38, 700–705. [Google Scholar] [CrossRef]
- Sing, A.; Pannell, D.; Karaiskakis, A.; Sturgeon, K.; Djabali, M.; Ellis, J.; Lipshitz, H.D.; Cordes, S.P. A Vertebrate Polycomb Response Element Governs Segmentation of the Posterior Hindbrain. Cell 2009, 138, 885–897. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833. [Google Scholar] [CrossRef]
- Ringrose, L.; Paro, R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 2007, 134, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct Repression of KNOX Loci by the asymmetric leaves 1 Complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Lodha, M.; Marco, C.F.; Timmermans, M.C. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex 2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef]
- Mu, Y.; Zou, M.; Sun, X.; He, B.; Xu, X.; Liu, Y.; Zhang, L.; Chi, W. Basic Pentacysteine Proteins Repress Abscisic Acid Insensitive4 Expression via Direct Recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis Root Development. Plant Cell Physiol. 2017, 58, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mohamed, D.; Dowhanik, S.; Petrella, R.; Gregis, V.; Li, J.; Wu, L.; Gazzarrini, S. Spatiotemporal Restriction of FUSCA3 Expression by Class I BPCs Promotes Ovule Development and Coordinates Embryo and Endosperm Growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [CrossRef] [PubMed]
- Petrella, R.; Caselli, F.; Roig-Villanova, I.; Vignati, V.; Chiara, M.; Ezquer, I.; Tadini, L.; Kater, M.M.; Gregis, V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020, 102, 582–599. [Google Scholar] [CrossRef]
- Marian, C.O.; Bordoli, S.J.; Goltz, M.; Santarella, R.A.; Jackson, L.P.; Danilevskaya, O.; Beckstette, M.; Meeley, R.; Bass, H.W. The Maize Single myb histone 1 Gene, Smh1, Belongs to a Novel Gene Family and Encodes a Protein That Binds Telomere DNA Repeats in Vitro. Plant Physiol. 2003, 133, 1336–1350. [Google Scholar] [CrossRef]
- Lindner, M.; Simonini, S.; Kooiker, M.; Gagliardini, V.; Somssich, M.; Hohenstatt, M.; Simon, R.; Grossniklaus, U.; Kater, M. TAF13 interacts with PRC2 members and is essential for Arabidopsis seed development. Dev. Biol. 2013, 379, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Xu, Y.; Ng, K.-H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef]
- Sun, B.; Zhou, Y.; Cai, J.; Shang, E.; Yamaguchi, N.; Xiao, J.; Looi, L.-S.; Wee, W.-Y.; Gao, X.; Wagner, D.; et al. Integration of Transcriptional Repression and Polycomb-Mediated Silencing of WUSCHEL in Floral Meristems. Plant Cell 2019, 31, 1488–1505. [Google Scholar] [CrossRef]
- Xu, Y.; Prunet, N.; Gan, E.S.; Wang, Y.; Stewart, D.; Wellmer, F.; Huang, J.; Yamaguchi, N.; Tatsumi, Y.; Kojima, M.; et al. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 2018, 37, e97499. [Google Scholar] [CrossRef] [PubMed]
- Milla, M.A.R.; Townsend, J.; Chang, I.-F.; Cushman, J.C. The Arabidopsis AtDi19 Gene Family Encodes a Novel Type of Cys2/His2 Zinc-finger Protein Implicated in ABA-independent Dehydration, High-salinity Stress and Light Signaling Pathways. Plant Mol. Biol. 2006, 61, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Gupta, P.; Rajabhoj, M.P.; Maruthachalam, R.; Nandi, A.K. The Polycomb-Group Repressor MEDEA Attenuates Pathogen Defense. Plant Physiol. 2018, 177, 1728–1742. