The Mutation Spectrum of Rare Variants in the Gene of Adenosine Triphosphate (ATP)-Binding Cassette Subfamily C Member 8 in Patients with a MODY Phenotype in Western Siberia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Sequencing of MODY-Associated Genes and Bioinformatic Analysis
2.3. ABCC8 Confirmation Analysis
3. Results
3.1. Variants in Genes GCK, HNF1A, HNF4A, and HNF1B
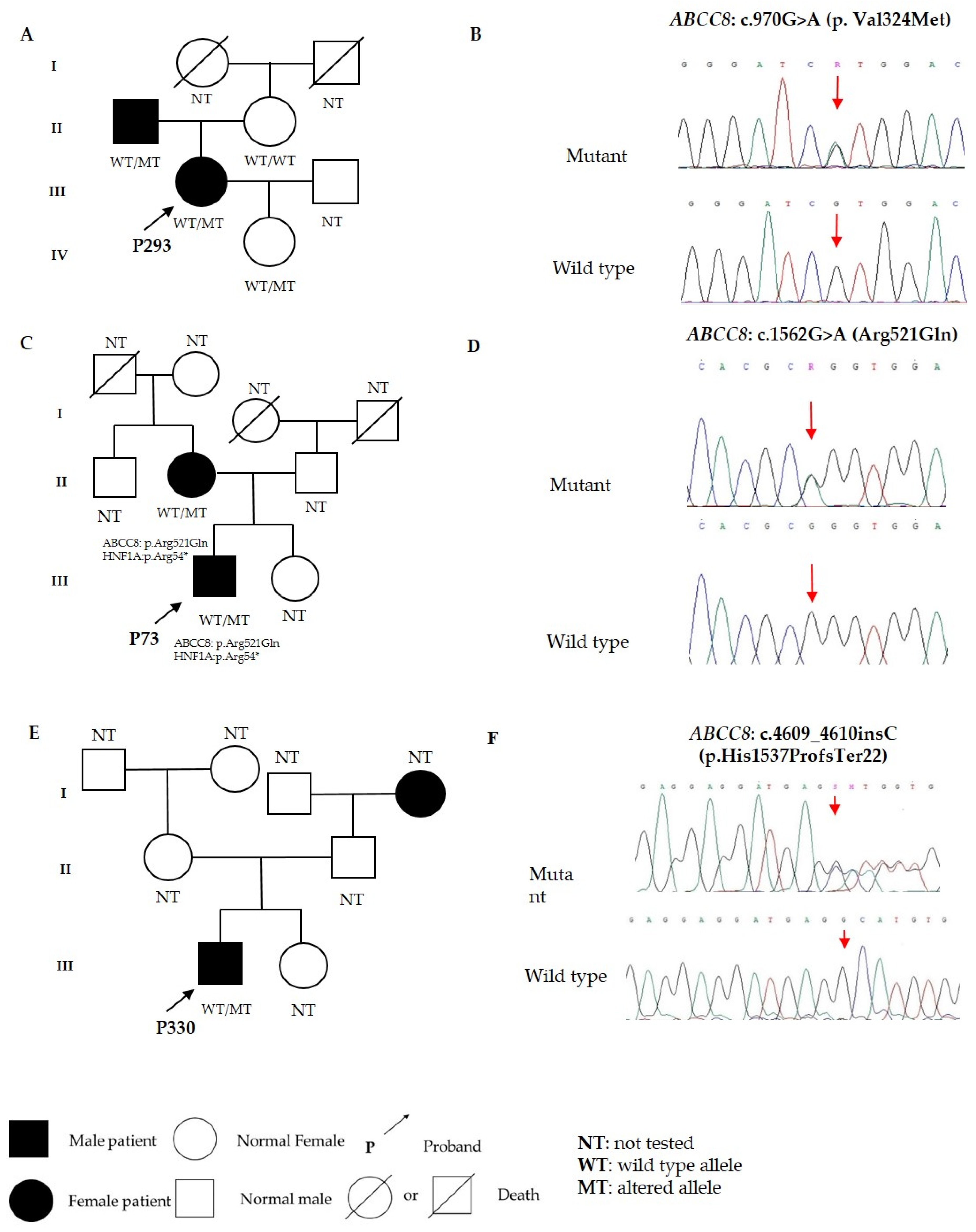
3.2. Variants in ABCC8
3.3. The Phenotype of Patients with MODY12
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Firdous, P.; Nissar, K.; Ali, S.; Ganai, B.A.; Shabir, U.; Hassan, T.; Masoodi, S.R. Genetic Testing of Maturity-Onset Diabetes of the Young Current Status and Future Perspectives. Front. Endocrinol. 2018, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Lachance, C.H. Practical Aspects of Monogenic Diabetes: A Clinical Point of View. Can. J. Diabetes 2016, 40, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Mohan, V.; Radha, V. Precision Diabetes Is Slowly Becoming a Reality. Med. Princ. Pract. 2019, 28, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shields, B.M.; Hicks, S.; Shepherd, M.H.; Colclough, K.; Hattersley, A.T.; Ellard, S. Maturity-onset diabetes of the young (MODY): How many cases are we missing? Diabetologia 2010, 53, 2504–2508. [Google Scholar] [CrossRef] [PubMed]
- Ellard, S.; Bellanné-Chantelot, C.; Hattersley, A.T. European Molecular Genetics Quality Network (EMQN) MODY group. Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia 2008, 51, 546–553. [Google Scholar] [CrossRef]
- Ellard, S.; Flanagan, S.E.; Girard, C.A.; Patch, A.M.; Harries, L.W.; Parrish, A.; Edghill, E.L.; Mackay, D.J.; Proks, P.; Shimomura, K.; et al. Permanent neonatal diabetes caused by dominant, recessive, or compound heterozygous SUR1 mutations with opposite functional effects. Am. J. Hum. Genet. 2007, 81, 375–382. [Google Scholar] [CrossRef]
- Bowman, P.; Flanagan, S.E.; Edghill, E.L.; Damhuis, A.; Shepherd, M.H.; Paisey, R.; Hattersley, A.T.; Ellard, S. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia 2012, 55, 123–127. [Google Scholar] [CrossRef]
- Nessa, A.; Rahman, S.A.; Hussain, K. Hyperinsulinemic Hypoglycemia—The Molecular Mechanisms. Front. Endocrinol. 2016, 7, 29. [Google Scholar] [CrossRef]
- Shyng, S.; Nichols, C.G. Octameric stoichiometry of the KATP channel complex. J. Gen. Physiol. 1997, 110, 655–664. [Google Scholar] [CrossRef]
