RNAi-Mediated Knockdown of Chitin Synthase 1 (CHS1) Gene Causes Mortality and Decreased Longevity and Fecundity in Aphis gossypii
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Culture
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Preparation of Double-Stranded RNA (dsRNA)
2.4. Dietary Delivery of the Double-Stranded RNA (dsRNA)
2.5. Quantitative Real-Time PCR (RT-qPCR)
2.6. Longevity and Fecundity Analysis
2.7. Data Analysis
3. Results
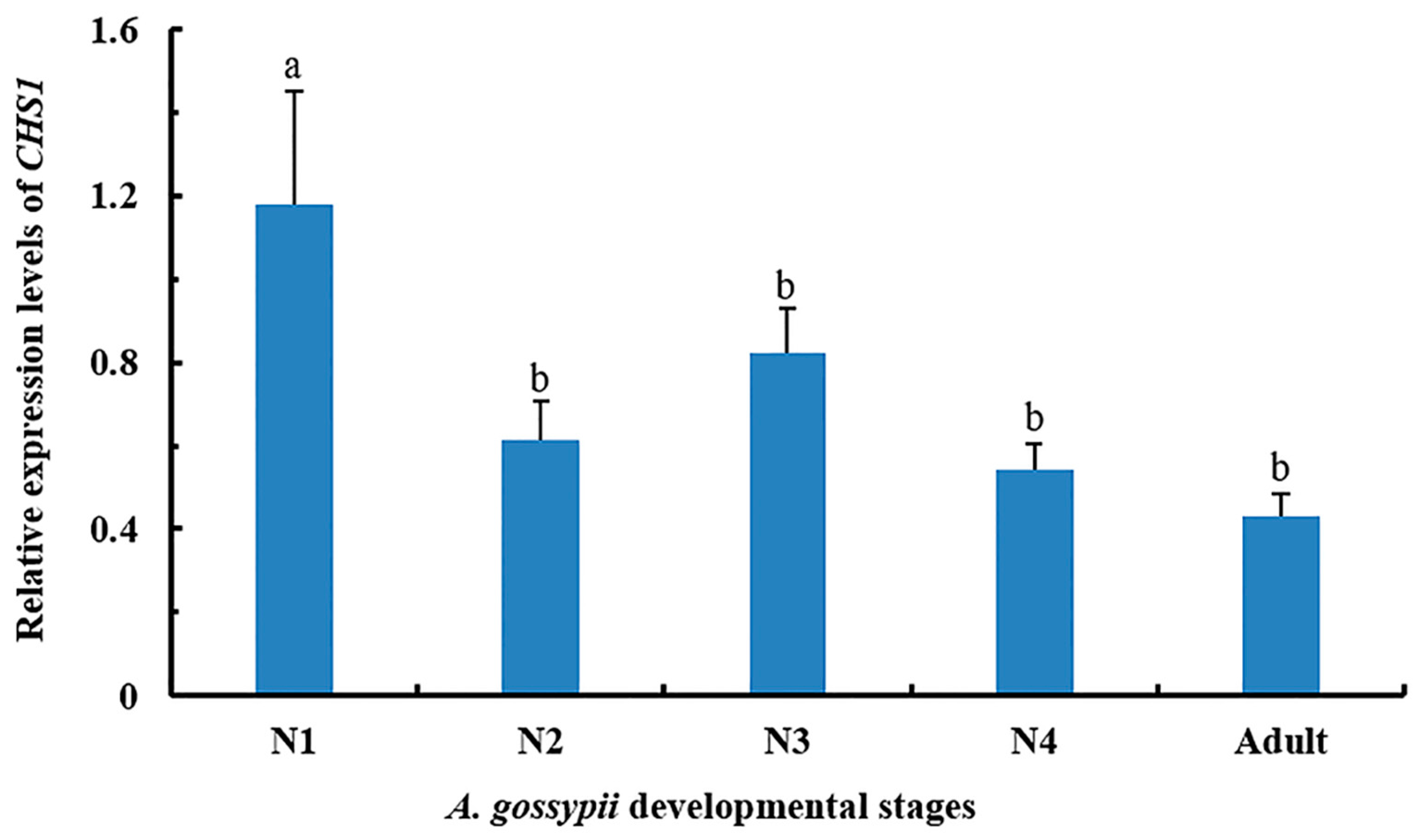
3.1. Expression of CHS1 Gene among Growth Stages of A. gossypii
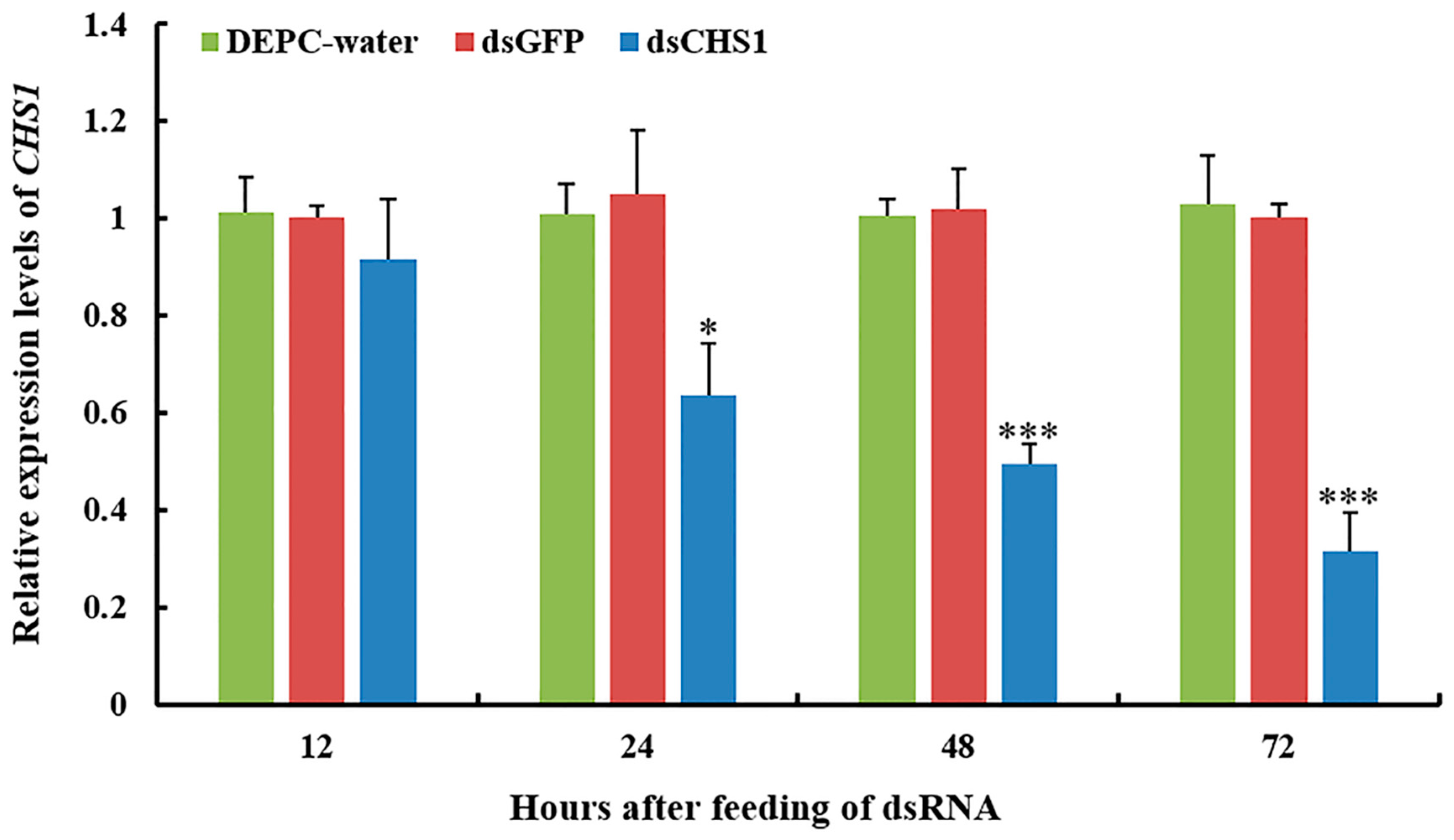
3.2. Functional Analysis of CHS1 by RNAi
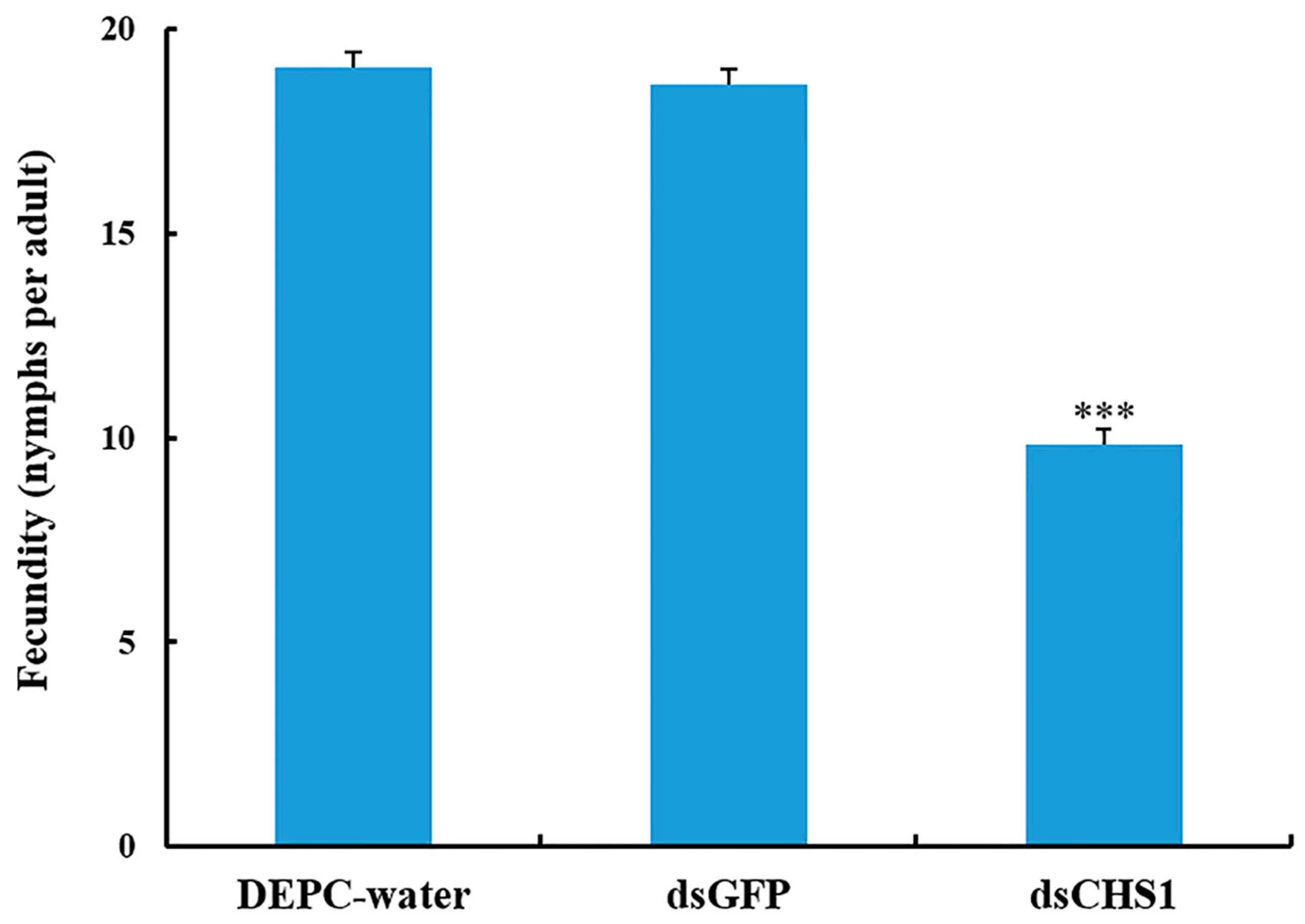
3.3. Effects of dsRNA-CHS1 on Aphid Mortality, Longevity, and Fecundity
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hullé, M.; Chaubet, B.; Turpeau, E.; Simon, J. Encyclop’Aphid: A website on aphids and their natural enemies. Entomol. Gen. 2019. [Google Scholar] [CrossRef]
- Van Emden, H.F.; Harrington, R. Aphids as Crop Pests; CABI: Wallingford, UK, 2017. [Google Scholar]
- Andrews, M.; Callaghan, A.; Field, L.; Williamson, M.; Moores, G. Identification of mutations conferring insecticide-insensitive AChE in the cotton-melon aphid, Aphis gossypii Glover. Insect Mol. Biol. 2004, 13, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.; Croft, P. Strategies for the control of Aphis gossypii Glover (Hom.: Aphididae) with Aphidius colemani Viereck (Hym.: Braconidae) in protected cucumbers. Biocontrol. Sci. Technol. 1998, 8, 377–387. [Google Scholar] [CrossRef]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops: An Identification and Information Guide; John Wiley & Sons Ltd.: New York, NY, USA, 2000. [Google Scholar]
- Ullah, F.; Gul, H.; Desneux, N.; Tariq, K.; Ali, A.; Gao, X.; Song, D. Clothianidin-induced sublethal effects and expression changes of vitellogenin and ecdysone receptors genes in the melon aphid, Aphis gossypii. Entomol. Gen. 2019, 39, 137–149. [Google Scholar] [CrossRef]
