Abstract
BDNF signalling in hypothalamic neuronal circuits is thought to regulate mammalian food intake. In light of this, we investigated how a lifestyle intervention influenced serum levels and DNA methylation of BDNF gene in fat tissue and buffy coat of NDH individuals. In total, 20 participants underwent anthropometric measurements/fasting blood tests and adipose tissue biopsy pre-/post-lifestyle (6 months) intervention. DNA was extracted from adipose tissue and buffy coat, bisulphite converted, and pyrosequencing was used to determine methylation levels in exon IV of the BDNF gene. RNA was extracted from buffy coat for gene expression analysis and serum BDNF levels were measured by ELISA. No differences were found in BDNF serum levels, but buffy coat mean BDNF gene methylation decreased post-intervention. There were correlations between BDNF serum levels and/or methylation and cardiometabolic markers. (i) Pre-intervention: for BDNF methylation, we found positive correlations between mean methylation in fat tissue and waist-hip ratio, and negative correlations between mean methylation in buffy coat and weight. (ii) Post-intervention: we found correlations between BDNF mean methylation in buffy coat and HbA1c, BDNF methylation in buffy coat and circulating IGFBP-2, and BDNF serum and insulin. Higher BDNF % methylation levels are known to reduce BNDF expression. The fall in buffy coat mean BDNF methylation plus the association between lower BDNF methylation (so potentially higher BDNF) and higher HbA1c and serum IGFBP-2 (as a marker of insulin sensitivity) and between lower serum BDNF and higher circulating insulin are evidence for the degree of BDNF gene methylation being implicated in insulinisation and glucose homeostasis, particularly after lifestyle change in NDH individuals.
1. Introduction
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family of growth factors that promote neuronal differentiation, adult neurogenesis, learning, memory [1,2] and neural plasticity [3,4]; it is produced and released by the brain into circulation. In addition to these roles, BDNF has also been identified as a key component of the hypothalamic pathway that controls body weight, food intake, and energy homeostasis in mammals [5].
There is substantial evidence linking BDNF to the progression of type 2 diabetes mellitus (T2DM). Plasma BDNF levels are decreased in T2DM compared to healthy controls and is thought to play a role in systemic inflammation and to interact with several inflammatory cytokines [4]. BDNF is widely expressed throughout the hippocampus and other brain areas as well as peripheral tissues important in the regulation of energy homeostasis, such as adipose tissue, skeletal and smooth muscle, and liver [6,7]. Furthermore, BDNF treatment in obese and diabetic rodents significantly reduces blood glucose level, attenuates body weight gain and food intake, and enhances energy and glucose metabolism [8,9].
Humans with mutations in the BDNF gene [10] or in the trkB receptor signal transduction pathway [11] exhibit severe obesity. Additionally, BDNF gene variants are associated with higher risk for increased body weight in both children and adults [12,13]. Similar findings have been shown in mice where reduced BDNF expression, either due to a brain specific conditional knockout or due to heterozygous gene expression, leads to hyperphagia, obesity, and insulin and leptin resistance [14,15].
Non-diabetic hyperglycaemia (NDH) refers to a metabolic state between normal glucose homeostasis and diabetes [16]. Additionally, referred to as pre-diabetes or impaired glucose regulation (IGR), NDH represents a high-risk state for the future development of T2DM. T2DM is thought to arise from a summation of genetic and environmental/epigenetic factors, which result in a decline in insulin activity, often on the background of longstanding elevation of body mass index (BMI) with attendant insulin resistance.
Biomarkers are necessary to better assess the trajectory towards T2DM, to monitor responses to treatment, and to personalise therapy, with the aim being to develop new strategies to prevent the progress from NDH to T2DM. BDNF is a potential biomarker. However, studies regarding DNA methylation alteration in key CpGs of the regulatory BDNF region in T2DM and obesity are still scarce and warrant further investigation.
In this study, we sought to determine whether a lifestyle intervention could influence DNA methylation of BDNF and circulating levels of BDNF in individuals with NDH. Additionally, we tested for possible correlations between BDNF methylation and/or serum levels and clinical variables, as BDNF is thought to play an important role in glucose metabolism.