- Ashcroft, F.M.; Rorsman, P. Electrophysiology of the pancreatic beta-cell. Prog. Biophys. Mol. Biol. 1989, 54, 87–143. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.J.; Gribble, F.M.; Zhao, C.; Trapp, S.; Ashcroft, F.M. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature 1997, 387, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Jha, R.M.; Koleck, T.A.; Puccio, A.M.; Okonkwo, D.O.; Park, S.Y.; Zusman, B.E.; Clark, R.S.B.; Shutter, L.A.; Wallisch, J.S.; Empey, P.E.; et al. Regionally clustered ABCC8 polymorphisms in a prospective cohort predict cerebral oedema and outcome in severe traumatic brain injury. J. Neurol. Neurosurg. Psychiatry 2018, 89, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Galcheva, S.; Demirbilek, H.; Al-Khawaga, S.; Hussain, K. The Genetic and Molecular Mechanisms of Congenital Hyperinsulinism. Front. Endocrinol. 2019, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- Ashcroft, F.M.; Puljung, M.C.; Vedovato, N. Neonatal Diabetes and the KATP Channel: From Mutation to Therapy. Trends Endocrinol. Metab. 2017, 28, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Patch, A.M.; Flanagan, S.E.; Boustred, C.; Hattersley, A.T.; Ellard, S. Mutations in the ABCC8 gene encoding the SUR1 subunit of the KATP channel cause transient neonatal diabetes, permanent neonatal diabetes or permanent diabetes diagnosed outside the neonatal period. Diabetes Obes. Metab. 2007, 9, 28–39. [Google Scholar] [CrossRef]
- Bonnefond, A.; Boissel, M.; Bolze, A.; Durand, E.; Toussaint, B.; Vaillant, E.; Gaget, S.; Graeve, F.; Dechaume, A.; Allegaert, F.; et al. Pathogenic variants in actionable MODY genes are associated with type 2 diabetes. Nat. Metab. 2020, 2, 1126–1134. [Google Scholar] [CrossRef]
- Koufakis, T.; Sertedaki, A.; Tatsi, E.-B.; Trakatelli, C.-M.; Karras, S.N.; Manthou, E.; Kanaka-Gantenbein, C.; Kotsa, K. First Report of Diabetes Phenotype due to a Loss-of-Function ABCC8 Mutation Previously Known to Cause Congenital Hyperinsulinism. Case Rep. Genet. 2019, 2019, 3654618. [Google Scholar] [CrossRef]
- Männikkö, R.; Flanagan, S.E.; Sim, X.; Segal, D.; Hussain, K.; Ellard, S.; Hattersley, A.T.; Ashcroft, F.M. Mutations of the same conserved glutamate residue in NBD2 of the sulfonylurea receptor 1 subunit of the KATP channel can result in either hyperinsulinism or neonatal diabetes. Diabetes 2011, 60, 1813–1822. [Google Scholar] [CrossRef]
- Işık, E.; Demirbilek, H.; Houghton, J.A.; Ellard, S.; Flanagan, S.E.; Hussain, K. Congenital Hyperinsulinism and Evolution to Sulfonylurearesponsive Diabetes Later in Life due to a Novel Homozygous p.L171F ABCC8 Mutation. J. Clin. Res. Pediatr. Endocrinol. 2019, 11, 82–87. [Google Scholar] [CrossRef]
- Abdulhadi-Atwan, M.; Bushman, J.; Tornovsky-Babaey, S.; Perry, A.; Abu-Libdeh, A.; Glaser, B.; Shyng, S.L.; Zangen, D.H. Novel de novo mutation in sulfonylurea receptor 1 presenting as hyperinsulinism in infancy followed by overt diabetes in early adolescence. Diabetes 2008, 57, 1935–4190. [Google Scholar] [CrossRef]