- Ullah, F.; Gul, H.; Yousaf, H.K.; Xiu, W.; Qian, D.; Gao, X.; Tariq, K.; Han, P.; Desneux, N.; Song, D. Impact of low lethal concentrations of buprofezin on biological traits and expression profile of chitin synthase 1 gene (CHS1) in melon aphid, Aphis gossypii. Sci. Rep. 2019, 9, 12291. [Google Scholar] [CrossRef]
- Ullah, F.; Gul, H.; Desneux, N.; Gao, X.; Song, D. Imidacloprid-induced hormetic effects on demographic traits of the melon aphid, Aphis gossypii. Entomol. Gen. 2019, 39, 325–337. [Google Scholar] [CrossRef]
- Ullah, F.; Gul, H.; Desneux, N.; Qu, Y.; Xiao, X.; Khattak, A.M.; Gao, X.; Song, D. Acetamiprid-induced hormetic effects and vitellogenin gene (Vg) expression in the melon aphid, Aphis gossypii. Entomol. Gen. 2019, 39, 259–270. [Google Scholar] [CrossRef]
- Gul, H.; Ullah, F.; Biondi, A.; Desneux, N.; Qian, D.; Gao, X.; Song, D. Resistance against clothianidin and associated fitness costs in the chive maggot, Bradysia odoriphaga. Entomol. Gen. 2019, 39, 81–92. [Google Scholar] [CrossRef]
- Desneux, N.; Decourtye, A.; Delpuech, J.M. The sublethal effects of pesticides on beneficial arthropods. Annu. Rev. Entomol. 2007, 52, 81–106. [Google Scholar] [CrossRef]
- O’Brien, P.; Graves, J. Insecticide resistance and reproductive biology of Aphis gossypii Glover. Southwest. Entomol. (USA) 1992, 17, 115–122. [Google Scholar]
- Koo, H.-N.; An, J.-J.; Park, S.-E.; Kim, J.-I.; Kim, G.-H. Regional susceptibilities to 12 insecticides of melon and cotton aphid, Aphis gossypii (Hemiptera: Aphididae) and a point mutation associated with imidacloprid resistance. Crop Prot. 2014, 55, 91–97. [Google Scholar] [CrossRef]
- Cui, L.; Qi, H.; Yang, D.; Yuan, H.; Rui, C. Cycloxaprid: A novel cis-nitromethylene neonicotinoid insecticide to control imidacloprid-resistant cotton aphid (Aphis gossypii). Pestic. Biochem. Physiol. 2016, 132, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009, 457, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Whangbo, J.S.; Hunter, C.P. Environmental RNA interference. Trends Genet. 2008, 24, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, G.; Wang-Pruski, G.; You, M. Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur. J. Entomol. 2008, 105. [Google Scholar] [CrossRef]