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Li, Z.; Luo, X.; Ou, Y.; Jiao, H.; Peng, L.; Fu, X.; Macho, A.P.; Liu, R.; He, Y. JASMONATE-ZIM DOMAIN proteins engage Polycomb chromatin modifiers to modulate Jasmonate signaling in Arabidopsis. Mol. Plant 2021, 14, 732–747. [Google Scholar] [CrossRef]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-Specific Transcription Factor NF-Y Displays Histone-like DNA Binding and H2B-like Ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Hu, Y.; Zhou, L.; Li, Y.; Hou, X. Temporal-Specific Interaction of NF-YC and CURLY LEAF during the Floral Transition Regulates Flowering. Plant Physiol. 2018, 177, 105–114. [Google Scholar] [CrossRef]
- Chen, N.; Veerappan, V.; Abdelmageed, H.; Kang, M.; Allen, R.D. HSI2/VAL1 Silences AGL15 to Regulate the Developmental Transition from Seed Maturation to Vegetative Growth in Arabidopsis. Plant Cell 2018, 30, 600–619. [Google Scholar] [CrossRef]
- Veerappan, V.; Chen, N.; Reichert, A.I.; Allen, R.D. HSI2/VAL1 PHD-like domain promotes H3K27 trimethylation to repress the expression of seed maturation genes and complex transgenes in Arabidopsis seedlings. BMC Plant Biol. 2014, 14, 293. [Google Scholar] [CrossRef]
- Schneider, A.; Aghamirzaie, D.; Elmarakeby, H.; Poudel, A.N.; Koo, A.; Heath, L.S.; Grene, R.; Collakova, E. Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 2016, 85, 305–319. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Morikami, A.; Nakamura, K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2007, 104, 2543–2547. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wang, H.; Abdelmageed, H.; Veerappan, V.; Tadege, M.; Allen, R.D. HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 2020, 227, 840–856. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Song, X.; Zhang, L.; Yu, Y.; Liang, Z.; Lei, Y.; Ruan, J.; Tan, B.; Liu, J.; Li, C. The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 2021, 49, 98–113. [Google Scholar] [CrossRef] [PubMed]
- Chanvivattana, Y.; Bishopp, A.; Schubert, D.; Stock, C.; Moon, Y.-H.; Sung, Z.R.; Goodrich, J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 2004, 131, 5263–5276. [Google Scholar] [CrossRef]
- Erokhin, M.; Georgiev, P.; Chetverina, D. Drosophila DNA-Binding Proteins in Polycomb Repression. Epigenomes 2018, 2, 1. [Google Scholar] [CrossRef]
- Calonje, M. PRC1 Marks the Difference in Plant PcG Repression. Mol. Plant 2014, 7, 459–471. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, L.; Liu, B.-Y.; Tan, C.-F.; Chen, D.-H.; Shen, W.-H.; Ruan, Y. Evolution and conservation of polycomb repressive complex 1 core components and putative associated factors in the green lineage. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Oktaba, K.; Scheuermann, J.C.; Gambetta, M.C.; Ly-Hartig, N.; Müller, J. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 2012, 139, 117–127. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, W.-H. Chromatin modulation and gene regulation in plants: Insight about PRC1 function. Biochem. Soc. Trans. 2018, 46, 957–966. [Google Scholar] [CrossRef]
- Calonje, M.; Sanchez, R.; Chen, L.; Sung, Z.R. EMBRYONIC FLOWER1 Participates in Polycomb Group–Mediated Gene Silencing in Arabidopsis. Plant Cell 2008, 20, 277–291. [Google Scholar] [CrossRef]
- Bratzel, F.; López-Torrejón, G.; Koch, M.; del Pozo, J.C.; Calonje, M. Keeping Cell Identity in Arabidopsis Requires PRC1 RING-Finger Homologs that Catalyze H2A Monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Beh, L.Y.; Colwell, L.J.; Francis, N.J. A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc. Natl. Acad. Sci. USA 2012, 109, E1063–E1071. [Google Scholar] [CrossRef] [PubMed]