2. Results
The clinical and demographic variables for all the participants were described previously [17,18]. In summary, the Care Call intervention resulted in weight reduction and BMI change (p < 0.05) but no other clinically significant alterations (i.e., insulin levels, total cholesterol, HbA1C, HOMA-S, HOMA-B, triglycerides).
BDNF serum levels or BDNF gene expression did not change pre- and post-Care Call intervention (t = −0.177, p = 0.862, for serum; t = 1.500, p = 0.154 for buffy coat gene expression) (Figure 1a). Regarding the DNA methylation data, we did find a reduction in mean BDNF methylation in buffy coat, (buffy coat: t = 2.306, p = 0.038) (Figure 1b). There was no significant alteration in methylation at any of the CpG sites or in mean methylation in adipose tissue between baseline and 6-month follow-up after the Care Call intervention (adipose tissue: t = −0.792, p = 0.438).
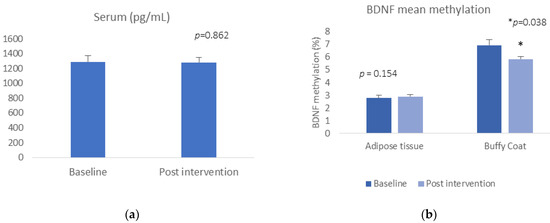
Figure 1.
(a) BDNF concentration pre- and post-intervention; (b) mean BDNF DNA methylation pre and post intervention.
We then tested for Pearson’s correlation the BDNF methylation data, buffy coat gene expression, BDNF serum levels and clinical variables at baseline (pre-intervention) and post-intervention (see below).
Pre-intervention results: We identified a positive correlation between BDNF mean methylation in adipose tissue and waist/hip ratio (r = 0.528, p = 0.017) (Figure 2a) and negative correlations between adipose BDNF mean methylation and total cholesterol (r = −0.552, p = 0.012) (Figure 2b).
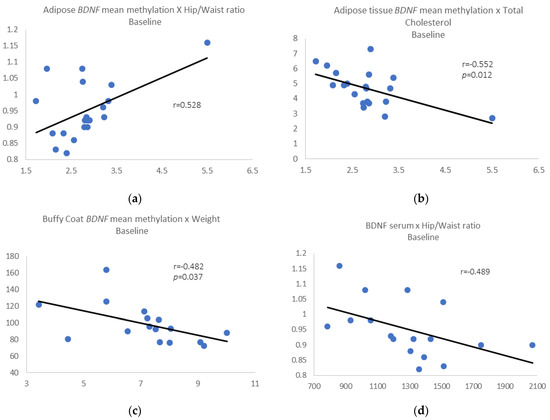
Figure 2.
(a) Adipose tissue BDNF mean methylation vs. waist/hip ratio; (b) adipose tissue BDNF mean methylation vs. total cholesterol; (c) buffy coat BDNF mean methylation vs. weight; (d) removing those individuals who gain 3% or more in weight, BDNF serum levels vs. waist/hip ratio.
We also found a negative correlation between BDNF mean methylation in buffy coat and weight (r = −0.482, p = 0.037) (Figure 2c). When we removed those individuals who gain 3% or more in weight, all the correlations described above were maintained, and we also found a negative correlation between BDNF serum levels and waist/hip ratio (r = −0.489, p = 0.046) (Figure 2d).
Post-intervention results: We found a negative correlation between buffy coat BDNF mean methylation and HbA1c levels (r = −0.510, p = 0.036; Figure 3a) and between serum BDNF levels and insulin (r = −0.629, p = 0.003; Figure 3b) for the whole group. Removing those individuals who gain 3% of weight or more, the above correlations were still significant, and we found a negative correlation between buffy coat BDNF mean methylation and serum insulin-like growth factor binding protein-2 gene (IGFBP-2) levels (r = −0.661, p = 0.014; Figure 3c). Additionally, we found a negative correlation between BDNF serum delta pre- and post-intervention and HOMA-S delta (r = −0.552, p = 0.027; Figure 3d), pre- and post-intervention.