- Walshe, D.; Lehane, S.; Lehane, M.; Haines, L. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009, 18, 11–19. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; Singh, P.K.; Tuli, R. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef]
- Burand, J.P.; Hunter, W.B. RNAi: Future in insect management. J. Invertebr. Pathol. 2013, 112, S68–S74. [Google Scholar] [CrossRef]
- Gu, L.; Knipple, D.C. Recent advances in RNA interference research in insects: Implications for future insect pest management strategies. Crop Prot. 2013, 45, 36–40. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Kola, V.S.R.; Renuka, P.; Madhav, M.S.; Mangrauthia, S.K. Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front. Physiol. 2015, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Tariq, K.; Ali, A.; Davies, T.E.; Naz, E.; Naz, L.; Sohail, S.; Hou, M.; Ullah, F. RNA interference-mediated knockdown of voltage-gated sodium channel (MpNa v) gene causes mortality in peach-potato aphid, Myzus persicae. Sci. Rep. 2019, 9, 5291. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.M.; Diab, M.R.; Abdelsattar, M.; Sayed, M. Characterization and RNAi-mediated knockdown of Chitin Synthase A in the potato tuber moth, Phthorimaea operculella. Sci. Rep. 2017, 7, 9502. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.; Davy, M.; MacDiarmid, R.; Plummer, K.; Birch, N.; Newcomb, R. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 2006, 15, 383–391. [Google Scholar] [CrossRef]
- Jaubert-Possamai, S.; Le Trionnaire, G.; Bonhomme, J.; Christophides, G.K.; Rispe, C.; Tagu, D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007, 7, 63. [Google Scholar] [CrossRef]