- Merini, W.; Romero-Campero, F.J.; Gomez-Zambrano, A.; Zhou, Y.; Turck, F.; Calonje, M. The Arabidopsis Polycomb Repressive Complex 1 (PRC1) Components AtBMI1A, B, and C Impact Gene Networks throughout All Stages of Plant Development. Plant Physiol. 2017, 173, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Shen, W.-H. Polycomb Silencing of KNOX Genes Confines Shoot Stem Cell Niches in Arabidopsis. Curr. Biol. 2008, 18, 1966–1971. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Cheng, J.; Liu, J.; Zhang, L.; He, C.; Shen, W.-H.; Jin, H.; Xu, L.; Zhang, Y. Arabidopsis Flower and Embryo Developmental Genes are Repressed in Seedlings by Different Combinations of Polycomb Group Proteins in Association with Distinct Sets of Cis-regulatory Elements. PLoS Genet. 2016, 12, e1005771. [Google Scholar] [CrossRef]
- Derkacheva, M.; Steinbach, Y.; Wildhaber, T.; Mozgova, I.; Mahrez, W.; Nanni, P.; Bischof, S.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013, 32, 2073–2085. [Google Scholar] [CrossRef]
- Mimida, N.; Kidou, S.-I.; Kotoda, N. Constitutive expression of two apple (Malus × domestica Borkh.) homolog genes of LIKE HETEROCHROMATIN PROTEIN1 affects flowering time and whole-plant growth in transgenic Arabidopsis. Mol. Genet. Genom. 2007, 278, 295–305. [Google Scholar] [CrossRef]
- Zemach, A.; Li, Y.; Ben-Meir, H.; Oliva, M.; Mosquna, A.; Kiss, V.; Avivi, Y.; Ohad, N.; Grafi, G. Different Domains Control the Localization and Mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis Nuclei. Plant Cell 2006, 18, 133–145. [Google Scholar] [CrossRef]
- Exner, V.; Aichinger, E.; Shu, H.; Wildhaber, T.; Alfarano, P.; Caflisch, A.; Gruissem, W.; Köhler, C.; Hennig, L. The Chromodomain of LIKE HETEROCHROMATIN PROTEIN1 Is Essential for H3K27me3 Binding and Function during Arabidopsis Development. PLoS ONE 2009, 4, e5335. [Google Scholar] [CrossRef]
- Wu, Z.; Gao, Y.; Fan, M.; Gao, Y. Gene cloning, expression pattern analysis, and subcellular localization of LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) homologs in chrysanthemum (Chrysanthemum morifolium Ramat.). Cell. Mol. Biol. 2019, 65, 25–31. [Google Scholar] [CrossRef]
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.-L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V. Arabidopsis TFL2/LHP1 Specifically Associates with Genes Marked by Trimethylation of Histone H3 Lysine 27. PLoS Genet. 2007, 3, e86. [Google Scholar] [CrossRef] [PubMed]
- Nakahigashi, K.; Jasencakova, Z.; Schubert, I.; Goto, K. The Arabidopsis HETEROCHROMATIN PROTEIN1 Homolog (TERMINAL FLOWER2) Silences Genes Within the Euchromatic Region but not Genes Positioned in Heterochromatin. Plant Cell Physiol. 2005, 46, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, B.; Liu, J.; Guo, Z.; Liu, Y.; Li, Y.; Shen, W.-H.; Huang, Y.; Huang, H.; Zhang, Y.; et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Veluchamy, A.; Jégu, T.; Ariel, F.; Latrasse, D.; Mariappan, K.G.; Kim, S.-K.; Crespi, M.; Hirt, H.; Bergounioux, C.; Raynaud, C.; et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS ONE 2016, 11, e0158936. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, V.; Libault, M.; Pouteau, S.; Juul, T.; Zhao, G.; Lefebvre, D.; Grandjean, O. Mutations in LIKE HETEROCHROMATIN PROTEIN1 affect flowering time and plant architecture in Arabidopsis. Development 2001, 128, 4847–4858. [Google Scholar] [CrossRef]