- Kapoor, R.R.; Flanagan, S.E.; James, C.T.; McKiernan, J.; Thomas, A.M.; Harmer, S.C.; Shield, J.P.; Tinker, A.; Ellard, S.; Hussain, K. Hyperinsulinaemic hypoglycaemia and diabetes mellitus due to dominant ABCC8/KCNJ11 mutations. Diabetologia 2011, 10, 2575–2583. [Google Scholar] [CrossRef]
- Qin, L.J.; Lv, Y.; Huang, Q.Y. Meta-analysis of association of common variants in the KCNJ11-ABCC8 region with type 2 diabetes. Genet. Mol. Res. 2013, 12, 2990–3002. [Google Scholar] [CrossRef] [PubMed]
- Delvecchio, M.; Pastore, C.; Giordano, P. Treatment Options for MODY Patients: A Systematic Review of Literature. Diabetes Ther. 2020, 11, 1667–1685. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2005, 28, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Stanik, J.; Dusatkova, P.; Cinek, O.; Valentinova, L.; Huckova, M.; Skopkova, M.; Dusatkova, L.; Stanikova, D.; Pura, M.; Klimes, I.; et al. De novo mutations of GCK, HNF1A and HNF4A may be more frequent in MODY than previously assumed. Diabetologia 2014, 57, 480–484. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Purification of nucleic acids by extraction with phenol: Chloroform. Cold Spring Harb. Protoc. 2006, 2006, 4455. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Ivanoshchuk, D.E.; Shakhtshneider, E.V.; Rymar, O.D.; Ovsyannikova, A.K.; Mikhailova, S.V.; Fishman, V.S.; Valeev, E.S.; Orlov, P.S.; Voevoda, M.I. The Mutation Spectrum of Maturity Onset Diabetes of the Young (MODY)-Associated Genes among Western Siberia Patients. J. Pers. Med. 2021, 11, 57. [Google Scholar] [CrossRef]
- In Supplimentary Wang, Z.; Diao, C.; Liu, Y.; Li, M.; Zheng, J.; Zhang, Q.; Yu, M.; Zhang, H.; Ping, F.; Li, M.; et al. Identification and functional analysis of GCK gene mutations in 12 Chinese families with hyperglycemia. J. Diabetes Investig. 2019, 10, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Ivanoshchuk, D.E.; Ovsyannikova, A.K.; Mikhailova, S.V.; Shakhtshneider, E.V.; Valeev, E.S.; Rymar, O.D.; Orlov, P.S.; Voevoda, M.I. Variants of the HNF4A and HNF1A genes in patients with impaired glucose metabolism and dyslipidemia. Ateroscleroz 2022, 17, 11–19. [Google Scholar] [CrossRef]
- Madariaga, L.; García-Castaño, A.; Ariceta, G.; Martínez-Salazar, R.; Aguayo, A.; Castaño, L.; Spanish Group for the Study of HNF1B Mutations. Variable phenotype in HNF1B mutations: Extrarenal manifestations distinguish affected individuals from the population with congenital anomalies of the kidney and urinary tract. Clin. Kidney J. 2018, 12, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Urrutia, I.; Martínez, R.; Rica, I.; Martínez de LaPiscina, I.; García-Castaño, A.; Aguayo, A.; Calvo, B.; Castaño, L. Spanish Pediatric Diabetes Collaborative Group. Negative autoimmunity in a Spanish pediatric cohort suspected of type 1 diabetes, could it be monogenic diabetes? PLoS ONE 2019, 14, e02206342019. [Google Scholar] [CrossRef]