- Nunes, F.M.F.; Simões, Z.L.P. A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem. Mol. Biol. 2009, 39, 157–160. [Google Scholar] [CrossRef]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef]
- Kramer, K.J.; Hopkins, T.L.; Schaefer, J. Applications of solids NMR to the analysis of insect sclerotized structures. Insect Biochem. Mol. Biol. 1995, 25, 1067–1080. [Google Scholar] [CrossRef]
- Kumar, N.S.; Tang, B.; Chen, X.; Tian, H.; Zhang, W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2008, 149, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Tellam, R.L.; Eisemann, C. Chitin is only a minor component of the peritrophic matrix from larvae of Lucilia cuprina. Insect Biochem. Mol. Biol. 2000, 30, 1189–1201. [Google Scholar] [CrossRef]
- Mansur, J.F.; Alvarenga, E.S.; Figueira-Mansur, J.; Franco, T.A.; Ramos, I.B.; Masuda, H.; Melo, A.C.; Moreira, M.F. Effects of chitin synthase double-stranded RNA on molting and oogenesis in the Chagas disease vector Rhodnius prolixus. Insect Biochem. Mol. Biol. 2014, 51, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Souza-Ferreira, P.S.; Mansur, J.F.; Berni, M.; Moreira, M.F.; dos Santos, R.E.; Araújo, H.M.M.; de Souza, W.; Ramos, I.B.; Masuda, H. Chitin deposition on the embryonic cuticle of Rhodnius prolixus: The reduction of CHS transcripts by CHS–dsRNA injection in females affects chitin deposition and eclosion of the first instar nymph. Insect Biochem. Mol. Biol. 2014, 51, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zimoch, L.; Merzendorfer, H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002, 308, 287–297. [Google Scholar] [CrossRef]
- Zhao, Y.; Sui, X.; Xu, L.; Liu, G.; Lu, L.; You, M.; Xie, C.; Li, B.; Ni, Z.; Liang, R. Plant-mediated RNAi of grain aphid CHS1 gene confers common wheat resistance against aphids. Pest Manag. Sci. 2018, 747, 2754–2760. [Google Scholar] [CrossRef]
- Shang, F.; Xiong, Y.; Xia, W.K.; Wei, D.D.; Wei, D.; Wang, J.J. Identification, characterization and functional analysis of a chitin synthase gene in the brown citrus aphid, Toxoptera citricida (Hemiptera, Aphididae). Insect Mol. Biol. 2016, 25, 422–430. [Google Scholar] [CrossRef]