- Liang, S.C.; Hartwig, B.; Perera, P.; Mora-Garcia, S.; De Leau, E.; Thornton, H.; De Alves, F.L.; Rapsilber, J.; Yang, S.; James, G.V.; et al. Kicking against the PRCs—A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2. PLoS Genet. 2015, 11, e1005660. [Google Scholar] [CrossRef]
- Sánchez, R.; Kim, M.Y.; Calonje, M.; Moon, Y.-H.; Sung, Z.R. Temporal and Spatial Requirement of EMF1 Activity for Arabidopsis Vegetative and Reproductive Development. Mol. Plant 2009, 2, 643–653. [Google Scholar] [CrossRef]
- Kinoshita, T.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 2001, 98, 14156–14161. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, J.; Eshed-Williams, L.; Zilberman, D.; Sung, Z.R. EMF1 and PRC2 Cooperate to Repress Key Regulators of Arabidopsis Development. PLoS Genet. 2012, 8, e1002512. [Google Scholar] [CrossRef]
- Kim, S.Y.; Zhu, T.; Sung, Z.R. Epigenetic Regulation of Gene Programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol. 2009, 152, 516–528. [Google Scholar] [CrossRef]
- Bloomer, R.H.; Hutchison, C.E.; Bäurle, I.; Walker, J.; Fang, X.; Perera, P.; Velanis, C.N.; Gümüs, S.; Spanos, C.; Rappsilber, J.; et al. The Arabidopsis epigenetic regulator ICU11 as an accessory protein of Polycomb Repressive Complex 2. Proc. Natl. Acad. Sci. USA 2020, 117, 16660–16666. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-Mediated H2Aub Initiate the Switch from Embryonic to Postgerminative Growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Molitor, A.; Shen, W.-H. The Polycomb Complex PRC1: Composition and Function in Plants. J. Genet. Genom. 2013, 40, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Romero-Campero, F.J.; Reyes, P.D.L.; Yan, P.; Yang, J.; Tian, G.; Yang, X.; Mo, X.; Zhao, S.; Calonje, M.; et al. H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Zhou, Y.; Romero-Campero, F.J.; Gómez-Zambrano, Á.; Turck, F.; Calonje, M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Merini, W.; Calonje, M. PRC1 is taking the lead in PcG repression. Plant J. 2015, 83, 110–120. [Google Scholar] [CrossRef]
- Yang, X.; Tong, A.; Yan, B.; Wang, X. Governing the Silencing State of Chromatin: The Roles of Polycomb Repressive Complex 1 in Arabidopsis. Plant Cell Physiol. 2017. [Google Scholar] [CrossRef]
- Callis, J. The Ubiquitination Machinery of the Ubiquitin System. Arab. Book 2014, 12, e0174. [Google Scholar] [CrossRef]
- March, E.; Farrona, S. Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones. Front. Plant Sci. 2018, 8, 2274. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.W.; Roh, H.; Dang, T.V.; Choi, Y.D.; Fischer, R.L.; Lee, J.S. An E3 ligase complex regulates SET-domain polycomb group protein activity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8036–8041. [Google Scholar] [CrossRef] [PubMed]
- Higa, L.A.; Zhang, H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2007, 2, 5. [Google Scholar] [CrossRef]
- Dumbliauskas, E.; Lechner, E.; Jaciubek, M.; Berr, A.; Pazhouhandeh, M.; Alioua, M.; Cognat, V.; Brukhin, V.; Koncz, C.; Grossniklaus, U.; et al. The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 2011, 30, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Pazhouhandeh, M.; Molinier, J.; Berr, A.; Genschik, P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 3430–3435. [Google Scholar] [CrossRef]
