Feeding Delivery of dsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Sample Preparation for Analysis of Hvsnf7 Expression
2.3. RNA Extraction and cDNA Synthesis
2.4. dsRNA Preparation
2.5. Effects of Oral Ingestion of dsRNAs on Neonate Development and HvSnf Gene Expression
HvSnf Gene Expression Analysis
2.6. Effects of Bacterially Expressed dsHvSnf7 on H. vigintioctopunctata Mortality
2.6.1. dsRNA Synthesis in Bacteria
2.6.2. Effects on H. vigintioctopunctata Mortality
2.7. Analysis of Larval Midgut Ultrastructure
2.8. Data Analysis
3. Results
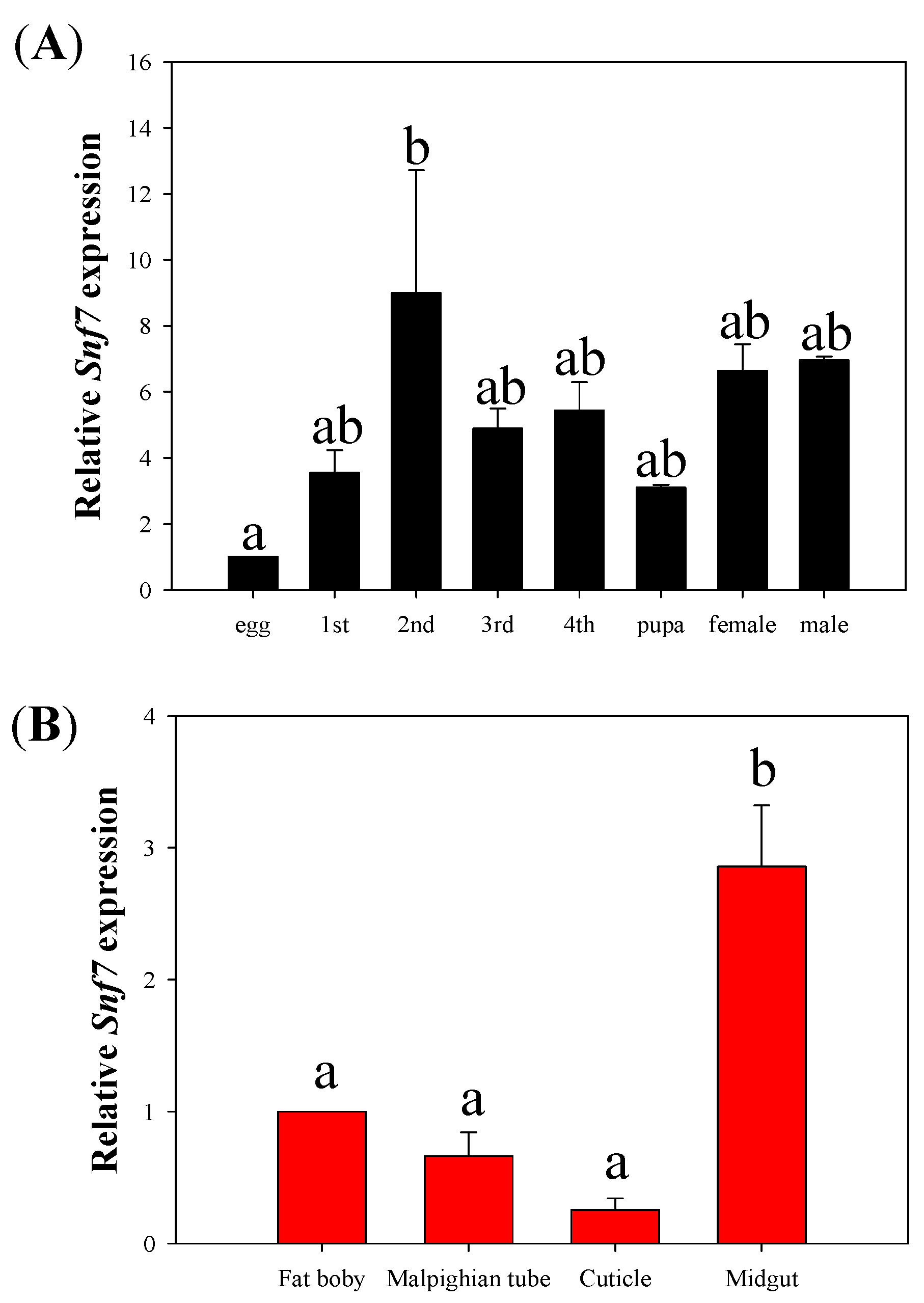
3.1. Temporal and Tissue Expression of HvSnf7
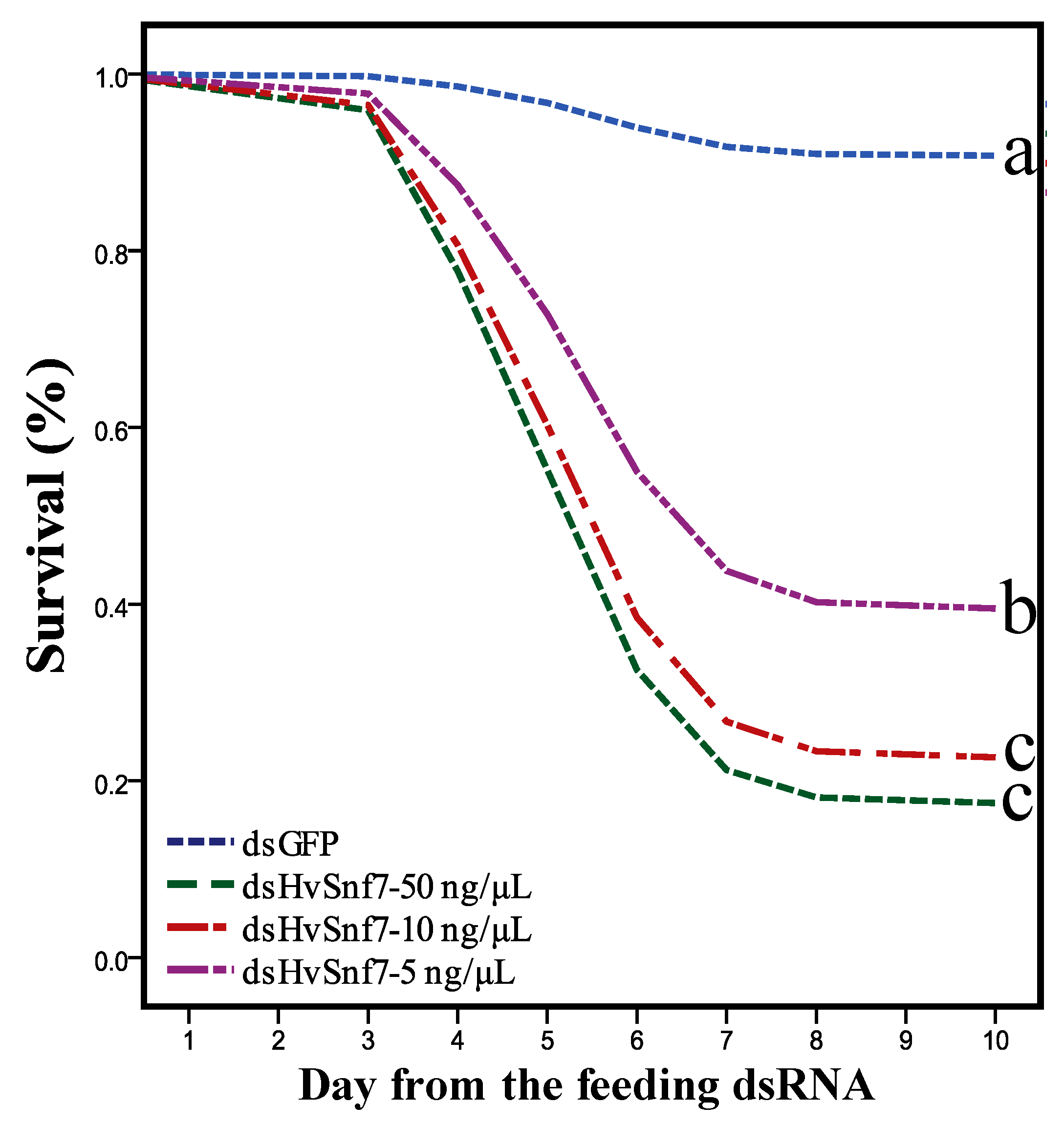
3.2. Effects of Consumption of In Vitro Synthesized dsHvSnf7
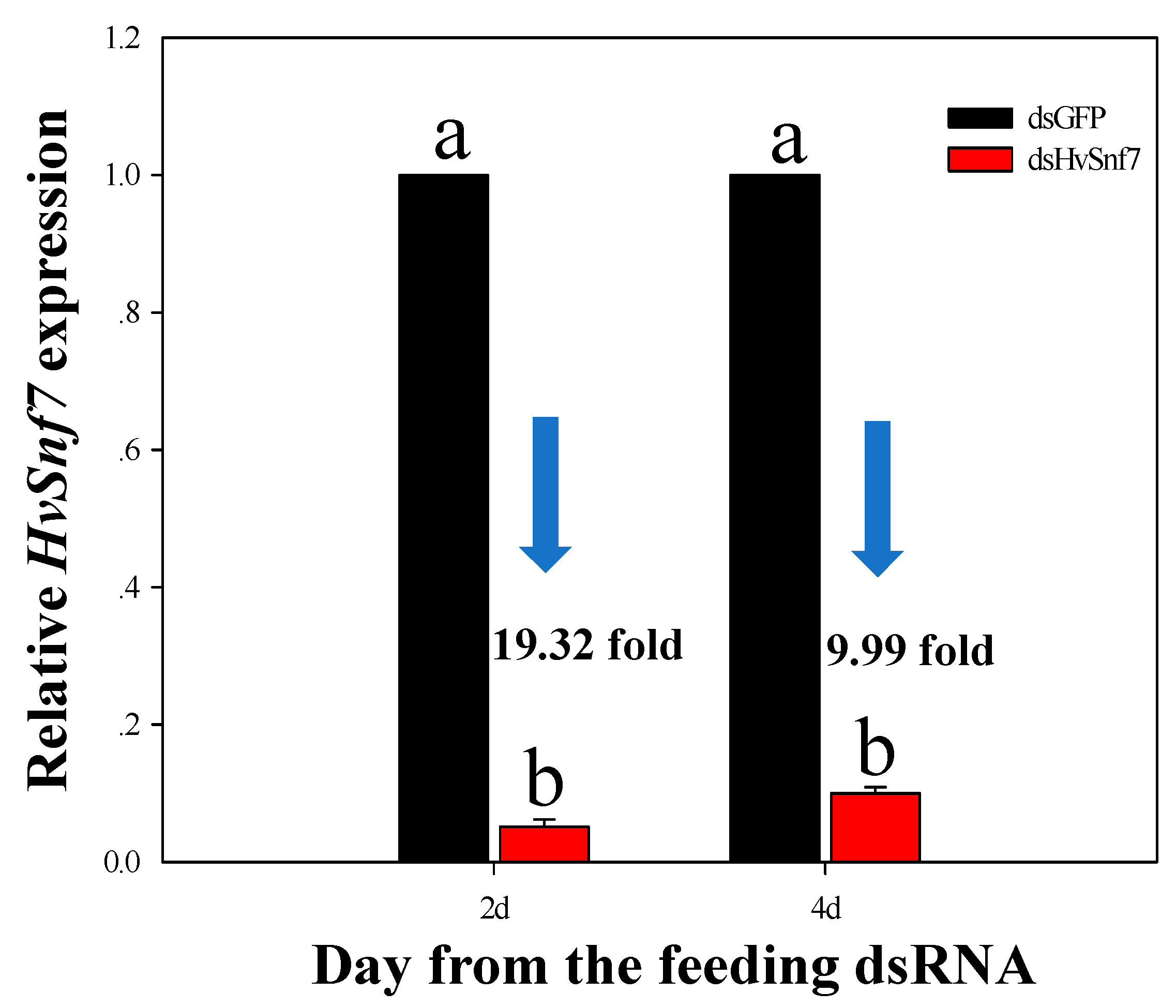
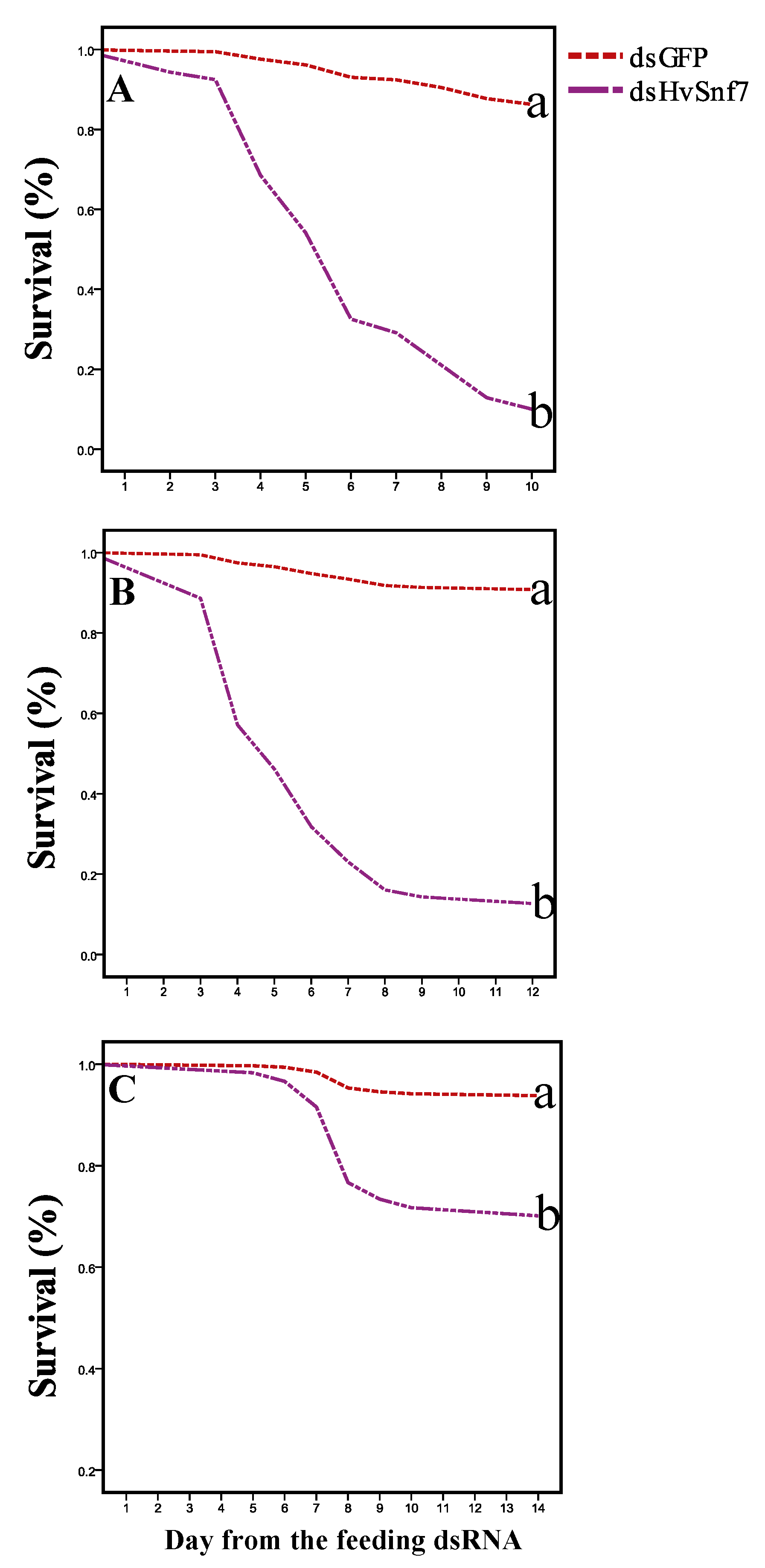
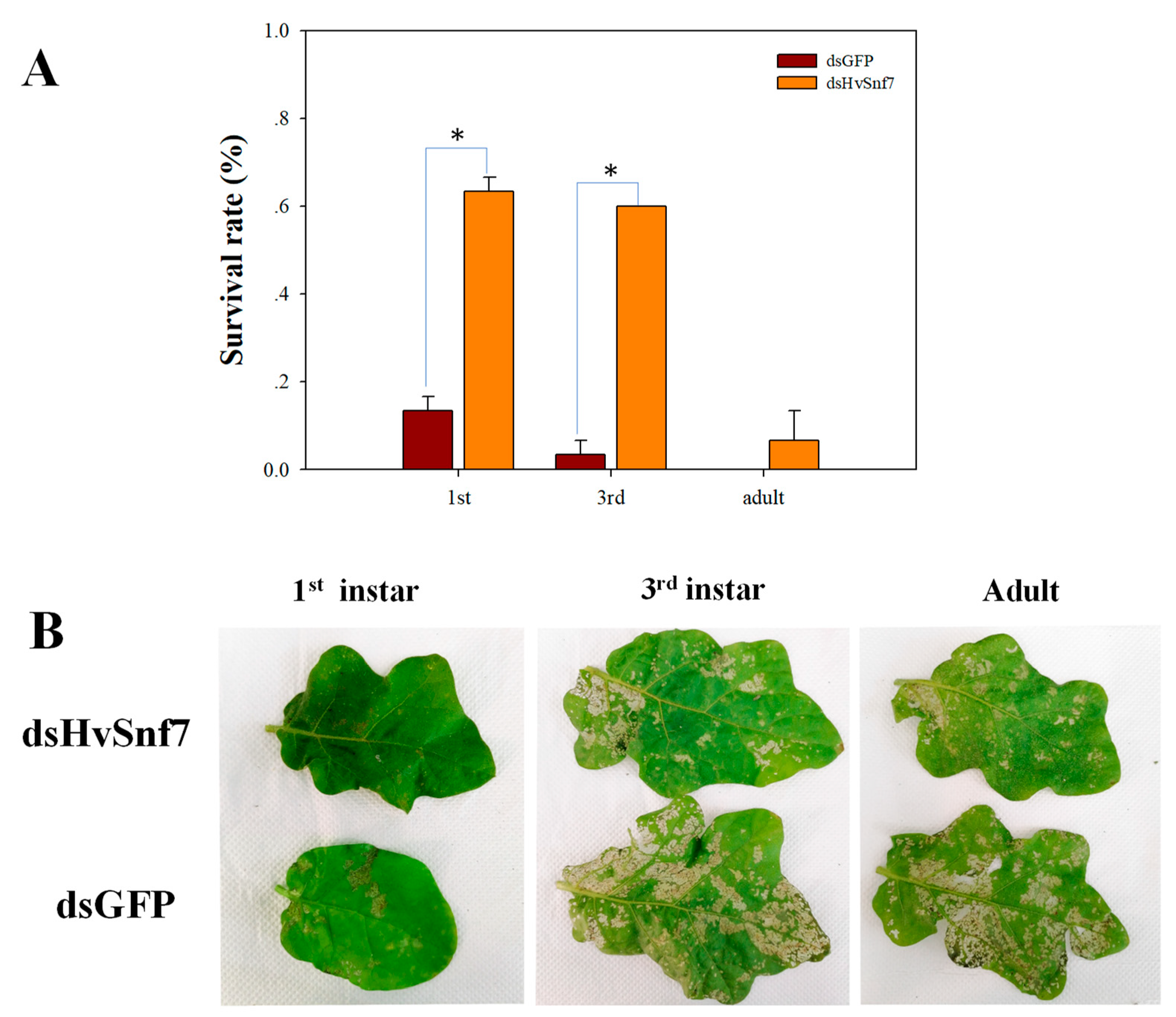
3.3. Impacts of Ingestion of Bacterially Expressed dsHvSnf7 on H. vigintioctopunctata Survival
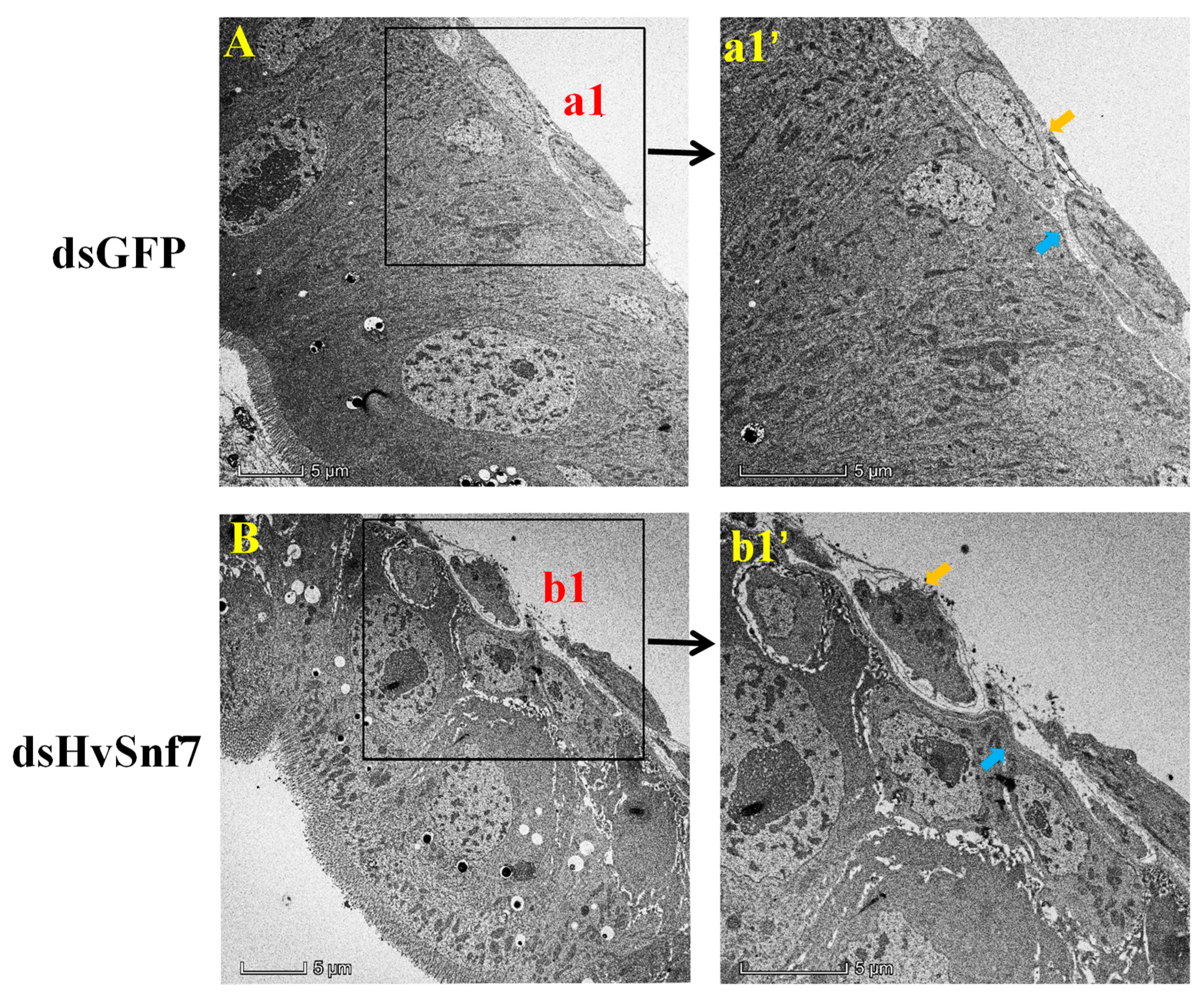
3.4. HvSnf7 Silencing Effects on Larval Midgut Ultrastructure