- Bansal, R.; Mian, M.R.; Mittapalli, O.; Michel, A.P. Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid, Aphis. glycines. Int. J. Biol. Sci. 2012, 8, 1323. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, H.-W.; Huang, H.-J.; Xue, J.; Wu, W.-J.; Bao, Y.-Y.; Xu, H.-J.; Zhu, Z.-R.; Cheng, J.-A.; Zhang, C.-X. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 2012, 42, 637–646. [Google Scholar] [CrossRef]
- Ma, K.; Tang, Q.; Zhang, B.; Liang, P.; Wang, B.; Gao, X. Overexpression of multiple cytochrome P450 genes associated with sulfoxaflor resistance in Aphis gossypii Glover. Pestic. Biochem. Physiol. 2019, 157, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Li, F.; Tang, Q.; Liang, P.; Liu, Y.; Zhang, B.; Gao, X. CYP4CJ1-mediated gossypol and tannic acid tolerance in Aphis gossypii Glover. Chemosphere 2019, 219, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.-S.; Li, F.; Liang, P.-Z.; Chen, X.-W.; Liu, Y.; Gao, X.-W. Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Kumar, N.S.; Tang, B.; Sun, X.; Qiu, X.; Hu, J.; Zhang, W. The class A chitin synthase gene of Spodoptera exigua: Molecular cloning and expression patterns. Insect Biochem. Mol. Biol. 2007, 37, 409–417. [Google Scholar] [CrossRef]
- Cohen, E. Chitin synthesis and inhibition: A revisit. Pest Manag. Sci. 2001, 57, 946–950. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Zhang, J.; Li, D.; Sun, Y.; Guo, Y.; Ma, E.; Zhu, K.Y. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem. Mol. Biol. 2010, 40, 824–833. [Google Scholar] [CrossRef]
- Tao, X.Y.; Xue, X.Y.; Huang, Y.P.; Chen, X.Y.; Mao, Y.B. Gossypol-enhanced P450 gene pool contributes to cotton bollworm tolerance to a pyrethroid insecticide. Mol. Ecol. 2012, 21, 4371–4385. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Gong, C.; Yao, X.; Jiang, C.; Yang, Q. Molecular identification of four novel cytochrome P450 genes related to the development of resistance of Spodoptera exigua (Lepidoptera: Noctuidae) to chlorantraniliprole. Pest Manag. Sci. 2018, 74, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-L.; Staehelin, C.; Xia, Q.