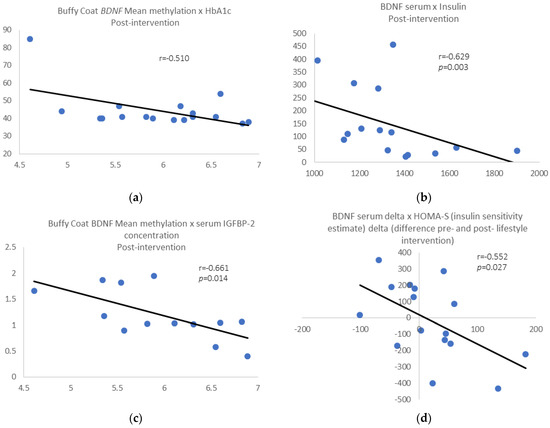
Figure 3.
(a) Buffy wCoat BDNF mean methylation vs. HbA1c; (b) serum BDNF levels vs. plasma insulin; (c) BDNF mean methylation vs. serum insulin-like growth factor binding protein-2 (IGFBP-2) levels. (d) BDNF serum delta pre- and post-intervention vs. HOMA-S delta pre- and post-intervention.
3. Discussion
In the present study, we analysed BDNF serum levels and BDNF DNA methylation (adipose tissue and buffy coat) in NDH individuals and the effects in these variables before and after a lifestyle intervention. We also tested these variables for correlations with clinical cardiometabolic data. We saw differences in buffy coat mean BDNF methylation but not BDNF serum levels after the intervention. We also potentially found clinically relevant correlations between BDNF DNA methylation and weight, total cholesterol, HbA1C levels, and waist/hip ratio, as well as between BDNF serum levels and insulin. The BDNF serum delta was negatively correlated with HOMA-S delta in relation to changes with the lifestyle intervention, revealing an important association between BDNF and insulin sensitivity in our sample.
These observed associations between BDNF DNA methylation patterns suggest that BDNF may be involved in the pathophysiological process of insulin resistance and T2DM. This is also supported by the link here between BDNF expression and circulating IGFBP-2, an important marker of insulin sensitivity [19].
It was demonstrated previously that BDNF affects glucose metabolism not only in central metabolic pathways but also in the peripheral glucagon secretion pathway [9]. Additionally, 3 weeks of BDNF administration significantly reduced blood glucose concentrations and glycated haemoglobin (HbA1c) in a db/db mice model [8]. BDNF not only temporarily reduced blood glucose concentrations but also ameliorated systemic glucose balance in this obese diabetic mouse model during the experimental period [8]. Given the positive association between higher circulating IGFBP-2 and greater insulin sensitivity in humans [19,20,21], the association between lower BDNF methylation (so potentially higher circulating BDNF) and higher circulating levels of IGFBP-2 is an important and relevant finding.
BDNF also plays an important role in fat accumulation in an animal model, since young rats under a hypercaloric diet showed an increase in adipose tissue concentration, and this was associated with reduced hippocampal BDNF expression [22]. Our results at baseline are in accordance with these findings, shown by the positive correlation we found between mean methylation in adipose tissue and waist/hip ratio, and the negative correlation between serum levels and waist/hip ratio, since if the methylation levels are high, we would expect lower BDNF expression accompanied by higher waist/hip ratio.
After the intervention, there was a reduction in BDNF mean methylation in buffy coat. Although we only observed a small number of individuals here, this finding suggests the idea that BDNF methylation may be influenced by dietary changes with consequent modulation of BDNF gene expression. We also found a correlation between the BDNF mean methylation in buffy coat and HbA1C levels. BDNF methylation is very poorly explored in the context of diabetes. So far, the DNA methylation of exon IV, a regulatory region of BDNF gene that contains a site for the transcription factor cAMP-response element binding protein (CREB), has been analysed by only one study [23]. CREB family transcription factors are required for the early induction of all the major BDNF transcripts [24]. The major findings were correlations with insulin levels and hip/waist ratio in specifics CpGs in newly diagnosed diabetes and pre-diabetes. Additionally, higher BDNF methylation was associated with high fasting insulin levels [23], supporting the hypothesis that BDNF methylation plays important role in diabetes progression.