- Hohendorff, J.; Kwiatkowska, M.; Pisarczyk-Wiza, D.; Ludwig-Słomczyńska, A.; Milcarek, M.; Kapusta, P.; Zapała, B.; Kieć-Wilk, B.; Trznadel-Morawska, I.; Szopa, M.; et al. Mutation search within monogenic diabetes genes in Polish patients with long-term type 1 diabetes and preserved kidney function. Pol. Arch. Intern. Med. 2022, 132, 16143. [Google Scholar] [CrossRef]
- Reilly, F.; Sanchez-Lechuga, B.; Clinton, S.; Crowe, G.; Burke, M.; Ng, N.; Colclough, K.; Byrne, M.M. Phenotype, genotype and glycaemic variability in people with activating mutations in the ABCC8 gene: Response to appropriate therapy. Diabet. Med. 2020, 37, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, A.K.; Rymar, O.D.; Shakhtshneider, E.V.; Klimontov, V.V.; Koroleva, E.A.; Myakina, N.E.; Voevoda, M.I. ABCC8-Related Maturity-Onset Diabetes of the Young (MODY12): Clinical Features and Treatment Perspective. Diabetes Ther. 2016, 7, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lai, Y.; Guan, T.; Zhan, J.; Pei, J.; Wu, D.; Ying, S.; Shen, Y. Associations of ATP-Sensitive Potassium Channel’s Gene Polymorphisms With Type 2 Diabetes and Related Cardiovascular Phenotypes. Front. Cardiovasc. Med. 2022, 23, 816847. [Google Scholar] [CrossRef]
- Globa, E.; Zelinska, N.; Mackay, D.J.; Temple, K.I.; Houghton, J.A.; Hattersley, A.T.; Flanagan, S.E.; Ellard, S. Neonatal diabetes in Ukraine: Incidence, genetics, clinical phenotype and treatment. J. Pediatr. Endocrinol. Metab. 2015, 28, 1279–1286. [Google Scholar] [CrossRef]
- Russo, L.; Iafusco, D.; Brescianini, S.; Nocerino, V.; Bizzarri, C.; Toni, S.; Cerutti, F.; Monciotti, C.; Pesavento, R.; Iughetti, L.; et al. Permanent diabetes during the first year of life: Multiple gene screening in 54 patients. Diabetologia 2011, 54, 1693–1701. [Google Scholar] [CrossRef]
- Vaxillaire, M.; Dechaume, A.; Busiah, K.; Cavé, H.; Pereira, S.; Scharfmann, R.; de Nanclares, G.P.; Castano, L.; Froguel, P.; Polak, M.; et al. New ABCC8 mutations in relapsing neonatal diabetes and clinical features. Diabetes 2007, 56, 1737–1741. [Google Scholar] [CrossRef]
- Rego, S.; Dagan-Rosenfeld, O.; Zhou, W.; Sailani, M.R.; Limcaoco, P.; Colbert, E.; Avina, M.; Wheeler, J.; Craig, C.; Salins, D.; et al. High-frequency actionable pathogenic exome variants in an average-risk cohort. Cold Spring Harb. Mol. Case Stud. 2018, 4, a003178. [Google Scholar] [CrossRef] [PubMed]
- De Franco, E.; Saint-Martin, C.; Brusgaard, K.; Knight Johnson, A.E.; Aguilar-Bryan, L.; Bowman, P.; Arnoux, J.B.; Larsen, A.R.; Sanyoura, M.; Greeley, S.A.W.; et al. Update of variants identified in the pancreatic β-cell KATP channel genes KCNJ11 and ABCC8 in individuals with congenital hyperinsulinism and diabetes. Hum. Mutat. 2020, 41, 884–905. [Google Scholar] [CrossRef] [PubMed]