- Meinke, D.W. Genome-wide identification of EMBRYO—DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2019, 226, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Chen, N.; Huang, Y.; Huang, S. Phase separation of Arabidopsis emb1579 controls transcription, mRNA splicing, and development. PLoS Biol. 2020, 18, e3000782. [Google Scholar] [CrossRef]
- Seif, E.; Kang, J.J.; Sasseville, C.; Senkovich, O.; Kaltashov, A.; Boulier, E.L.; Kapur, I.; Kim, C.A.; Francis, N.J. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat. Commun. 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Santos, A.P.; Gaudin, V.; Mozgová, I.; Pontvianne, F.; Schubert, D.; Tek, A.L.; Dvořáčková, M.; Liu, C.; Fransz, P.; Rosa, S.; et al. Tidying-up the plant nuclear space: Domains, functions, and dynamics. J. Exp. Bot. 2020, 71, 5160–5178. [Google Scholar] [CrossRef]
- Gaillard, P.-H.; Martini, E.M.-D.; Kaufman, P.; Stillman, B.; Moustacchi, E.; Almouzni, G. Chromatin Assembly Coupled to DNA Repair: A New Role for Chromatin Assembly Factor I. Cell 1996, 86, 887–896. [Google Scholar] [CrossRef]
- Hammond, C.; Strømme, C.B.; Huang, H.; Patel, H.H.D.J.; Groth, C.M.H.C.B.S.A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.; Shibahara, K.-I.; Taoka, K.-I.; Iwabuchi, M.; Stillman, B.; Araki, T. FASCIATA Genes for Chromatin Assembly Factor-1 in Arabidopsis Maintain the Cellular Organization of Apical Meristems. Cell 2001, 104, 131–142. [Google Scholar] [CrossRef]
- Exner, V.; Taranto, P.; Schönrock, N.; Gruissem, W.; Hennig, L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 2006, 133, 4163–4172. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Berger, F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 2017, 357, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Simon, A.C.; Bazan, M.A.O.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef]
- Zhou, Y.; Tergemina, E.; Cui, H.; Förderer, A.; Hartwig, B.; James, G.V.; Schneeberger, K.; Turck, F. Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 4833–4838. [Google Scholar] [CrossRef]
- Del Olmo, I.; López-González, L.; Martín-Trillo, M.M.; Martínez-Zapater, J.M.; Piñeiro, M.; Jarillo, J.A. Early in short days 7(ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010, 61, 623–636. [Google Scholar] [CrossRef]
- Jenik, P.D.; Jurkuta, R.E.; Barton, M.K. Interactions between the Cell Cycle and Embryonic Patterning in Arabidopsis Uncovered by a Mutation in DNA Polymerase ε. Plant Cell 2005, 17, 3362–3377. [Google Scholar] [CrossRef]
- Del Olmo, I.; López, J.A.; Vázquez, J.; Raynaud, C.; Piñeiro, M.; Jarillo, J.A. Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 2016, 44, 5597–5614. [Google Scholar] [CrossRef]
- Pedroza-Garcia, J.-A.; De Veylder, L.; Raynaud, C. Plant DNA Polymerases. Int. J. Mol. Sci. 2019, 20, 4814. [Google Scholar] [CrossRef]
- Dodd, I.; Micheelsen, M.A.; Sneppen, K.; Thon, G. Theoretical Analysis of Epigenetic Cell Memory by Nucleosome Modification. Cell 2007, 129, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Sneppen, K.; Ringrose, L. Theoretical analysis of Polycomb-Trithorax systems predicts that poised chromatin is bistable and not bivalent. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- Yang, H.; Howard, M.; Dean, C. Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. USA 2016, 113, 9369–9374. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis Homolog of Trithorax, Activates Flower Homeotic Genes. Curr. Biol. 2003, 13, 627–637. [Google Scholar] [CrossRef]
