Genetic and Epigenetic Inheritance at Telomeres
Abstract
:1. Introduction
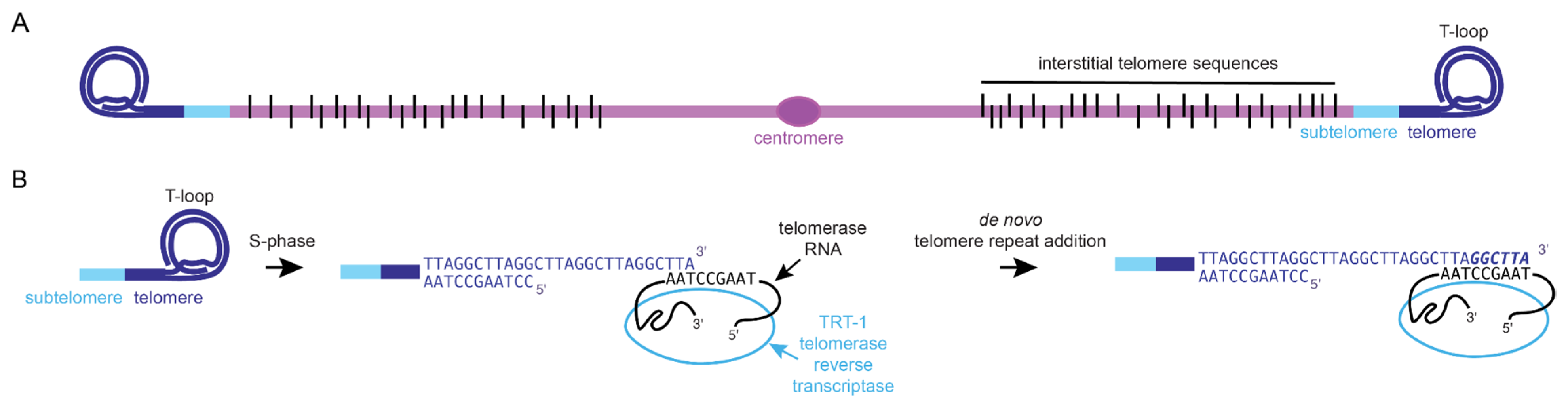
2. Genetic Inheritance of Mammalian Telomeres and Their Health Impact
3. Epigenetics of Telomeres
4. Transgenerational Epigenetic Inheritance
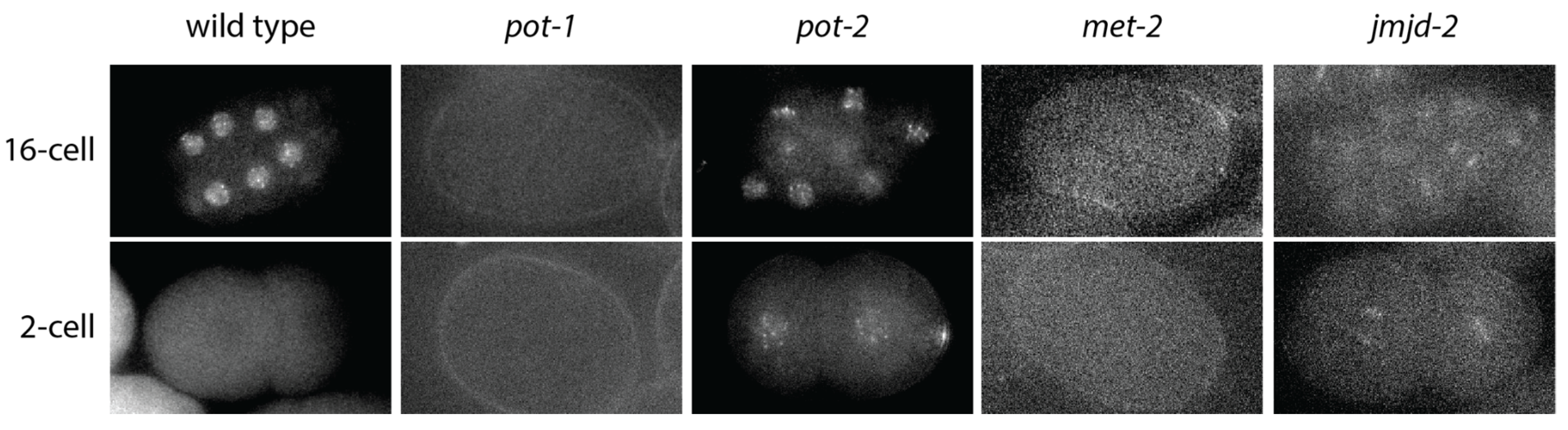
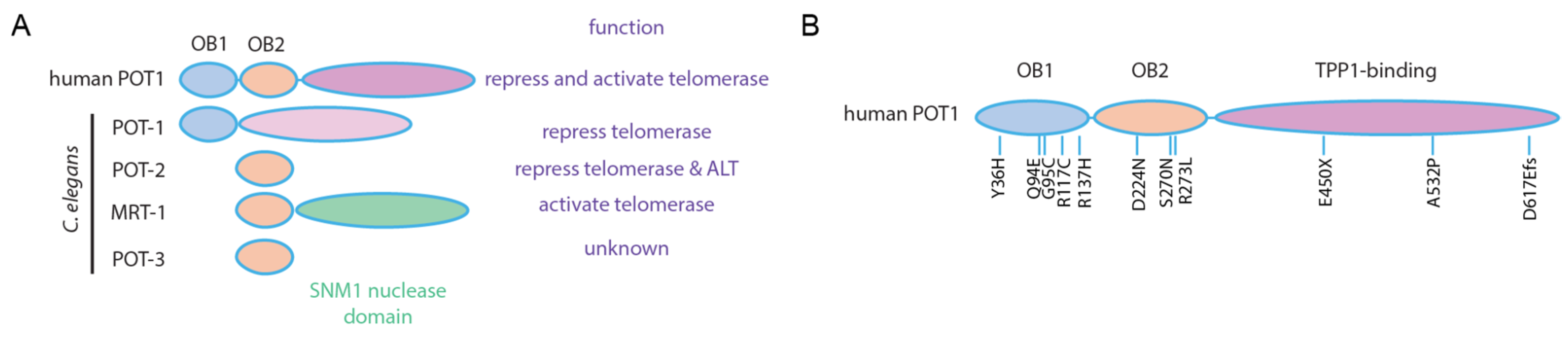
5. Transgenerational Epigenetic Inheritance at C. elegans Telomeres
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian Telomeres End in a Large Duplex Loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef] [Green Version]
- Tomáška, Ľ.; Cesare, A.J.; AlTurki, T.M.; Griffith, J.D. Twenty Years of T-Loops: A Case Study for the Importance of Collaboration in Molecular Biology. DNA Repair 2020, 94, 102901. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Martin, G.M.; Sprague, C.A.; Epstein, C.J. Others Replicative Life-Span of Cultivated Human Cells. Lab. Investig. 1970, 23, 86–92. [Google Scholar]
- Olovnikov, A.M. A Theory of Marginotomy. The Incomplete Copying of Template Margin in Enzymic Synthesis of Polynucleotides and Biological Significance of the Phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Lingner, J.; Cooper, J.P.; Cech, T.R. Telomerase and DNA End Replication: No Longer a Lagging Strand Problem? Science 1995, 269, 1533–1534. [Google Scholar] [CrossRef]
- Olovnikov, A.M. Telomeres, Telomerase, and Aging: Origin of the Theory. Exp. Gerontol. 1996, 31, 443–448. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Gall, J.G. A Tandemly Repeated Sequence at the Termini of the Extrachromosomal Ribosomal RNA Genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Gorovsky, M.A. Genome Organization and Reorganization in Tetrahymena. Annu. Rev. Genet. 1980, 14, 203–239. [Google Scholar] [CrossRef]
- Hamilton, E.P.; Kapusta, A.; Huvos, P.E.; Bidwell, S.L.; Zafar, N.; Tang, H.; Hadjithomas, M.; Krishnakumar, V.; Badger, J.H.; Caler, E.V.; et al. Structure of the Germline Genome of Tetrahymena Thermophila and Relationship to the Massively Rearranged Somatic Genome. Elife 2016, 5, e19090. [Google Scholar] [CrossRef]
- Blackburn, E.H. Telomeres and Telomerase: The Means to the End (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2010, 49, 7405–7421. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. The Telomere Terminal Transferase of Tetrahymena is a Ribonucleoprotein Enzyme with Two Kinds of Primer Specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Blasco, M.A.; Funk, W.; Villeponteau, B.; Greider, C.W. Functional Characterization and Developmental Regulation of Mouse Telomerase RNA. Science 1995, 269, 1267–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Funk, W.D.; Wang, S.S.; Weinrich, S.L.; Avilion, A.A.; Chiu, C.P.; Adams, R.R.; Chang, E.; Allsopp, R.C.; Yu, J. The RNA Component of Human Telomerase. Science 1995, 269, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Collins, K. The Biogenesis and Regulation of Telomerase Holoenzymes. Nat. Rev. Mol. Cell Biol. 2006, 7, 484–494. [Google Scholar] [CrossRef] [Green Version]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Kim, N.; Piatyszek, M.; Prowse, K.; Harley, C.; West, M.; Ho, P.; Coviello, G.; Wright, W.; Weinrich, S.; Shay, J. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of Life-Span by Introduction of Telomerase into Normal Human Cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef] [Green Version]
- Counter, C.M.; Hahn, W.C.; Wei, W.; Caddle, S.D.; Beijersbergen, R.L.; Lansdorp, P.M.; Sedivy, J.M.; Weinberg, R.A. Dissociation among in Vitro Telomerase Activity, Telomere Maintenance, and Cellular Immortalization. Proc. Natl. Acad. Sci. USA 1998, 95, 14723–14728. [Google Scholar] [CrossRef] [Green Version]
- d’Adda di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; Von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA Damage Checkpoint Response in Telomere-Initiated Senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Herbig, U.; Ferreira, M.; Condel, L.; Carey, D.; Sedivy, J.M. Cellular Senescence in Aging Primates. Science 2006, 311, 1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shay, J.W.; Wright, W.E. Hayflick, His Limit, and Cellular Ageing. Nat. Rev. Mol. Cell Biol. 2000, 1, 72–76. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Van Ly, D.; Low, R.R.J.; Frölich, S.; Bartolec, T.K.; Kafer, G.R.; Pickett, H.A.; Gaus, K.; Cesare, A.J. Telomere Loop Dynamics in Chromosome End Protection. Mol. Cell 2018, 71, 510–525.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stansel, R.M.; de Lange, T.; Griffith, J.D. T-Loop Assembly in Vitro Involves Binding of TRF2 near the 3′ Telomeric Overhang. EMBO J. 2001, 20, 5532–5540. [Google Scholar] [CrossRef] [Green Version]
- Karlseder, J.; Smogorzewska, A.; de Lange, T. Senescence Induced by Altered Telomere State, Not Telomere Loss. Science 2002, 295, 2446–2449. [Google Scholar] [CrossRef] [Green Version]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; Bucci, G.; Dobreva, M.; Matti, V.; Beausejour, C.M.; et al. Telomeric DNA Damage is Irreparable and Causes Persistent DNA-Damage-Response Activation. Nat. Cell Biol. 2012, 14, 355–365. [Google Scholar] [CrossRef] [Green Version]
- Micco, R.D.; Di Micco, R.; Krizhanovsky, V.; Baker, D.; di Fagagna, F.D. Cellular Senescence in Ageing: From Mechanisms to Therapeutic Opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomeres and Telomerase: Three Decades of Progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef]