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Burand, J.P.; Hunter, W.B. RNAi: Future in insect management. J. Invertebr. Pathol. 2013, 112, S68–S74. [Google Scholar] [CrossRef]
- Katoch, R.; Sethi, A.; Thakur, N.; Murdock, L.L. RNAi for insect control: Current perspective and future challenges. Appl. Biochem. Biotech. 2013, 171, 847–873. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef]
- Cagliari, D.; Dias, N.P.; Galdeano, D. Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 2019, 10, 1319. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 34, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Osman, G.H.; Assem, S.K.; Alreedy, R.M.; El-Ghareeb, D.K.; Basry, M.A.; Rastogi, A.; Kalaji, H.M. Erratum: Development of insect resistant maize plants expressing a chitinase gene from the cotton leaf worm, Spodoptera littoralis. Sci. Rep. 2016, 6, 20449. [Google Scholar] [CrossRef]
- Guan, R.; Li, H.; Miao, X. RNAi pest control and enhanced BT insecticidal efficiency achieved by dsRNA of chymotrypsin-like genes in Ostrinia furnacalis. J. Pest Sci. 2017, 90, 745–757. [Google Scholar] [CrossRef]
- Ni, M.; Ma, W.; Wang, X.F.; Gao, M.J.; Dai, Y.; Wei, X.L.; Zhang, L.; Peng, Y.G.; Chen, S.Y.; Ding, L.Y. Next-generation transgenic cotton: Pyramiding RNAi and Bt counters insect resistance. Plant Biotechnol. J. 2017, 15, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Z.; Yang, B.; Zhang, A.H.; Ding, D.R.; Wang, G.R. Plant-mediated RNAi for controlling Apolygus lucorum. Front. Plant Sci. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Knipple, D.C. Recent advances in RNA interference research in insects: Implications for future insect pest management strategies. Crop Prot. 2013, 45, 36–40. [Google Scholar] [CrossRef]
- Yoon, J.S.; Mogilicherla, K.; Gurusamy, D.; Chen, X.; Cheredy, S.C.R.R.; Palli, S.R. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc. Natl. Acad. Sci. USA 2018, 115, 8334–8339. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, C.R.; Yuan, J.J.; Li, S.X.; Wang, X.P.; Chi, H. Demographic Comparison of Henosepilachna vigintioctopunctata (F.) (Coleoptera: Coccinellidae) reared on three cultivars of Solanum melongena L. and a wild host plant Solanum nigrum L. J. Econ. Entomol. 2017, 110, 2084–2091. [Google Scholar] [CrossRef]