In this study, we found that BDNF serum levels correlated with insulin post-intervention, and the delta values of BDNF serum and HOMA-S, an indicator for insulin sensitivity, were also inversely correlated. These findings reinforce the role of BDNF in glucose regulation [25,26] and the evidence showing negative interaction between impaired glucose tolerance and circulating BDNF levels [27], which could contribute to our understanding of the frequently seen association between diabetes and cognitive impairments [1,28].
Considering that epigenetic markers are modified by external factors, such as age, physical activity, in utero environment, and availability of nutrients, the transient and reversible nature of epigenetic modifications provides a vast field for discovery of targets for future prediction and therapeutic concepts in NDH and T2DM.
Our findings in this paper contribute to the knowledge about the epigenetic influence on the trajectory over time in relation to T2DM and add important evidence related to BDNF methylation supporting its role in glucose metabolism and in modification of eating behaviour. Future studies involving BDNF methylation, NDH, and T2DM patients are necessary to the development of new therapeutic targets and for future preventive, earlier interventions, such as more specific diets and tailored exercise regimes.
We accept that these findings are only based on a small number of people, and that there was no change in serum BDNF concentration with the lifestyle intervention. However, the group that we studied was fully characterised metabolically, and we were able to analyse BDNF methylation in both buffy coat samples and adipose tissue.
In conclusion, higher BDNF % methylation levels are known to reduce BNDF expression. The association between lower BDNF methylation (so potentially higher BDNF) and higher HbA1c/circulating IGFBP-2 (as a marker of insulin sensitivity) and between lower serum BDNF and higher circulating insulin suggests that the degree of BDNF methylation may be indirectly implicated in insulinisation and glucose homeostasis, particularly after lifestyle change in NDH individuals.
4. Materials and Methods
4.1. Samples
Serum, fat tissue, and whole blood were collected from 20 NDH individuals (10 male, 10 female) at baseline and following a 6-month telephone-led structured lifestyle intervention (Care Call programme) where they received exercise and nutritional advice with the aim of improving glycaemia and reducing weight. All 20 individuals participated in the Care Call programme. The Care Call programme is a modular telephone-based intervention programme utilising motivational support techniques, lifestyle education, and one-to-one and peer discussion and encouragement of progress with goals and signposting/referral to relevant services, with tailoring of content to individual needs (Supplementary File S1). The complete description of sample collecting and Care Call programme details were described previously by our group [17,18].
4.2. DNA Methylation
Genomic DNA was extracted from adipose tissue and buffy coat from all 20 individuals before and 6 months after the intervention using DNA Qiamp mini kit (Qiagen, Manchester, UK). Afterwards, the DNA was bisulfite converted using EpiTec Fast DNA Bisulphite Kit (Qiagen) to convert unmethylated cytosine residues to uracil with a calculated mean conversion of 99%. A pyrosequencing method was used for the determination of methylation at four CpG sites within the BDNF IV exon, as previously described by our group [29]. The sequence was amplified by PCR, using the primers described on Table 1. Pyrosequence setup and data reading were conducted by PyroMark Q48 using the version 2.4.2 software (Qiagen). The samples underwent PCR and pyrosequencing in duplicate; any inconsistencies between samples were resolved following further repetition.

Table 1.
List of Forward (F) and biotinylated Reverse (R) primers used in PCR reactions, and Sequencing (Seq) primer for pyrosequencing.
4.3. Gene Expression
RNA was extracted from buffy coat using the RNeasy mini kit (Qiagen) and reverse-transcribed using the QuantiTect reverse transcription kit (Qiagen). The cDNA concentration and purity were determined using a NanoDrop Lite spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). Relative gene expression was determined using a QuantiStudio5 (Applied Biosystems, Waltham, MA, USA) and TaqMan® reagents, with β-actin (ACTB) as housekeeping gene. The experiments were assayed in triplicate, and 50 ng of total cDNA was used per reaction for BDNF and ACTB. The PCR protocol was as follows: pre-incubation—1 cycle at 95 °C for 10 min, amplification—50 cycles at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 1 s, and cooling—1 cycle, 40 °C for 30 s.