- Saleh, A.; Al-Abdallat, A.; Ndamukong, I.; Alvarez-Venegas, R.; Avramova, Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007, 35, 6290–6296. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef]
- Jang, I.-C.; Chung, P.J.; Hemmes, H.; Jung, C.; Chua, N.-H. Rapid and Reversible Light-Mediated Chromatin Modifications of Arabidopsis Phytochrome A Locus. Plant Cell 2011, 23, 459–470. [Google Scholar] [CrossRef]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef]
- Mehdi, S.; Derkacheva, M.; Ramström, M.; Kralemann, L.; Bergquist, J.; Hennig, L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell 2015, 28, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Baile, F.; Merini, W.; Hidalgo, I.; Calonje, M. EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis. Plant Cell 2021, 33, 2701–2715. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Gao, Z.; Jiang, C.; Yang, Y.; Liu, R.; He, Y. HISTONE DEACETYLASE 9 Functions with Polycomb Silencing to Repress FLOWERING LOCUS C Expression. Plant Physiol. 2019, 182, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Chhun, T.; Chong, S.Y.; Park, B.S.; Wong, E.C.C.; Yin, J.-L.; Kim, M.; Chua, N.-H. HSI2 Repressor Recruits MED13 and HDA6 to Down-Regulate Seed Maturation Gene Expression Directly During Arabidopsis Early Seedling Growth. Plant Cell Physiol. 2016, 57, 1689–1706. [Google Scholar] [CrossRef]
- Nevins, J.R. E2F: A Link Between the Rb Tumor Suppressor Protein and Viral Oncoproteins. Science 1992, 258, 424–429. [Google Scholar] [CrossRef]
- Gruissem, W. Function of the Retinoblastoma-related Protein in Plants. Annu. Plant Rev. Cell Cycle Control Plant Dev. 2007, 32, 164–186. [Google Scholar]
- Gutzat, R.; Borghi, L.; Fütterer, J.; Bischof, S.; Laizet, Y.; Hennig, L.; Feil, R.; Lunn, J.; Gruissem, W. Retinoblastoma-Related protein controls the transition to autotrophic plant development. Development 2011, 138, 2977–2986. [Google Scholar] [CrossRef]
- Mosquna, A.; Katz, A.; Shochat, S.; Grafi, G.; Ohad, N. Interaction of FIE, a Polycomb protein, with pRb: A possible mechanism regulating endosperm development. Mol. Genet. Genom. 2004, 271, 651–657. [Google Scholar] [CrossRef]
- Johnston, A.J.; Matveeva, E.; Kirioukhova, O.; Grossniklaus, U.; Gruissem, W. A Dynamic Reciprocal RBR-PRC2 Regulatory Circuit Controls Arabidopsis Gametophyte Development. Curr. Biol. 2008, 18, 1680–1686. [Google Scholar] [CrossRef]
- Jullien, P.E.; Mosquna, A.; Ingouff, M.; Sakata, T.; Ohad, N.; Berger, F. Retinoblastoma and Its Binding Partner MSI1 Control Imprinting in Arabidopsis. PLOS Biol. 2008, 6, e194. [Google Scholar] [CrossRef]
- Jackson, J.P.; Lindroth, A.; Cao, X.; Jacobsen, S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002, 416, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Wöhrmann, H.J.P.; Raissig, M.T.; Arand, J.; Gheyselinck, J.; Gagliardini, V.; Heichinger, C.; Walter, J.; Grossniklaus, U. The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J. 2012, 73, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.M.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.-M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2005, 439, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Fitz Gerald, J.; Luo, M.; Chaudhury, A.; Berger, F. DNA Methylation Causes Predominant Maternal Controls of Plant Embryo Growth. PLoS ONE 2008, 3, e2298. [Google Scholar] [CrossRef]