- Heidenreich, B.; Nagore, E.; Rachakonda, P.S.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Becker, J.; Soufir, N.; Hemminki, K.; Kumar, R. Telomerase Reverse Transcriptase Promoter Mutations in Primary Cutaneous Melanoma. Nat. Commun. 2014, 5, 3401. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [Green Version]
- Pickett, H.A.; Reddel, R.R. Molecular Mechanisms of Activity and Derepression of Alternative Lengthening of Telomeres. Nat. Struct. Mol. Biol. 2015, 22, 875–880. [Google Scholar] [CrossRef]
- Gomes, N.M.V.; Ryder, O.A.; Houck, M.L.; Charter, S.J.; Walker, W.; Forsyth, N.R.; Austad, S.N.; Venditti, C.; Pagel, M.; Shay, J.W.; et al. Comparative Biology of Mammalian Telomeres: Hypotheses on Ancestral States and the Roles of Telomeres in Longevity Determination. Aging Cell 2011, 10, 761–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seluanov, A.; Chen, Z.; Hine, C.; Sasahara, T.H.C.; Ribeiro, A.A.C.M.; Catania, K.C.; Presgraves, D.C.; Gorbunova, V. Telomerase Activity Coevolves with Body Mass not Lifespan. Aging Cell 2007, 6, 45–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risques, R.A.; Promislow, D.E.L. All’s Well That Ends Well: Why Large Species Have Short Telomeres. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160448. [Google Scholar] [CrossRef] [Green Version]
- Gorbunova, V.; Seluanov, A. Coevolution of Telomerase Activity and Body Mass in Mammals: From Mice to Beavers. Mech. Ageing Dev. 2009, 130, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Whittemore, K.; Vera, E.; Martínez-Nevado, E.; Sanpera, C.; Blasco, M.A. Telomere Shortening Rate Predicts Species Life Span. Proc. Natl. Acad. Sci. USA 2019, 116, 15122–15127. [Google Scholar] [CrossRef] [Green Version]
- Tricola, G.M.; Simons, M.J.P.; Atema, E.; Boughton, R.K.; Brown, J.L.; Dearborn, D.C.; Divoky, G.; Eimes, J.A.; Huntington, C.E.; Kitaysky, A.S.; et al. The Rate of Telomere Loss is Related to Maximum Lifespan in Birds. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160445. [Google Scholar] [CrossRef]
- Ruis, P.; Van Ly, D.; Borel, V.; Kafer, G.R.; McCarthy, A.; Howell, S.; Blassberg, R.; Snijders, A.P.; Briscoe, J.; Niakan, K.K.; et al. TRF2-Independent Chromosome End Protection during Pluripotency. Nature 2021, 589, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Bekaert, S.; Derradji, H.; Baatout, S. Telomere Biology in Mammalian Germ Cells and during Development. Dev. Biol. 2004, 274, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, E.; Hiyama, K. Telomere and Telomerase in Stem Cells. Br. J. Cancer 2007, 96, 1020–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decker, M.L.; Chavez, E.; Vulto, I.; Lansdorp, P.M. Telomere Length in Hutchinson-Gilford Progeria Syndrome. Mech. Ageing Dev. 2009, 130, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Risques, R.A.; Martin, G.M.; Rabinovitch, P.S.; Oshima, J. Accelerated Telomere Shortening and Replicative Senescence in Human Fibroblasts Overexpressing Mutant and Wild-Type Lamin A. Exp. Cell Res. 2008, 314, 82–91. [Google Scholar] [CrossRef] [Green Version]
- Burla, R.; La Torre, M.; Saggio, I. Mammalian Telomeres and Their Partnership with Lamins. Nucleus 2016, 7, 187–202. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Lee, L.; Kudlow, B.A.; Dos Santos, H.G.; Sletvold, O.; Shafeghati, Y.; Botha, E.G.; Garg, A.; Hanson, N.B.; Martin, G.M.; et al. LMNA Mutations in Atypical Werner’s Syndrome. Lancet 2003, 362, 440–445. [Google Scholar] [CrossRef]
- Lombard, D.B.; Beard, C.; Johnson, B.; Marciniak, R.A.; Dausman, J.; Bronson, R.; Buhlmann, J.E.; Lipman, R.; Curry, R.; Sharpe, A.; et al. Mutations in the WRN Gene in Mice Accelerate Mortality in a p53-Null Background. Mol. Cell. Biol. 2000, 20, 3286–3291. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.; Multani, A.S.; Cabrera, N.G.; Naylor, M.L.; Laud, P.; Lombard, D.; Pathak, S.; Guarente, L.; DePinho, R.A. Essential Role of Limiting Telomeres in the Pathogenesis of Werner Syndrome. Nat. Genet. 2004, 36, 877–882. [Google Scholar] [CrossRef]
- Campisi, J. Cellular Senescence: Putting the Paradoxes in Perspective. Curr. Opin. Genet. Dev. 2011, 21, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Ju, Z.; Rudolph, K.L. Telomere Shortening and Ageing. Z. Gerontol. Geriatr. 2007, 40, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Du, H.-Y.; Idol, R.; Robledo, S.; Ivanovich, J.; An, P.; Londono-Vallejo, A.; Wilson, D.B.; Mason, P.J.; Bessler, M. Telomerase Reverse Transcriptase Haploinsufficiency and Telomere Length in Individuals with 5p-Syndrome. Aging Cell 2007, 6, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armanios, M.Y.; Chen, J.J.-L.; Cogan, J.D.; Alder, J.K.; Ingersoll, R.G.; Markin, C.; Lawson, W.E.; Xie, M.; Vulto, I.; Phillips, J.A., 3rd; et al. Telomerase Mutations in Families with Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2007, 356, 1317–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman, F.; Bouarich, R.; Kulkarni, S.; Freeman, S.; Du, H.-Y.; Harrington, L.; Mason, P.J.; Londoño-Vallejo, A.; Bessler, M. The Effect of TERC Haploinsufficiency on the Inheritance of Telomere Length. Proc. Natl. Acad. Sci. USA 2005, 102, 17119–17124. [Google Scholar] [CrossRef] [Green Version]
- Chiang, Y.J.; Calado, R.T.; Hathcock, K.S.; Lansdorp, P.M.; Young, N.S.; Hodes, R.J. Telomere Length is Inherited with Resetting of the Telomere Set-Point. Proc. Natl. Acad. Sci. USA 2010, 107, 10148–10153. [Google Scholar] [CrossRef] [Green Version]