4.4. Serum Protein and Other Assays
Serum quantitative determination of BDNF was performed by ELISA using the Human Free BDNF Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. The detection range was 62.5–4000 pg/mL, and the sensitivity was 20 pg/mL. Serum insulin-like growth factor binding protein-2 (IGFBP-2) concentration was determined by ELISA. The measurement of all other analytes reported here was described in previous papers [17,18].
4.5. Statistical Analysis
All analyses were carried out using the Statistical Package for Social Sciences (SPSS version 20.0, Armonk, NY, USA). Descriptive analyses were performed to evaluate socio-demographic and clinical characteristics and have been published elsewhere [17,18].
Gene expression was quantified using the comparative threshold (Ct) method (ΔΔCt method), and the amount of BDNF (target gene) was normalised to the housekeeping gene ACTB and determined by 2−ΔΔCt, with relative expression levels reported as fold change. Changes in BDNF gene expression, serum levels, and BDNF DNA methylation for each CpG or mean methylation before and after intervention were compared by paired t-test followed by Bonferroni correction for multiple comparisons as a post hoc test and were considered significant when p ≤ 0.025. Normality of distribution was assessed by the Kolmogorov–Smirnov test, and all the variables presented normal distribution (p > 0.05); thus, we performed parametric tests for our analysis. The BDNF serum levels, gene expression, and DNA methylation were then correlated with clinical cardiometabolic parameters by Pearson’s correlation considering r ≥ 0.35 and p ≤ 0.05 significant. As we did not see any effect of age and sex for any of the variables, pre and post intervention, we carried out our correlations without age and sex adjustment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/epigenomes6010007/s1, File S1. Care Call: Preventing type 2 diabetes.
Author Contributions
Conceptualisation, H.A.F. and A.H.H.; methodology, H.A.F., N.M., G.Y.C. and K.S.; validation, H.A.F., K.S., J.M.G., R.D., C.F.D. and A.H.H.; formal analysis, H.A.F. and K.S.; investigation, H.A.F., N.M., G.Y.C. and K.S.; resources, J.M.G., R.D. and A.H.H.; data curation, R.D., J.M.G. and A.H.H.; writing—original draft preparation, H.A.F. and A.H.H.; writing—review and editing, H.A.F., J.M.G., C.F.D., R.D. and A.H.H.; visualisation, C.F.D. and R.D.; supervision, R.D., C.F.D., J.M.G. and A.H.H.; project administration, A.H.H. and J.M.G.; funding acquisition, A.H.H. and J.M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by NOVO Nordisk 2016.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Salford Royal NHS Foundation Trust (protocol code 2014/164Endo, 18 November 2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Request can be made to adrian.heald@manchester.ac.uk.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gunstad, J.; Benitez, A.; Smith, J.; Glickman, E.; Spitznagel, M.B.; Alexander, T.; Juvancic-Heltzel, J.; Murray, L. Serum brain-derived neurotrophic factor is associated with cognitive function in healthy older adults. J. Geriatr. Psychiatry Neurol. 2008, 21, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Erickson, K.I.; Voss, M.W.; Prakash, R.S.; Basak, C.; Szabo, A.; Chaddock, L.; Kim, J.S.; Heo, S.; Alves, H.; White, S.M.; et al. Exercise training increases size of hippocampus and improves memory. Proc. Natl. Acad. Sci. USA 2011, 108, 3017–3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leibrock, J.; Lottspeich, F.; Hohn, A.; Hofer, M.; Hengerer, B.; Masiakowski, P.; Thoenen, H.; Barde, Y. Molecular cloning and expression of brain-derived neurotrophic factor. Nature 1989, 341, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Eyileten, C.; Kaplon-Cieslicka, A.; Mirowska-Guzel, D.; Małek, L.; Postula, M. Antidiabetic Effect of Brain-Derived Neurotrophic Factor and Its Association with Inflammation in Type 2 Diabetes Mellitus. J. Diabetes Res. 2017, 2017, 2823671. [Google Scholar] [CrossRef] [PubMed]