- Hohenstatt, M.L.; Mikulski, P.; Komarynets, O.; Klose, C.; Kycia, I.; Jeltsch, A.; Farrona, S.; Schubert, D. Pwwp-domain interactor of polycombs1 Interacts with Polycomb-Group Proteins and Histones and Regulates Arabidopsis Flowering and Development. Plant Cell 2018, 30, 117–133. [Google Scholar] [CrossRef]
- Mikulski, P.; Hohenstatt, M.L.; Farrona, S.; Smaczniak, C.; Stahl, Y.; Kalyanikrishna; Kaufmann, K.; Angenent, G.; Schubert, D. The Chromatin-Associated Protein PWO1 Interacts with Plant Nuclear Lamin-like Components to Regulate Nuclear Size. Plant Cell 2019, 31, 1141–1154. [Google Scholar] [CrossRef]
- Tan, L.; Zhang, C.; Hou, X.; Shao, C.; Lu, Y.; Zhou, J.; Li, Y.; Li, L.; Chen, S.; He, X. The PEAT protein complexes are required for histone deacetylation and heterochromatin silencing. EMBO J. 2018, 37, e98770. [Google Scholar] [CrossRef]
- Sinzelle, L.; Izsvak, Z.; Ivics, Z. Molecular domestication of transposable elements: From detrimental parasites to useful host genes. Cell. Mol. Life Sci. 2009, 66, 1073–1093. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, N.; Feschotte, C.; Wessler, S.R. PIF- and Pong-Like Transposable Elements: Distribution, Evolution and Relationship with Tourist-Like Miniature Inverted-Repeat Transposable Elements. Genetics 2004, 166, 971–986. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Harbinger Transposons and an Ancient HARBI1 Gene Derived from a Transposase. DNA Cell Biol. 2004, 23, 311–324. [Google Scholar] [CrossRef]
- Velanis, C.N.; Perera, P.; Thomson, B.; De Leau, E.; Liang, S.C.; Hartwig, B.; Förderer, A.; Thornton, H.; Arede, P.; Chen, J.; et al. The domesticated transposase ALP2 mediates formation of a novel Polycomb protein complex by direct interaction with MSI1, a core subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 2020, 16, e1008681. [Google Scholar] [CrossRef] [PubMed]
- Schatlowski, N.; Stahl, Y.; Hohenstatt, M.L.; Goodrich, J.; Schubert, D. The CURLY LEAF Interacting Protein BLISTER Controls Expression of Polycomb-Group Target Genes and Cellular Differentiation of Arabidopsis thaliana. Plant Cell 2010, 22, 2291–2305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hong, Z.-H.; Qing, T.; Schubert, D.; Kleinmanns, J.A.; Liu, J.-X. BLISTER-regulated vegetative growth is dependent on the protein kinase domain of ER stress modulator IRE1A in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1008563. [Google Scholar] [CrossRef] [PubMed]
- Kleinmanns, J.A.; Schatlowski, N.; Heckmann, D.; Schubert, D. BLISTER Regulates Polycomb-Target Genes, Represses Stress-Regulated Genes and Promotes Stress Responses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Purdy, S.J.; Bussell, J.D.; Nelson, D.C.; Villadsen, D.; Smith, S.M. A nuclear-localized protein, KOLD SENSITIV-1, affects the expression of cold-responsive genes during prolonged chilling in Arabidopsis. J. Plant Physiol. 2011, 168, 263–269. [Google Scholar] [CrossRef]
- Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 2013, 19, 429–442. [Google Scholar] [CrossRef]
- Trotman, J.B.; Braceros, K.C.A.; Cherney, R.E.; Murvin, M.M.; Calabrese, J.M. The control of polycomb repressive complexes by long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2021, 12, e1657. [Google Scholar] [CrossRef]