- de Souza Costa, D.; Rosa, D.V.F.; Barros, A.G.A.; Romano-Silva, M.A.; Malloy-Diniz, L.F.; Mattos, P.; de Miranda, D.M. Telomere Length is Highly Inherited and Associated with Hyperactivity-Impulsivity in Children with Attention Deficit/Hyperactivity Disorder. Front. Mol. Neurosci. 2015, 8, 28. [Google Scholar]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human Telomere Biology: A Contributory and Interactive Factor in Aging, Disease Risks, and Protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [Green Version]
- Vaiserman, A.; Krasnienkov, D. Telomere Length as a Marker of Biological Age: State-of-the-Art, Open Issues, and Future Perspectives. Front. Genet. 2020, 11, 630186. [Google Scholar] [CrossRef]
- Chan, K.L.; Lo, C.K.M.; Ho, F.K.; Leung, W.C.; Yee, B.K.; Ip, P. The Association between Intimate Partner Violence against Women and Newborn Telomere Length. Transl. Psychiatry 2019, 9, 239. [Google Scholar] [CrossRef]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef]
- Arsenis, N.C.; You, T.; Ogawa, E.F.; Tinsley, G.M.; Zuo, L. Physical Activity and Telomere Length: Impact of Aging and Potential Mechanisms of Action. Oncotarget 2017, 8, 45008–45019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luxton, J.J.; McKenna, M.J.; Lewis, A.; Taylor, L.E.; George, K.A.; Dixit, S.M.; Moniz, M.; Benegas, W.; Mackay, M.J.; Mozsary, C.; et al. Telomere Length Dynamics and DNA Damage Responses Associated with Long-Duration Spaceflight. Cell Rep. 2020, 33, 108457. [Google Scholar] [CrossRef] [PubMed]
- Krishna, B.H.; Keerthi, G.S.; Kumar, C.K.; Reddy, N.M. Association of Leukocyte Telomere Length with Oxidative Stress in Yoga Practitioners. J. Clin. Diagn. Res. 2015, 9, CC01–CC03. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.R.; Cheng, J.; Collins, K. A Box H/ACA Small Nucleolar RNA-like Domain at the Human Telomerase RNA 3′ End. Mol. Cell. Biol. 1999, 19, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellodi, C.; McMahon, M.; Contreras, A.; Juliano, D.; Kopmar, N.; Nakamura, T.; Maltby, D.; Burlingame, A.; Savage, S.A.; Shimamura, A.; et al. H/ACA Small RNA Dysfunctions in Disease Reveal Key Roles for Noncoding RNA Modifications in Hematopoietic Stem Cell Differentiation. Cell Rep. 2013, 3, 1493–1502. [Google Scholar] [CrossRef] [Green Version]
- Vulliamy, T.; Marrone, A.; Goldman, F.; Dearlove, A.; Bessler, M.; Mason, P.J.; Dokal, I. The RNA Component of Telomerase is Mutated in Autosomal Dominant Dyskeratosis Congenita. Nature 2001, 413, 432–435. [Google Scholar] [CrossRef]
- Dokal, I.; Vulliamy, T. Dyskeratosis Congenita: Its Link to Telomerase and Aplastic Anaemia. Blood Rev. 2003, 17, 217–225. [Google Scholar] [CrossRef]
- Tsakiri, K.D.; Cronkhite, J.T.; Kuan, P.J.; Xing, C.; Raghu, G.; Weissler, J.C.; Rosenblatt, R.L.; Shay, J.W.; Garcia, C.K. Adult-Onset Pulmonary Fibrosis Caused by Mutations in Telomerase. Proc. Natl. Acad. Sci. USA 2007, 104, 7552–7557. [Google Scholar] [CrossRef] [Green Version]
- Armanios, M.; Blackburn, E.H. The Telomere Syndromes. Nat. Rev. Genet. 2012, 13, 693–704. [Google Scholar] [CrossRef]
- Glousker, G.; Touzot, F.; Revy, P.; Tzfati, Y.; Savage, S.A. Unraveling the Pathogenesis of Hoyeraal-Hreidarsson Syndrome, a Complex Telomere Biology Disorder. Br. J. Haematol. 2015, 170, 457–471. [Google Scholar] [CrossRef]
- Savage, S.A. Beginning at the Ends: Telomeres and Human Disease. F1000Research 2018, 7, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannengiesser, C.; Borie, R.; Ménard, C.; Réocreux, M.; Nitschké, P.; Gazal, S.; Mal, H.; Taillé, C.; Cadranel, J.; Nunes, H.; et al. Heterozygous RTEL1 Mutations are Associated with Familial Pulmonary Fibrosis. Eur. Respir. J. 2015, 46, 474–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannier, J.-B.; Sarek, G.; Boulton, S.J. RTEL1: Functions of a Disease-Associated Helicase. Trends Cell Biol. 2014, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Cech, T.R. Publisher Correction: Shaping Human Telomeres: From Shelterin and CST Complexes to Telomeric Chromatin Organization. Nat. Rev. Mol. Cell Biol. 2021, 22, 299. [Google Scholar] [CrossRef]
- Lue, N.F. Evolving Linear Chromosomes and Telomeres: A C-Strand-Centric View. Trends Biochem. Sci. 2018, 43, 314–326. [Google Scholar] [CrossRef]
- Kirwan, M.; Dokal, I. Dyskeratosis Congenita: A Genetic Disorder of Many Faces. Clin. Genet. 2008, 73, 103–112. [Google Scholar] [CrossRef]
- Walne, A.J.; Vulliamy, T.; Kirwan, M.; Plagnol, V.; Dokal, I. Constitutional Mutations in RTEL1 Cause Severe Dyskeratosis Congenita. Am. J. Hum. Genet. 2013, 92, 448–453. [Google Scholar] [CrossRef] [Green Version]
- Jullien, L.; Kannengiesser, C.; Kermasson, L.; Cormier-Daire, V.; Leblanc, T.; Soulier, J.; Londono-Vallejo, A.; de Villartay, J.-P.; Callebaut, I.; Revy, P. Mutations of the RTEL1 Helicase in a Hoyeraal-Hreidarsson Syndrome Patient Highlight the Importance of the ARCH Domain. Hum. Mutat. 2016, 37, 469–472. [Google Scholar] [CrossRef]
- Gramatges, M.M.; Qi, X.; Sasa, G.S.; Chen, J.J.-L.; Bertuch, A.A. A Homozygous Telomerase T-Motif Variant Resulting in Markedly Reduced Repeat Addition Processivity in Siblings with Hoyeraal Hreidarsson Syndrome. Blood J. Am. Soc. Hematol. 2013, 121, 3586–3593. [Google Scholar] [CrossRef] [Green Version]