- Wisse, B.E.; Schwartz, M.W. The skinny on neurotrophins. Nat. Neurosci. 2003, 6, 655–656. [Google Scholar] [CrossRef]
- Ukropec, J.; Ukropcova, B.; Kurdiova, T.; Gasperikova, D.; Klimeš, I. Adipose tissue and skeletal muscle plasticity modulates metabolic health. Arch. Physiol. Biochem. 2008, 114, 357–368. [Google Scholar] [CrossRef]
- Noble, E.E.; Billington, C.J.; Kotz, C.M.; Wang, C. The lighter side of BDNF. Am. J. Physiol. Integr. Comp. Physiol. 2011, 300, R1053–R1069. [Google Scholar] [CrossRef]
- Ono, M.; Itakura, Y.; Nonomura, T.; Nakagawa, T.; Nakayama, C.; Taiji, M.; Noguchi, H. Intermittent administration of brain-derived neurotrophic factor ameliorates glucose metabolism in obese diabetic mice. Metabolism 2000, 49, 129–133. [Google Scholar] [CrossRef]
- Hanyu, O.; Yamatani, K.; Ikarashi, T.; Soda, S.; Maruyama, S.; Kamimura, T.; Kaneko, S.; Hirayama, S.; Suzuki, K.; Nakagawa, O.; et al. Brain-derived neurotrophic factor modulates glucagon secretion from pancreatic alpha cells: Its contribution to glucose metabolism. Diabetes Obes. Metab. 2003, 5, 27–37. [Google Scholar] [CrossRef]
- Gray, J.; Yeo, G.S.; Cox, J.J.; Morton, J.; Adlam, A.-L.R.; Keogh, J.M.; Yanovski, J.A.; El Gharbawy, A.; Han, J.C.; Tung, Y.L.; et al. Hyperphagia, Severe Obesity, Impaired Cognitive Function, and Hyperactivity Associated with Functional Loss of One Copy of the Brain-Derived Neurotrophic Factor (BDNF) Gene. Diabetes 2006, 55, 3366–3371. [Google Scholar] [CrossRef] [Green Version]
- Yeo, G.S.H.; Hung, C.-C.C.; Rochford, J.; Keogh, J.; Gray, J.; Sivaramakrishnan, S.; O’Rahilly, S.; Farooqi, S. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 2004, 7, 1187–1189. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xi, B.; Zhang, M.; Shen, Y.; Zhao, X.; Cheng, H.; Hou, D.; Sun, D.; Ott, J.; Wang, X.; et al. Associations of Six Single Nucleotide Polymorphisms in Obesity-Related Genes with BMI and Risk of Obesity in Chinese Children. Diabetes 2010, 59, 3085–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skledar, M.; Nikolac, M.; Dodig-Curkovic, K.; Curkovic, M.; Borovecki, F.; Pivac, N. Association between brain-derived neurotrophic factor Val66Met and obesity in children and adolescents. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2012, 36, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kernie, S.; Liebl, D.J.; Parada, L.F. BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000, 19, 1290–1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios, M.; Fan, G.; Fekete, C.; Kelly, J.; Bates, B.; Kuehn, R.; Lechan, R.M.; Jaenisch, R. Conditional Deletion of Brain-Derived Neurotrophic Factor in the Postnatal Brain Leads to Obesity and Hyperactivity. Mol. Endocrinol. 2001, 15, 1748–1757. [Google Scholar] [CrossRef] [PubMed]