-Q.; Su, Y.-J.; Zeng, R.-S. Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 2015, 16, 22606–22620. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-L.; He, Y.-N.; Staehelin, C.; Liu, S.-W.; Su, Y.-J.; Zhang, J.-E. Identification of Two Cytochrome Monooxygenase P450 Genes, CYP321A7 and CYP321A9, from the Tobacco Cutworm Moth (Spodoptera Litura) and Their Expression in Response to Plant Allelochemicals. Int. J. Mol. Sci. 2017, 18, 2278. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, J.; Shen, G.; Xu, Z.; Xu, Q.; He, L. Collaborative contribution of six cytochrome P450 monooxygenase genes to fenpropathrin resistance in Tetranychus cinnabarinus (Boisduval). Insect Mol. Biol. 2016, 25, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, M.; Liu, S.; Jan, S.; Shi, L.; Fernández-Grandon, G.M.; Gulzar, A.; Ali, B.; Rehman, M.; Wang, M. Knock-down of gossypol-inducing cytochrome P450 genes reduced deltamethrin sensitivity in Spodoptera exigua (Hübner). Int. J. Mol. Sci. 2019, 20, 2248. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-L.; Liu, S.-W.; Baerson, S.; Qin, Z.; Ma, Z.-H.; Su, Y.-J.; Zhang, J.-E. Identification and functional analysis of a novel cytochrome P450 gene CYP9A105 associated with pyrethroid detoxification in Spodoptera exigua hübner. Int. J. Mol. Sci. 2018, 19, 737. [Google Scholar] [CrossRef]
- Liu, S.; Hafeez, M.; Zhang, X.; Dawar, F.U.; Guo, J.; Gao, C.; Wang, M. Isolation and functional identification of three cuticle protein genes during metamorphosis of the beet armyworm, Spodoptera exigua. Sci. Rep. 2017, 7, 16061. [Google Scholar]
- Mao, Y.-B.; Cai, W.-J.; Wang, J.-W.; Hong, G.-J.; Tao, X.-Y.; Wang, L.-J.; Huang, Y.-P.; Chen, X.-Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307. [Google Scholar] [CrossRef]