- Negishi, M.; Wongpalee, S.; Sarkar, S.; Park, J.; Lee, K.Y.; Shibata, Y.; Reon, B.J.; Abounader, R.; Suzuki, Y.; Sugano, S.; et al. A New lncRNA, APTR, Associates with and Represses the CDKN1A/p21 Promoter by Recruiting Polycomb Proteins. PLoS ONE 2014, 9, e95216. [Google Scholar] [CrossRef]
- Kanhere, A.; Viiri, K.; Araújo, C.C.; Rasaiyaah, J.; Bouwman, R.D.; Whyte, W.A.; Pereira, C.F.; Brookes, E.; Walker, K.; Bell, G.W.; et al. Short RNAs Are Transcribed from Repressed Polycomb Target Genes and Interact with Polycomb Repressive Complex-2. Mol. Cell 2010, 38, 675–688. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef]
- Macknight, R.; Bancroft, I.; Page, T.; Lister, C.; Schmidt, R.; Love, K.; Westphal, L.; Murphy, G.; Sherson, S.; Cobbett, C.; et al. FCA, a Gene Controlling Flowering Time in Arabidopsis, Encodes a Protein Containing RNA-Binding Domains. Cell 1997, 89, 737–745. [Google Scholar] [CrossRef]
- Liu, F.; Quesada, V.; Crevillén, P.; Bäurle, I.; Swiezewski, S.; Dean, C. The Arabidopsis RNA-Binding Protein FCA Requires a Lysine-Specific Demethylase 1 Homolog to Downregulate FLC. Mol. Cell 2007, 28, 398–407. [Google Scholar] [CrossRef]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, eaau7246. [Google Scholar] [CrossRef]
- Wei, C.; Xiao, R.; Chen, L.; Cui, H.; Zhou, Y.; Xue, Y.; Hu, J.; Zhou, B.; Tsutsui, T.; Qiu, J.; et al. RBFox2 Binds Nascent RNA to Globally Regulate Polycomb Complex 2 Targeting in Mammalian Genomes. Mol. Cell 2016, 62, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Liebeskind, B.J.; Aldrich, R.W.; Marcotte, E.M. Ancestral reconstruction of protein interaction networks. PLoS Comput. Biol. 2019, 15, e1007396. [Google Scholar] [CrossRef]
- Bontinck, M.; Van Leene, J.; Gadeyne, A.; De Rybel, B.; Eeckhout, D.; Nelissen, H.; De Jaeger, G. Recent Trends in Plant Protein Complex Analysis in a Developmental Context. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Alfaro, J.A.; Bohländer, P.; Dai, M.; Filius, M.; Howard, C.J.; van Kooten, X.F.; Ohayon, S.; Pomorski, A.; Schmid, S.; Aksimentiev, A.; et al. The emerging landscape of single-molecule protein sequencing technologies. Nat. Methods 2021, 18, 604–617. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, X.; Zhou, C.; Zhou, Q.; Zhao, Y.; Li, G.; Zhou, D.-X. Cooperation between the H3K27me3 chromatin marker and non-CG methylation in epigenetic regulation. Plant Physiol. 2016, 172, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, D.-H.; Liu, B.-Y.; Shen, W.-H.; Ruan, Y. Conservation and diversification of polycomb repressive complex 2 (PRC2) proteins in the green lineage. Brief. Funct. Genom. 2016, 16, 106–119. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godwin, J.; Farrona, S. The Importance of Networking: Plant Polycomb Repressive Complex 2 and Its Interactors. Epigenomes 2022, 6, 8. https://doi.org/10.3390/epigenomes6010008
Godwin J, Farrona S. The Importance of Networking: Plant Polycomb Repressive Complex 2 and Its Interactors. Epigenomes. 2022; 6(1):8. https://doi.org/10.3390/epigenomes6010008
Chicago/Turabian StyleGodwin, James, and Sara Farrona. 2022. "The Importance of Networking: Plant Polycomb Repressive Complex 2 and Its Interactors" Epigenomes 6, no. 1: 8. https://doi.org/10.3390/epigenomes6010008
APA StyleGodwin, J., & Farrona, S. (2022). The Importance of Networking: Plant Polycomb Repressive Complex 2 and Its Interactors. Epigenomes, 6(1), 8. https://doi.org/10.3390/epigenomes6010008