- Zhou, L.; Xie, B.G.; Wang, X.P. Population dynamic of Henosepilachna vigintioctopunctatain different host plants in Jianghan plain. China J. North. Hort. 2015, 11, 103–105. [Google Scholar]
- Tu, X.Y.; Wang, G.H. Research progress of bio-control of Henosepilachna vigintioctopunctata. Chin. Plant Prot. 2010, 30, 13–16. [Google Scholar]
- Michelet, X.; Djeddi, A.; Legouis, R. Developmental and cellular functions of the ESCRT machinery in pluricellular organisms. Biol. Cell 2010, 102, 191–202. [Google Scholar] [CrossRef]
- Li, Z.; Blissard, G. The vacuolar protein sorting genes in insects: A comparative genome view. Insect Biochem. Mol. Biol. 2015, 62, 211–225. [Google Scholar] [CrossRef]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J.H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.G.; Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science 2007, 316, 1908–1912. [Google Scholar] [CrossRef] [PubMed]
- Dukes, J.D.; Richardson, J.D.; Simmons, R.; Whitley, P. A dominant-negative ESCRT-III protein perturbs cytokinesis and trafficking to lysosomes. Biochem. J. 2008, 411, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Elia, N.; Sougrat, R.; Spurlin, T.A.; Hurley, J.H.; Lippincott-Schwartz, J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA 2011, 108, 4846–4851. [Google Scholar] [CrossRef] [PubMed]
- Guizetti, J.; Schermelleh, L.; Mantler, J.; Maar, S.; Poser, I.; Leonhardt, H.; Muller-Reichert, T.; Gerlich, D.W. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 2011, 331, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Ramaseshadri, P.; Segers, G.; Flannagan, R.; Wiggins, E.; Clinton, W.; Ilagan, O.; McNulty, B.; Clark, T.; Bolognesi, R. Physiological and cellular responses caused by RNAi-mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS ONE 2013, 8, e54270. [Google Scholar] [CrossRef]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef]
- Pereira, A.E.; Vélez, A.M.; Meinke, L.J.; Siegfried, B.D. Sublethal effects of vATPase-A and Snf7 dsRNAs on biology of Southern corn rootworm, Diabrotica undecimpunctata howardi Barber. J. Econ. Entomol. 2017, 110, 2545–2553. [Google Scholar] [CrossRef]