- Odgerel, Z.; Lee, H.S.; Erdenebileg, N.; Gandbold, S.; Luvsanjamba, M.; Sambuughin, N.; Sonomtseren, S.; Sharavdorj, P.; Jodov, E.; Altaisaikhan, K.; et al. Genetic variants in potassium channels are associated with type 2 diabetes in a Mongolian population. J. Diabetes 2012, 4, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Engwa, G.A.; Nwalo, F.N.; Chikezie, C.C.; Onyia, C.O.; Ojo, O.O.; Mbacham, W.F.; Ubi, B.E. Possible association between ABCC8 C49620T polymorphism and type 2 diabetes in a Nigerian population. BMC Med. Genet. 2018, 19, 78. [Google Scholar] [CrossRef]
- Matharoo, K.; Arora, P.; Bhanwer, A.J. Association of adiponectin (AdipoQ) and sulphonylurea receptor (ABCC8) gene polymorphisms with Type 2 Diabetes in North Indian population of Punjab. Gene 2013, 527, 228–234. [Google Scholar] [CrossRef]
- Reis, A.F.; Ye, W.Z.; Dubois-Laforgue, D.; Bellanné-Chantelot, C.; Timsit, J.; Velho, G. Association of a variant in exon 31 of the sulfonylurea receptor 1 (SUR1) gene with type 2 diabetes mellitus in French Caucasians. Hum. Genet. 2000, 107, 138–144. [Google Scholar] [CrossRef]
- Flanagan, S.E.; Patch, A.M.; Mackay, D.; Edghill, E.L.; Gloyn, A.L.; Robinson, D.; Shield, J.P.; Temple, K.; Ellard, S.; Hattersley, A.T. Mutations in ATP-sensitive K+ channel genes cause transient neonatal diabetes and permanent diabetes in childhood or adulthood. Diabetes 2007, 56, 1930–1937. [Google Scholar] [CrossRef]
- Zhou, Q.; Garin, I.; Castaño, L.; Argente, J.; Muñoz-Calvo, M.; Perez de Nanclares, G.; Shyng, S.L. Neonatal diabetes caused by mutations in sulfonylurea receptor 1: Interplay between expression and Mg-nucleotide gating defects of ATP-sensitive potassium channels. J. Clin. Endocrinol. Metab. 2010, 95, 473–478. [Google Scholar] [CrossRef]
- Edghill, E.L.; Flanagan, S.E.; Ellard, S. Permanent neonatal diabetes due to activating mutations in ABCC8 and KCNJ11. Rev. Endocr. Metab. Disord. 2010, 11, 193–198. [Google Scholar] [CrossRef]
| SNV | Forward Primer 5′-3′ | Revers Primer 5′-3′ | Product Length |
|---|---|---|---|
| c.970G>A (p.Val324Met) | GCCCAGCCGTGAATTAGCC | CCTCTGGCATTTCTGTTGACCA | 429 |
| c.1562G>A (p.Arg521Gln) | CTTTGAGTAGGCCACTTCACCT | CAGAGCCAGTTTGAGGCTCC | 501 |
| c.4609_4610insC (p.His1537Profs*22) | CCTGTCCCAAGGCCTTATATGT | GTATGGGCAGGGTCCGAATG | 502 |
| dbSNP ID | Substitution (NM_000352.6) | Nucleotide Changes (NM_000352.6) | Minor Allele Frequency (Our Study) | Minor Allele Frequency (gnomADv3.1.2) | Minor Allele Frequency (RUSeq) | Associated Phenotype [Reference *] |
|---|---|---|---|---|---|---|
| rs1048099 | p.Pro69= | c.207T>C | 0.468 | 0.459 | 0.478 | - |
| rs8192695 | p.Ala110= | c.330C>T | 0.046 | 0.065 | 0.043 | - |