- Marrone, A.; Walne, A.; Tamary, H.; Masunari, Y.; Kirwan, M.; Beswick, R.; Vulliamy, T.; Dokal, I. Telomerase Reverse-Transcriptase Homozygous Mutations in Autosomal Recessive Dyskeratosis Congenita and Hoyeraal-Hreidarsson Syndrome. Blood 2007, 110, 4198–4205. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, K.E.; Bohnsack, M.T. Uncovering the Assembly Pathway of Human Ribosomes and Its Emerging Links to Disease. EMBO J. 2019, 38, e100278. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-W.; Blasco, M.A.; Gottlieb, G.J.; Horner, J.W.; Greider, C.W.; DePinho, R.A. Essential Role of Mouse Telomerase in Highly Proliferative Organs. Nature 1998, 392, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, K.L.; Chang, S.; Lee, H.W.; Blasco, M.; Gottlieb, G.J.; Greider, C.; DePinho, R.A. Longevity, Stress Response, and Cancer in Aging Telomerase-Deficient Mice. Cell 1999, 96, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Blasco, M.A.; Lee, H.W.; Hande, M.P.; Samper, E.; Lansdorp, P.M.; DePinho, R.A.; Greider, C.W. Telomere Shortening and Tumor Formation by Mouse Cells Lacking Telomerase RNA. Cell 1997, 91, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Armanios, M. Telomeres and Age-Related Disease: How Telomere Biology Informs Clinical Paradigms. J. Clin. Investig. 2013, 123, 996–1002. [Google Scholar] [CrossRef] [Green Version]
- Stanley, S.E.; Merck, S.J.; Armanios, M. Telomerase and the Genetics of Emphysema Susceptibility. Implications for Pathogenesis Paradigms and Patient Care. Ann. Am. Thorac. Soc. 2016, 13 (Suppl. 5), S447–S451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anic, G.M.; Sondak, V.K.; Messina, J.L.; Fenske, N.A.; Zager, J.S.; Cherpelis, B.S.; Lee, J.-H.; Fulp, W.J.; Epling-Burnette, P.K.; Park, J.Y.; et al. Telomere Length and Risk of Melanoma, Squamous Cell Carcinoma, and Basal Cell Carcinoma. Cancer Epidemiol. 2013, 37, 434–439. [Google Scholar] [CrossRef] [Green Version]
- Rode, L.; Nordestgaard, B.G.; Bojesen, S.E. Long Telomeres and Cancer Risk among 95,568 Individuals from the General Population. Int. J. Epidemiol. 2016, 45, 1634–1643. [Google Scholar] [CrossRef] [Green Version]
- Telomeres Mendelian Randomization Collaboration; Haycock, P.C.; Burgess, S.; Nounu, A.; Zheng, J.; Okoli, G.N.; Bowden, J.; Wade, K.H.; Timpson, N.J.; Evans, D.M.; et al. Association between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017, 3, 636–651. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere Position Effect: Regulation of Gene Expression with Progressive Telomere Shortening over Long Distances. Genes Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position Effect at S. Cerevisiae Telomeres: Reversible Repression of Pol II Transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef]
- Aparicio, O.M.; Billington, B.L.; Gottschling, D.E. Modifiers of Position Effect are Shared between Telomeric and Silent Mating-Type Loci in S. Cerevisiae. Cell 1991, 66, 1279–1287. [Google Scholar] [CrossRef]
- Sussel, L.; Shore, D. Separation of Transcriptional Activation and Silencing Functions of the RAP1-Encoded Repressor/Activator Protein 1: Isolation of Viable Mutants Affecting both Silencing and Telomere Length. Proc. Natl. Acad. Sci. USA 1991, 88, 7749–7753. [Google Scholar] [CrossRef] [Green Version]
- Tham, W.-H.; Zakian, V.A. Transcriptional Silencing at Saccharomyces Telomeres: Implications for Other Organisms. Oncogene 2002, 21, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, B.K.; Austriaco, N.R., Jr.; Zhang, J.; Guarente, L. Mutation in the Silencing Gene SIR4 Can Delay Aging in S. Cerevisiae. Cell 1995, 80, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, B.K.; Gotta, M.; Sinclair, D.A.; Mills, K.; McNabb, D.S.; Murthy, M.; Pak, S.M.; Laroche, T.; Gasser, S.M.; Guarente, L. Redistribution of Silencing Proteins from Telomeres to the Nucleolus is Associated with Extension of Life Span in S. Cerevisiae. Cell 1997, 89, 381–391. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA Circles--A Cause of Aging in Yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef] [Green Version]
- Renauld, H.; Aparicio, O.M.; Zierath, P.D.; Billington, B.L.; Chhablani, S.K.; Gottschling, D.E. Silent Domains are Assembled Continuously from the Telomere and are Defined by Promoter Distance and Strength, and by SIR3 Dosage. Genes Dev. 1993, 7, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Henderson, K.A.; Gottschling, D.E. A Mother’s Sacrifice: What is She Keeping for Herself? Curr. Opin. Cell Biol. 2008, 20, 723–728. [Google Scholar] [CrossRef] [Green Version]
- Pryde, F.E.; Louis, E.J. Limitations of Silencing at Native Yeast Telomeres. EMBO J. 1999, 18, 2538–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zill, O.A.; Scannell, D.; Teytelman, L.; Rine, J. Co-Evolution of Transcriptional Silencing Proteins and the DNA Elements Specifying Their Assembly. PLoS Biol. 2010, 8, e1000550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radman-Livaja, M.; Ruben, G.; Weiner, A.; Friedman, N.; Kamakaka, R.; Rando, O.J. Dynamics of Sir3 Spreading in Budding Yeast: Secondary Recruitment Sites and Euchromatic Localization. EMBO J. 2011, 30, 1012–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourel, G.; Revardel, E.; Koering, C.E.; Gilson, E. Cohabitation of Insulators and Silencing Elements in Yeast Subtelomeric Regions. EMBO J. 1999, 18, 2522–2537. [Google Scholar] [CrossRef] [Green Version]
- Ellahi, A.; Thurtle, D.M.; Rine, J. The Chromatin and Transcriptional Landscape of Native Saccharomyces Cerevisiae Telomeres and Subtelomeric Domains. Genetics 2015, 200, 505–521. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J. The Nature of Heterochromatin. Cold Spring Harb. Symp. Quant. Biol. 1947, 12, 179–191. [Google Scholar] [CrossRef]