- Head, G.P.; Carroll, M.W.; Evans, S.P.; Rule, D.M.; Willse, A.R.; Clark, T.L.; Storer, N.P.; Flannagan, R.D.; Samuel, L.W.; Meinke, L.J. Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: Efficacy and resistance management. Pest Manag. Sci. 2017, 73, 1883–1899. [Google Scholar] [CrossRef]
- Bachman, P.M.; Bolognesi, R.; Moar, W.J.; Mueller, G.M.; Paradise, M.S.; Ramaseshadri, P.; Tan, J.; Uffman, J.P.; Warren, J.; Elizabeth Wiggins, B.; et al. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res. 2013, 22, 1207–1222. [Google Scholar] [CrossRef]
- Lü, J.; Chen, S.M.; Guo, M.J.; Ye, C.Y.; Qiu, B.L.; Wu, J.H.; Yang, C.X.; Pan, H.P. Corrigendum: Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctopunctata. Front. Physiol. 2019, 10, 981. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Yang, X.; Bidne, K.; Hellmich, R.L.; Siegfried, B.D.; Zhou, X. Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio-indicator. PLoS ONE 2015, 10, e0129482. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Koči, J.; Ramaseshadri, P.; Bolognesi, R.; Segers, G.; Flannagan, R.; Park, Y. Ultrastructural changes caused by Snf7 RNAi in larval enterocytes of western corn rootworm (Diabrotica virgifera virgifera Le Conte). PLoS ONE 2014, 9, e83985. [Google Scholar] [CrossRef]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Roberts, A.F.; Devos, Y.; Lemgo, G.N.; Zhou, X. Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front. Plant Sci. 2015, 6, 958. [Google Scholar] [CrossRef]
| Gene | Primer Sequences (5′–3′) |
|---|---|
| HvSnf7-RT-qPCR-F | CAGAGAGGAACACTAGAGGAA |
| HvSnf7-RT-qPCR-R | GGTCAACGTTCATGTGTTTATG |
| dsHvSnf7-F | TAATACGACTCACTATAGGGACCCTTACAACCCTTGAATTAC |
| dsHvSnf7-R | TAATACGACTCACTATAGGGCTTCATCATCTTCCACTGCTT |
| dsGFP-F | TAATACGACTCACTATAGGGCTTGAAGTTGACCTTGATGCC |
| dsGFP-R | TAATACGACTCACTATAGGGTGGTCCCAATTCTCGTGGAAC |
| L4440-HvSnf7-F | ATCATCGATGAATTCACCCTTACAACCCTTGAATTAC |
| L4440-HvSnf7-R | TTCCTGCAGCCCGGGCTTCATCATCTTCCACTGCTT |
| L4440-GFP-F | ATCATCGATGAATTCCTTGAAGTTGACCTTGATGCC |