- Kim, Y.H.; Issa, M.S.; Cooper, A.M.; Zhu, K.Y. RNA interference: Applications and advances in insect toxicology and insect pest management. Pestic. Biochem. Physiol. 2015, 120, 109–117. [Google Scholar] [CrossRef]
- Nunes, F.; Aleixo, A.; Barchuk, A.; Bomtorin, A.; Grozinger, C.; Simões, Z. Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 2013, 4, 90–103. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequences |
|---|---|
| dsCHS1-F | TAATACGACTCACTATAGGGACCATTTTAGGACCGGGAAC |
| dsCHS1-R | TAATACGACTCACTATAGGGCTTTTCTGACTCCTCAGCGG |
| dsGFP-F | TAATACGACTCACTATAGGGTGACCACCCTGACCTAC |
| dsGFP-R | TAATACGACTCACTATAGGGTTGATGCCGTTCTTCTGC |
| CHS1-F | ATTGCGTCACGATGATCCTT |
| CHS1-R | TGGTCGCTAGACGTTCACAC |
| EF1α-F | GAAGCCTGGTATGGTTGTCGT |
| EF1α-R | GGGTGGGTTGTTCTTTGTG |
| β-Actin-F | GGGAGTCATGGTTGGTATGG |
| β-Actin-R | TCCATATCGTCCCAGTTGGT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, F.; Gul, H.; Wang, X.; Ding, Q.; Said, F.; Gao, X.; Desneux, N.; Song, D. RNAi-Mediated Knockdown of Chitin Synthase 1 (CHS1) Gene Causes Mortality and Decreased Longevity and Fecundity in Aphis gossypii. Insects 2020, 11, 22. https://doi.org/10.3390/insects11010022
Ullah F, Gul H, Wang X, Ding Q, Said F, Gao X, Desneux N, Song D. RNAi-Mediated Knockdown of Chitin Synthase 1 (CHS1) Gene Causes Mortality and Decreased Longevity and Fecundity in Aphis gossypii. Insects. 2020; 11(1):22. https://doi.org/10.3390/insects11010022
Chicago/Turabian StyleUllah, Farman, Hina Gul, Xiu Wang, Qian Ding, Fazal Said, Xiwu Gao, Nicolas Desneux, and Dunlun Song. 2020. "RNAi-Mediated Knockdown of Chitin Synthase 1 (CHS1) Gene Causes Mortality and Decreased Longevity and Fecundity in Aphis gossypii" Insects 11, no. 1: 22. https://doi.org/10.3390/insects11010022
APA StyleUllah, F., Gul, H., Wang, X., Ding, Q., Said, F., Gao, X., Desneux, N., & Song, D. (2020). RNAi-Mediated Knockdown of Chitin Synthase 1 (CHS1) Gene Causes Mortality and Decreased Longevity and Fecundity in Aphis gossypii. Insects, 11(1), 22. https://doi.org/10.3390/insects11010022