- George, J.A.; DeBaryshe, P.G.; Traverse, K.L.; Celniker, S.E.; Pardue, M.-L. Genomic Organization of the Drosophila Telomere Retrotransposable Elements. Genome Res. 2006, 16, 1231–1240. [Google Scholar] [CrossRef] [Green Version]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef]
- García-Cao, M.; O’Sullivan, R.; Peters, A.H.F.; Jenuwein, T.; Blasco, M.A. Epigenetic Regulation of Telomere Length in Mammalian Cells by the Suv39h1 and Suv39h2 Histone Methyltransferases. Nat. Genet. 2004, 36, 94–99. [Google Scholar] [CrossRef]
- Koering, C.E.; Pollice, A.; Zibella, M.P.; Bauwens, S.; Puisieux, A.; Brunori, M.; Brun, C.; Martins, L.; Sabatier, L.; Pulitzer, J.F.; et al. Human Telomeric Position Effect is Determined by Chromosomal Context and Telomeric Chromatin Integrity. EMBO Rep. 2002, 3, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Schoeftner, S.; Blasco, M.A. Developmentally Regulated Transcription of Mammalian Telomeres by DNA-Dependent RNA Polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Hartlieb, S.A.; Sieverling, L.; Nadler-Holly, M.; Ziehm, M.; Toprak, U.H.; Herrmann, C.; Ishaque, N.; Okonechnikov, K.; Gartlgruber, M.; Park, Y.-G.; et al. Alternative Lengthening of Telomeres in Childhood Neuroblastoma from Genome to Proteome. Nat. Commun. 2021, 12, 1269. [Google Scholar] [CrossRef] [PubMed]
- Achrem, M.; Szućko, I.; Kalinka, A. The Epigenetic Regulation of Centromeres and Telomeres in Plants and Animals. Comp. Cytogenet. 2020, 14, 265–311. [Google Scholar] [CrossRef]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Schotta, G.; Klatt, P.; Jenuwein, T.; Blasco, M.A. Suv4-20h Deficiency Results in Telomere Elongation and Derepression of Telomere Recombination. J. Cell Biol. 2007, 178, 925–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered Telomeres in Tumors with ATRX and DAXX Mutations. Science 2011, 333, 6041. [Google Scholar] [CrossRef] [Green Version]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, Genome Instability, and an Altered DNA Damage Response are Hallmarks of the Alternative Lengthening of Telomeres Pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef]
- Episkopou, H.; Draskovic, I.; Van Beneden, A.; Tilman, G.; Mattiussi, M.; Gobin, M.; Arnoult, N.; Londoño-Vallejo, A.; Decottignies, A. Alternative Lengthening of Telomeres is Characterized by Reduced Compaction of Telomeric Chromatin. Nucleic Acids Res. 2014, 42, 4391–4405. [Google Scholar] [CrossRef]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of Non-Telomeric Sites of the (TTAGGG)n Telomeric Sequence in Vertebrate Chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic Features of Human Telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef] [Green Version]
- Frenk, S.; Lister-Shimauchi, E.H.; Ahmed, S. Corrigendum: Telomeric Small RNAs in the Genus Caenorhabditis. RNA 2019, 25, 1405. [Google Scholar] [CrossRef] [Green Version]
- McMurchy, A.N.; Stempor, P.; Gaarenstroom, T.; Wysolmerski, B.; Dong, Y.; Aussianikava, D.; Appert, A.; Huang, N.; Kolasinska-Zwierz, P.; Sapetschnig, A.; et al. Correction: A Team of Heterochromatin Factors Collaborates with Small RNA Pathways to Combat Repetitive Elements and Germline Stress. Elife 2017, 6, e21666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towbin, B.D.; González-Aguilera, C.; Sack, R.; Gaidatzis, D.; Kalck, V.; Meister, P.; Askjaer, P.; Gasser, S.M. Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell 2012, 150, 934–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrbsky, J.; Akimcheva, S.; Matthew Watson, J.; Turner, T.L.; Daxinger, L.; Vyskot, B.; Aufsatz, W.; Riha, K. siRNA–Mediated Methylation of Arabidopsis Telomeres. PLoS Genet. 2010, 6, e1000986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaquero-Sedas, M.I.; Luo, C.; Vega-Palas, M.A. Analysis of the Epigenetic Status of Telomeres by Using ChIP-Seq Data. Nucleic Acids Res. 2012, 40, e163. [Google Scholar] [CrossRef] [Green Version]
- Elgin, S.C.R.; Reuter, G. Position-Effect Variegation, Heterochromatin Formation, and Gene Silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef]
- Spielmann, M.; Lupiáñez, D.G.; Mundlos, S. Structural Variation in the 3D Genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef] [Green Version]
- Baur, J.A.; Zou, Y.; Shay, J.W.; Wright, W.E. Telomere Position Effect in Human Cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef]
- Pearson, C.E. FSHD: A Repeat Contraction Disease Finally Ready to Expand (Our Understanding of Its Pathogenesis). PLoS Genet. 2010, 6, e1001180. [Google Scholar] [CrossRef]
- Stadler, G.; Rahimov, F.; King, O.D.; Chen, J.C.J.; Robin, J.D.; Wagner, K.R.; Shay, J.W.; Emerson, C.P.; Wright, W.E. Telomere Position Effect Regulates DUX4 in Human Facioscapulohumeral Muscular Dystrophy. Nat. Struct. Mol. Biol. 2013, 20, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Tichy, E.D.; Sidibe, D.K.; Tierney, M.T.; Stec, M.J.; Sharifi-Sanjani, M.; Hosalkar, H.; Mubarak, S.; Johnson, F.B.; Sacco, A.; Mourkioti, F. Single Stem Cell Imaging and Analysis Reveals Telomere Length Differences in Diseased Human and Mouse Skeletal Muscles. Stem Cell Rep. 2017, 9, 1328–1341. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.R.; Armstrong, D.; Caskey, C.T. Recovery of Induced Mutations for X Chromosome-Linked Muscular Dystrophy in Mice. Proc. Natl. Acad. Sci. USA 1989, 86, 1292–1296. [Google Scholar]