- Gabir, M.M.; Hanson, R.L.; Dabelea, D.; Imperatore, G.; Roumain, J.; Bennett, P.H.; Knowler, W.C. The 1997 American Diabetes Association and 1999 World Health Organization criteria for hyperglycemia in the diagnosis and prediction of diabetes. Diabetes Care 2000, 23, 1108–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malipatil, N.; Fachim, H.A.; Siddals, K.; Geary, B.; Wark, G.; Porter, N.; Anderson, S.; Donn, R.; Harvie, M.; Whetton, A.D.; et al. Data Independent Acquisition Mass Spectrometry Can Identify Circulating Proteins That Predict Future Weight Loss with a Diet and Exercise Programme. J. Clin. Med. 2019, 8, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fachim, H.A.; Loureiro, C.M.; Siddals, K.; Dalton, C.F.; Reynolds, G.P.; Gibson, J.M.; Chen, Z.B.; Heald, A.H. Circulating microRNA changes in patients with impaired glucose regulation. Adipocyte 2020, 9, 443–453. [Google Scholar] [CrossRef]
- Heald, A.H.; Kaushal, K.; Siddals, K.W.; Rudenski, A.S.; Anderson, S.G.; Gibson, J.M. Insulin-like Growth Factor Binding Protein-2 (IGFBP-2) is a Marker for the Metabolic Syndrome. Exp. Clin. Endocrinol. Diabetes 2006, 114, 371–376. [Google Scholar] [CrossRef]
- Kang, H.S.; Cho, H.C.; Lee, J.H.; Oh, G.T.; Koo, S.H.; Park, B.H.; Lee, I.K.; Choi, H.S.; Song, D.K.; Im, S.S. Metformin stimulates IGFBP-2 gene expression through PPARalpha in diabetic states. Sci. Rep. 2016, 6, 23665. [Google Scholar] [CrossRef]
- Ceccarini, G.; Pelosini, C.; Ferrari, F.; Magno, S.; Vitti, J.; Salvetti, G.; Moretto, C.; Marioni, A.; Buccianti, P.; Piaggi, P.; et al. Serum IGF-binding protein 2 (IGFBP-2) concentrations change early after gastric bypass bariatric surgery revealing a possible marker of leptin sensitivity in obese subjects. Endocrine 2019, 65, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Malheiros, R.T.; Delgado, H.O.; Felber, D.T.; Kraus, S.I.; Dos Santos, A.R.S.; Manfredini, V.; Da Silva, M.D. Mood disorders are associated with the reduction of brain derived neurotrophic factor in the hypocampus in rats submitted to the hipercaloric diet. Metab. Brain Dis. 2020, 36, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Han, X.; Zhou, X.; Zhang, S.; Cai, X.; Zhang, L.; Li, Y.; Li, M.; Gong, S.; Ji, L. Brain derived neurotrophic factor in newly diagnosed diabetes and prediabetes. Mol. Cell. Endocrinol. 2016, 429, 106–113. [Google Scholar] [CrossRef]
- Esvald, E.-E.; Tuvikene, J.; Sirp, A.; Patil, S.; Bramham, C.R.; Timmusk, T. CREB Family Transcription Factors Are Major Mediators of BDNF Transcriptional Autoregulation in Cortical Neurons. J. Neurosci. 2020, 40, 1405–1426. [Google Scholar] [CrossRef] [PubMed]
- Marosi, K.; Mattson, M.P. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 2013, 25, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Guzzardi, M.; Sanguinetti, E.; Bartoli, A.; Kemeny, A.; Panetta, D.; Salvadori, P.A.; Burchielli, S.; Iozzo, P. Elevated glycemia and brain glucose utilization predict BDNF lowering since early life. J. Cereb. Blood Flow Metab. 2017, 38, 447–455. [Google Scholar] [CrossRef]
- Krabbe, K.S.; Nielsen, A.R.; Krogh-Madsen, R.; Plomgaard, P.; Rasmussen, P.; Erikstrup, C.; Fischer, C.; Lindegaard, B.; Petersen, A.M.W.; Taudorf, S.; et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia 2006, 50, 431–438. [Google Scholar] [CrossRef]
- Zhen, Y.F.; Zhang, J.; Liu, X.Y.; Fang, H.; Tian, L.B.; Zhou, D.H.; Kosten, T.R.; Zhang, X.Y. Low BDNF is associated with cognitive deficits in patients with type 2 diabetes. Psychopharmacology 2012, 227, 93–100. [Google Scholar] [CrossRef]
- Fachim, H.A.; Corsi-Zuelli, F.; Loureiro, C.M.; Iamjan, S.-A.; Shuhama, R.; Joca, S.; Menezes, P.R.; Heald, A.; Louzada-Junior, P.; Dalton, C.F.; et al. Early-life stress effects on BDNF DNA methylation in first-episode psychosis and in rats reared in isolation. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 108, 110188. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).