| L4440-GFP-R | TTCCTGCAGCCCGGGTGGTCCCAATTCTCGTGGAAC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lü, J.; Liu, Z.; Guo, W.; Guo, M.; Chen, S.; Li, H.; Yang, C.; Zhang, Y.; Pan, H. Feeding Delivery of dsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insects 2020, 11, 34. https://doi.org/10.3390/insects11010034
Lü J, Liu Z, Guo W, Guo M, Chen S, Li H, Yang C, Zhang Y, Pan H. Feeding Delivery of dsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insects. 2020; 11(1):34. https://doi.org/10.3390/insects11010034
Chicago/Turabian StyleLü, Jing, Zhuoqi Liu, Wei Guo, Mujuan Guo, Shimin Chen, Huali Li, Chunxiao Yang, Youjun Zhang, and Huipeng Pan. 2020. "Feeding Delivery of dsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae)" Insects 11, no. 1: 34. https://doi.org/10.3390/insects11010034
APA StyleLü, J., Liu, Z., Guo, W., Guo, M., Chen, S., Li, H., Yang, C., Zhang, Y., & Pan, H. (2020). Feeding Delivery of dsHvSnf7 Is a Promising Method for Management of the Pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insects, 11(1), 34. https://doi.org/10.3390/insects11010034