- Bulfield, G.; Siller, W.G.; Wight, P.A.; Moore, K.J. X Chromosome-Linked Muscular Dystrophy (mdx) in the Mouse. Proc. Natl. Acad. Sci. USA 1984, 81, 1189–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Hathcock, K.S.; Hande, P.; Lansdorp, P.M.; Seldin, M.F.; Hodes, R.J. Telomere Length Regulation in Mice is Linked to a Novel Chromosome Locus. Proc. Natl. Acad. Sci. USA 1998, 95, 8648–8653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, A.; Mourkioti, F.; Tran, R.; Choi, J.; Llewellyn, M.; Kraft, P.; Shkreli, M.; Delp, S.; Pomerantz, J.H.; Artandi, S.E.; et al. Short Telomeres and Stem Cell Exhaustion Model Duchenne Muscular Dystrophy in mdx/mTR Mice. Cell 2010, 143, 1059–1071. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Maekawa, T.; Yoshida, K.; Ly, N.H.; Inoue, K.; Hasegawa, A.; Chatton, B.; Ogura, A.; Ishii, S. Telomere Shortening by Transgenerational Transmission of TNF-α-Induced TERRA via ATF7. Nucleic Acids Res. 2019, 47, 283–298. [Google Scholar] [CrossRef] [Green Version]
- Jordan, J.M.; Hibshman, J.D.; Webster, A.K.; Kaplan, R.E.W.; Leinroth, A.; Guzman, R.; Maxwell, C.S.; Chitrakar, R.; Bowman, E.A.; Fry, A.L.; et al. Insulin/IGF Signaling and Vitellogenin Provisioning Mediate Intergenerational Adaptation to Nutrient Stress. Curr. Biol. 2019, 29, 2380–2388.e5. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent Epigenetic Differences Associated with Prenatal Exposure to Famine in Humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zelner, J.L.; Remais, J.V. Prenatal and Early-Life Exposure to the Great Chinese Famine Increased the Risk of Tuberculosis in Adulthood across Two Generations. Proc. Natl. Acad. Sci. USA 2020, 117, 27549–27555. [Google Scholar]
- Ideraabdullah, F.Y.; Belenchia, A.M.; Rosenfeld, C.S.; Kullman, S.W.; Knuth, M.; Mahapatra, D.; Bereman, M.; Levin, E.D.; Peterson, C.A. Maternal Vitamin D Deficiency and Developmental Origins of Health and Disease (DOHaD). J. Endocrinol. 2019, 241, R65–R80. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.-F.; Lin, R.C.Y.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic High-Fat Diet in Fathers Programs β-Cell Dysfunction in Female Rat Offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally Induced Transgenerational Environmental Reprogramming of Metabolic Gene Expression in Mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and Function of tRNA Fragments during Sperm Maturation and Fertilization in Mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conine, C.C.; Sun, F.; Song, L.; Rivera-Pérez, J.A.; Rando, O.J. Small RNAs Gained during Epididymal Transit of Sperm are Essential for Embryonic Development in Mice. Dev. Cell 2018, 46, 470–480.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cauwenbergh, O.; Di Serafino, A.; Tytgat, J.; Soubry, A. Transgenerational Epigenetic Effects from Male Exposure to Endocrine-Disrupting Compounds: A Systematic Review on Research in Mammals. Clin. Epigenet. 2020, 12, 65. [Google Scholar] [CrossRef]
- Martini, M.; Corces, V.G.; Rissman, E.F. Mini-Review: Epigenetic Mechanisms That Promote Transgenerational Actions of Endocrine Disrupting Chemicals: Applications to Behavioral Neuroendocrinology. Horm. Behav. 2020, 119, 104677. [Google Scholar] [CrossRef] [PubMed]
- Lister-Shimauchi, E.H.; Dinh, M.; Maddox, P.; Ahmed, S. Gametes Deficient for Pot1 Telomere Binding Proteins Alter Levels of Telomeric Foci for Multiple Generations. Commun. Biol. 2021, 4, 158. [Google Scholar] [CrossRef]
- Shtessel, L.; Lowden, M.R.; Cheng, C.; Simon, M.; Wang, K.; Ahmed, S. Caenorhabditis Elegans POT-1 and POT-2 Repress Telomere Maintenance Pathways. G3 2013, 3, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Meier, B.; Barber, L.J.; Liu, Y.; Shtessel, L.; Boulton, S.J.; Gartner, A.; Ahmed, S. The MRT-1 Nuclease Is Required for DNA Crosslink Repair and Telomerase Activity in Vivo in Caenorhabditis Elegans. EMBO J. 2009, 28, 3549–3563. [Google Scholar] [CrossRef] [Green Version]
- Perez, M.F.; Lehner, B. Intergenerational and Transgenerational Epigenetic Inheritance in Animals. Nat. Cell Biol. 2019, 21, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Duempelmann, L.; Skribbe, M.; Bühler, M. Small RNAs in the Transgenerational Inheritance of Epigenetic Information. Trends Genet. 2020, 36, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Skvortsova, K.; Iovino, N.; Bogdanović, O. Functions and Mechanisms of Epigenetic Inheritance in Animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, F.; Li, X.; Hiew, S.; Brady, H.; Liu, Y.; Dou, Y. Dicer Independent Small RNAs Associate with Telomeric Heterochromatin. RNA 2009, 15, 1274–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossiello, F.; Aguado, J.; Sepe, S.; Iannelli, F.; Nguyen, Q.; Pitchiaya, S.; Carninci, P.; d’Adda di Fagagna, F. DNA Damage Response Inhibition at Dysfunctional Telomeres by Modulation of Telomeric DNA Damage Response RNAs. Nat. Commun. 2017, 8, 13980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalinava, N.; Ni, J.Z.; Gajic, Z.; Ushakov, H.; Gu, S.G. Caenorhabditis Elegans Heterochromatin Factor SET-32 Plays an Essential Role in Transgenerational Establishment of Nuclear RNAi-Mediated Epigenetic Silencing. Cell Rep. 2018, 25, 2273–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashe, A.; Sapetschnig, A.; Weick, E.-M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.-L.; Goldstein, L.D.; Lehrbach, N.J.; Le Pen, J.; et al. piRNAs can Trigger a Multigenerational Epigenetic Memory in the Germline of C. Elegans. Cell 2012, 150, 88–99. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Niu, R.; Huang, T.; Shao, L.-W.; Peng, Y.; Ding, W.; Wang, Y.; Jia, G.; He, C.; Li, C.-Y.; et al. N6-Methyldeoxyadenine is a Transgenerational Epigenetic Signal for Mitochondrial Stress Adaptation. Nat. Cell Biol. 2019, 21, 319–327. [Google Scholar] [CrossRef]