| rs137873871 | p.Val118= | c.354C>T | 0.008 | 0.004 | 0.009 | - |
| rs2301703 | - | c.579 + 14C>T | 0.390 | 0.470 | 0.384 | - |
| rs1328072266 | p. Val324Met | c.970G>A | 0.002 | - | - | ND/TND [38,39,40] |
| rs368114790 | p.Arg521Gln | c.1562G>A | 0.002 | 0.000 | 0.000 | DM [41,42] |
| rs2074308 | - | c.1672-74G>A | 0.159 | 0.121 | 0.153 | T2DM [43] |
| rs4148619 | p.Val560Met | c.1678G>A | 0.002 | 0.000 | 0.000 | - |
| rs1799857 | p.His562= | c.1686C>T | 0.390 | 0.441 | 0.408 | - |
| rs1799858 | p.Lys649= | c.1947G>A | 0.131 | 0.166 | 0.142 | T2DM [43] |
| rs1799854 | - | c.2117-3C>T | 0.523 | 0.372 | 0.480 | T2DM [44] |
| rs761258571 | p.Ala758= | c.2274G>A | 0.002 | 0.000 | - | - |
| rs1801261 | p.Thr759= | c.2277C>T | 0.002 | 0.028 | - | T2DM [45] |
| rs1805036 | p.Leu829= | c.2485C>T | 0.079 | 0.140 | 0.099 | - |
| rs1799859 | p.Arg1273= | c.3819G>A | 0.295 | 0.387 | 0.273 | T2DM [46] |
| rs757110 | p.Ala1369Ser | c.4105G>T | 0.605 | 0.712 | 0.622 | T2DM [22] |
| rs72559717 | p.Ala1457Thr | c.4369G>A | 0.002 | 0.000 | - | MODY [36] |
| New | p.His1537Profs*22 | c.4609_4610insC | 0.002 | - | - | - |
| rs8192690 | p.Val1572Ile | c.4714G>A | 0.055 | 0.051 | 0.069 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanoshchuk, D.; Shakhtshneider, E.; Mikhailova, S.; Ovsyannikova, A.; Rymar, O.; Valeeva, E.; Orlov, P.; Voevoda, M. The Mutation Spectrum of Rare Variants in the Gene of Adenosine Triphosphate (ATP)-Binding Cassette Subfamily C Member 8 in Patients with a MODY Phenotype in Western Siberia. J. Pers. Med. 2023, 13, 172. https://doi.org/10.3390/jpm13020172
Ivanoshchuk D, Shakhtshneider E, Mikhailova S, Ovsyannikova A, Rymar O, Valeeva E, Orlov P, Voevoda M. The Mutation Spectrum of Rare Variants in the Gene of Adenosine Triphosphate (ATP)-Binding Cassette Subfamily C Member 8 in Patients with a MODY Phenotype in Western Siberia. Journal of Personalized Medicine. 2023; 13(2):172. https://doi.org/10.3390/jpm13020172
Chicago/Turabian StyleIvanoshchuk, Dinara, Elena Shakhtshneider, Svetlana Mikhailova, Alla Ovsyannikova, Oksana Rymar, Emil Valeeva, Pavel Orlov, and Mikhail Voevoda. 2023. "The Mutation Spectrum of Rare Variants in the Gene of Adenosine Triphosphate (ATP)-Binding Cassette Subfamily C Member 8 in Patients with a MODY Phenotype in Western Siberia" Journal of Personalized Medicine 13, no. 2: 172. https://doi.org/10.3390/jpm13020172
APA StyleIvanoshchuk, D., Shakhtshneider, E., Mikhailova, S., Ovsyannikova, A., Rymar, O., Valeeva, E., Orlov, P., & Voevoda, M. (2023). The Mutation Spectrum of Rare Variants in the Gene of Adenosine Triphosphate (ATP)-Binding Cassette Subfamily C Member 8 in Patients with a MODY Phenotype in Western Siberia. Journal of Personalized Medicine, 13(2), 172. https://doi.org/10.3390/jpm13020172