- Jobson, M.A.; Jordan, J.M.; Sandrof, M.A.; Hibshman, J.D.; Lennox, A.L.; Baugh, L.R. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis Elegans. Genetics 2015, 201, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Klosin, A.; Casas, E.; Hidalgo-Carcedo, C.; Vavouri, T.; Lehner, B. Transgenerational Transmission of Environmental Information in C. Elegans. Science 2017, 356, 320–323. [Google Scholar] [CrossRef] [Green Version]
- Webster, A.K.; Jordan, J.M.; Hibshman, J.D.; Chitrakar, R.; Baugh, L.R. Transgenerational Effects of Extended Dauer Diapause on Starvation Survival and Gene Expression Plasticity in Caenorhabditis Elegans. Genetics 2018, 210, 263–274. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, S.; Uno, M.; Okabe, E.; Nono, M.; Nishida, E. Environmental Stresses Induce Transgenerationally Inheritable Survival Advantages via Germline-to-Soma Communication in Caenorhabditis Elegans. Nat. Commun. 2017, 8, 14031. [Google Scholar] [CrossRef] [Green Version]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational Epigenetic Inheritance of Longevity in Caenorhabditis Elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.W.; David, H.S.; Engstrom, A.K.; Carpenter, B.S.; Katz, D.J. H3K9me2 Protects Lifespan against the Transgenerational Burden of Germline Transcription in C. Elegans. Elife 2019, 8, e48498. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Kandula, V.; Kosuru, R.; Ye, X.; Irwin, M.G.; Xia, Z. Decoding Telomere Protein Rap1: Its Telomeric and Nontelomeric Functions and Potential Implications in Diabetic Cardiomyopathy. Cell Cycle 2017, 16, 1765–1773. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Renault, V.M.; Jamet, K.; Gilson, E. Transcriptional Outcome of Telomere Signalling. Nat. Rev. Genet. 2014, 15, 491–503. [Google Scholar] [CrossRef]
- Shakirov, E.V.; Perroud, P.-F.; Nelson, A.D.; Cannell, M.E.; Quatrano, R.S.; Shippen, D.E. Protection of Telomeres 1 is Required for Telomere Integrity in the Moss Physcomitrella Patens. Plant Cell 2010, 22, 1838–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, P.; Cech, T.R. Pot1, the Putative Telomere End-Binding Protein in Fission Yeast and Humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Beilstein, M.A.; Shippen, D.E. Evolution of Arabidopsis Protection of Telomeres 1 Alters Nucleic Acid Recognition and Telomerase Regulation. Nucleic Acids Res. 2016, 44, 9821–9830. [Google Scholar] [CrossRef]
- Renfrew, K.B.; Song, X.; Lee, J.R.; Arora, A.; Shippen, D.E. POT1a and Components of CST Engage Telomerase and Regulate Its Activity in Arabidopsis. PLoS Genet. 2014, 10, e1004738. [Google Scholar] [CrossRef] [Green Version]
- Surovtseva, Y.V.; Shakirov, E.V.; Vespa, L.; Osbun, N.; Song, X.; Shippen, D.E. Arabidopsis POT1 Associates with the Telomerase RNP and is Required for Telomere Maintenance. EMBO J. 2007, 26, 3653–3661. [Google Scholar] [CrossRef]
- Shakirov, E.V.; Surovtseva, Y.V.; Osbun, N.; Shippen, D.E. The Arabidopsis Pot1 and Pot2 Proteins Function in Telomere Length Homeostasis and Chromosome End Protection. Mol. Cell. Biol. 2005, 25, 7725–7733. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, C.R.; Castillo-González, C.; Survotseva, Y.; Canal, E.; Nelson, A.D.L.; Shippen, D.E. Recent Emergence and Extinction of the Protection of Telomeres 1c Gene in Arabidopsis Thaliana. Plant Cell Rep. 2019, 38, 1081–1097. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, R.; Horikawa, I.; Fujita, K.; Afshar, Y.; Kokko, A.; Laiho, P.; Aaltonen, L.A.; Harris, C.C. Functional Diversity of Human Protection of Telomeres 1 Isoforms in Telomere Protection and Cellular Senescence. Cancer Res. 2007, 67, 11677–11686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, P.; Gómez-López, G.; García, F.; Mercken, E.; Mitchell, S.; Flores, J.M.; de Cabo, R.; Blasco, M.A. RAP1 Protects from Obesity through Its Extratelomeric Role Regulating Gene Expression. Cell Rep. 2013, 3, 2059–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, F.; Ramírez, C.M.; Mateos-Gomez, P.A.; Pinzaru, A.; Ceccarini, G.; Kabir, S.; Fernández-Hernando, C.; Sfeir, A. Nontelomeric Role for Rap1 in Regulating Metabolism and Protecting against Obesity. Cell Rep. 2013, 3, 1847–1856. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Multani, A.S.; He, H.; Cosme-Blanco, W.; Deng, Y.; Deng, J.M.; Bachilo, O.; Pathak, S.; Tahara, H.; Bailey, S.M.; et al. Pot1 Deficiency Initiates DNA Damage Checkpoint Activation and Aberrant Homologous Recombination at Telomeres. Cell 2006, 126, 49–62. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Wang, Y.; Guo, X.; Ramchandani, S.; Ma, J.; Shen, M.-F.; Garcia, D.A.; Deng, Y.; Multani, A.S.; You, M.J.; et al. Pot1b Deletion and Telomerase Haploinsufficiency in Mice Initiate an ATR-Dependent DNA Damage Response and Elicit Phenotypes Resembling Dyskeratosis Congenita. Mol. Cell. Biol. 2009, 29, 229–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockemeyer, D.; Daniels, J.-P.; Takai, H.; de Lange, T. Recent Expansion of the Telomeric Complex in Rodents: Two Distinct POT1 Proteins Protect Mouse Telomeres. Cell 2006, 126, 63–77. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.; Bisht, K.; Savage, S.A.; Nandakumar, J.; Keegan, C.E.; Maillard, I. The Shelterin Complex and Hematopoiesis. J. Clin. Investig. 2016, 126, 1621–1629. [Google Scholar] [CrossRef] [Green Version]
- Celli, G.B.; de Lange, T. DNA Processing is not Required for ATM-Mediated Telomere Damage Response after TRF2 Deletion. Nat. Cell Biol. 2005, 7, 712–718. [Google Scholar] [CrossRef]
- Karlseder, J.; Kachatrian, L.; Takai, H.; Mercer, K.; Hingorani, S.; Jacks, T.; de Lange, T. Targeted Deletion Reveals an Essential Function for the Telomere Length Regulator Trf1. Mol. Cell. Biol. 2003, 23, 6533–6541. [Google Scholar] [CrossRef] [Green Version]
- Chiang, Y.J.; Kim, S.-H.; Tessarollo, L.; Campisi, J.; Hodes, R.J. Telomere-Associated Protein TIN2 is Essential for Early Embryonic Development through a Telomerase-Independent Pathway. Mol. Cell. Biol. 2004, 24, 6631–6634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denchi, E.L.; de Lange, T. Protection of Telomeres through Independent Control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; de Lange, T. Removal of Shelterin Reveals the Telomere End-Protection Problem. Science 2012, 336, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Calvete, O.; Garcia-Pavia, P.; Domínguez, F.; Bougeard, G.; Kunze, K.; Braeuninger, A.; Teule, A.; Lasa, A.; Cajal, T.R.Y.; Llort, G.; et al. The Wide Spectrum of POT1 Gene Variants Correlates with Multiple Cancer Types. Eur. J. Hum. Genet. 2017, 25, 1278–1281. [Google Scholar] [CrossRef] [Green Version]
- Bainbridge, M.N.; Armstrong, G.N.; Gramatges, M.M.; Bertuch, A.A.; Jhangiani, S.N.; Doddapaneni, H.; Lewis, L.; Tombrello, J.; Tsavachidis, S.; Liu, Y.; et al. Germline Mutations in Shelterin Complex Genes are Associated with Familial Glioma. J. Natl. Cancer Inst. 2015, 107, 384. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.R.; Ballew, B.; Rotunno, M.; Calista, D.; Fargnoli, M.C.; Ghiorzo, P.; Bressac-de Paillerets, B.; Nagore, E.; Avril, M.F.; et al. Rare Missense Variants in POT1 Predispose to Familial Cutaneous Malignant Melanoma. Nat. Genet. 2014, 46, 482–486. [Google Scholar] [CrossRef]
- Robles-Espinoza, C.D.; Harland, M.; Ramsay, A.J.; Aoude, L.G.; Quesada, V.; Ding, Z.; Pooley, K.A.; Pritchard, A.L.; Tiffen, J.C.; Petljak, M.; et al. POT1 Loss-of-Function Variants Predispose to Familial Melanoma. Nat. Genet. 2014, 46, 478–481. [Google Scholar] [CrossRef] [Green Version]
- McMaster, M.L.; Sun, C.; Landi, M.T.; Savage, S.A.; Rotunno, M.; Yang, X.R.; Jones, K.; Vogt, A.; Hutchinson, A.; Zhu, B.; et al. Germline Mutations in Protection of Telomeres 1 in Two Families with Hodgkin Lymphoma. Br. J. Haematol. 2018, 181, 372–377. [Google Scholar] [CrossRef] [Green Version]
- Speedy, H.E.; Kinnersley, B.; Chubb, D.; Broderick, P.; Law, P.J.; Litchfield, K.; Jayne, S.; Dyer, M.J.S.; Dearden, C.; Follows, G.A.; et al. Germ Line Mutations in Shelterin Complex Genes are Associated with Familial Chronic Lymphocytic Leukemia. Blood 2016, 128, 2319–2326. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, A.J.; Quesada, V.; Foronda, M.; Conde, L.; Martínez-Trillos, A.; Villamor, N.; Rodríguez, D.; Kwarciak, A.; Garabaya, C.; Gallardo, M.; et al. POT1 Mutations Cause Telomere Dysfunction in Chronic Lymphocytic Leukemia. Nat. Genet. 2013, 45, 526–530. [Google Scholar] [CrossRef]
- Kim, W.; Hennick, K.; Johnson, J.; Finnerty, B.; Choo, S.; Short, S.B.; Drubin, C.; Forster, R.; McMaster, M.L.; Hockemeyer, D. Cancer-associated POT1 Mutations Lead to Telomere Elongation without Induction of a DNA Damage Response. EMBO J. 2021, 40, e107346. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Poulos, R.C.; Reddel, R.R. Role of POT1 in Human Cancer. Cancers 2020, 12, 2739. [Google Scholar] [CrossRef] [PubMed]
- Chang, S. Cancer Chromosomes Going to POT1. Nat. Genet. 2013, 45, 473–475. [Google Scholar] [CrossRef] [Green Version]
- Astuti, Y.; Wardhana, A.; Watkins, J.; Wulaningsih, W. PILAR Research Network Cigarette Smoking and Telomere Length: A Systematic Review of 84 Studies and Meta-Analysis. Environ. Res. 2017, 158, 480–489. [Google Scholar] [CrossRef]
- Dixit, S.; Whooley, M.A.; Vittinghoff, E.; Roberts, J.D.; Heckbert, S.R.; Fitzpatrick, A.L.; Lin, J.; Leung, C.; Mukamal, K.J.; Marcus, G.M. Alcohol Consumption and Leukocyte Telomere Length. Sci. Rep. 2019, 9, 1404. [Google Scholar] [CrossRef] [PubMed]
- Galiè, S.; Canudas, S.; Muralidharan, J.; García-Gavilán, J.; Bulló, M.; Salas-Salvadó, J. Impact of Nutrition on Telomere Health: Systematic Review of Observational Cohort Studies and Randomized Clinical Trials. Adv. Nutr. 2020, 11, 576–601. [Google Scholar] [CrossRef] [PubMed]
- Ridout, K.K.; Levandowski, M.; Ridout, S.J.; Gantz, L.; Goonan, K.; Palermo, D.; Price, L.H.; Tyrka, A.R. Early Life Adversity and Telomere Length: A Meta-Analysis. Mol. Psychiatry 2018, 23, 858–871. [Google Scholar] [CrossRef]
- Lin, J.; Epel, E. Stress and Telomere Shortening: Insights from Cellular Mechanisms. Ageing Res. Rev. 2022, 73, 101507. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lister-Shimauchi, E.H.; McCarthy, B.; Lippincott, M.; Ahmed, S. Genetic and Epigenetic Inheritance at Telomeres. Epigenomes 2022, 6, 9. https://doi.org/10.3390/epigenomes6010009
Lister-Shimauchi EH, McCarthy B, Lippincott M, Ahmed S. Genetic and Epigenetic Inheritance at Telomeres. Epigenomes. 2022; 6(1):9. https://doi.org/10.3390/epigenomes6010009
Chicago/Turabian StyleLister-Shimauchi, Evan H., Benjamin McCarthy, Michael Lippincott, and Shawn Ahmed. 2022. "Genetic and Epigenetic Inheritance at Telomeres" Epigenomes 6, no. 1: 9. https://doi.org/10.3390/epigenomes6010009
APA StyleLister-Shimauchi, E. H., McCarthy, B., Lippincott, M., & Ahmed, S. (2022). Genetic and Epigenetic Inheritance at Telomeres. Epigenomes, 6(1), 9. https://doi.org/10.3390/epigenomes6010009





