Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy
Abstract
:1. Introduction
2. Materials and Methods
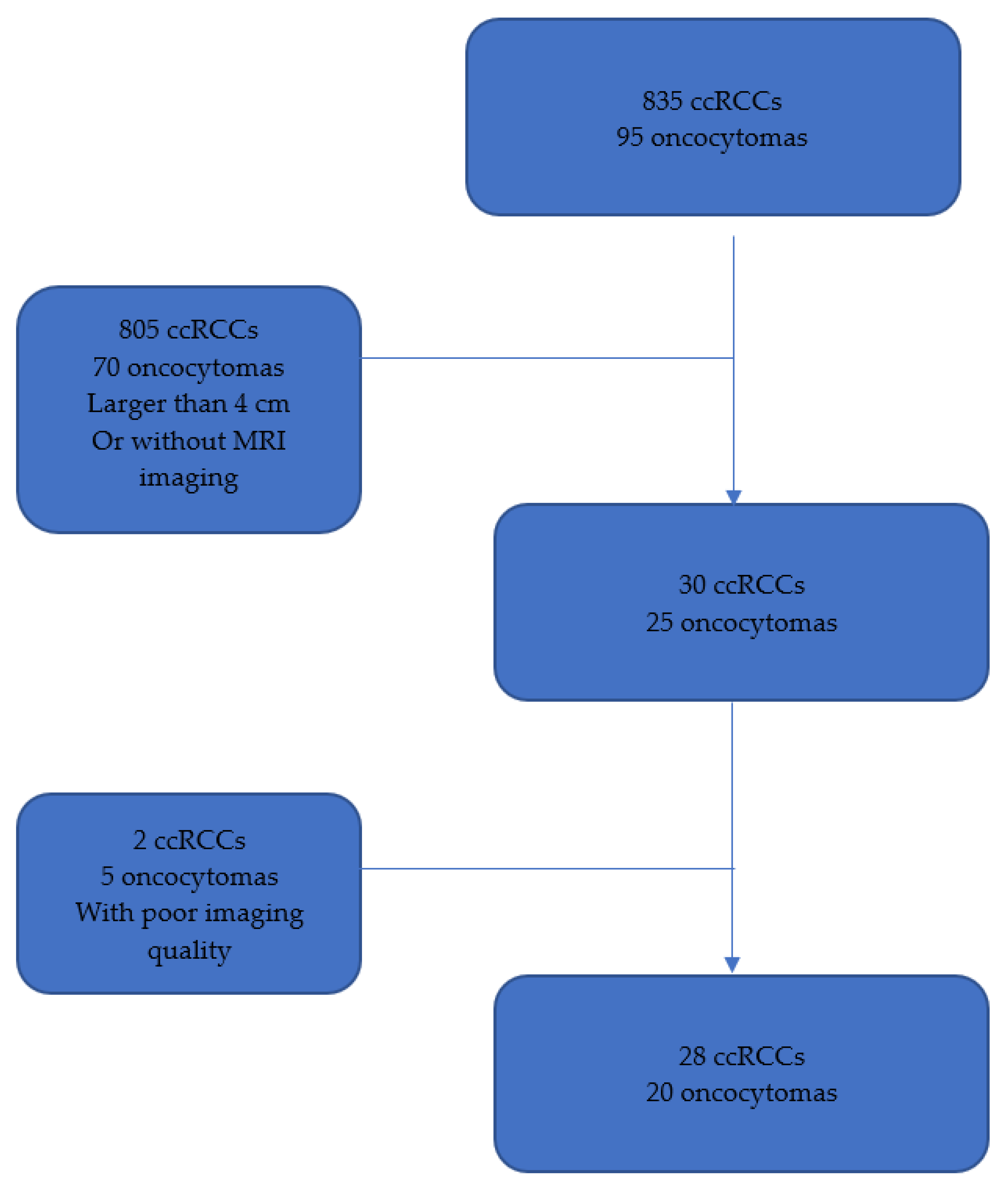
2.1. Study Design
2.2. MRI Protocol
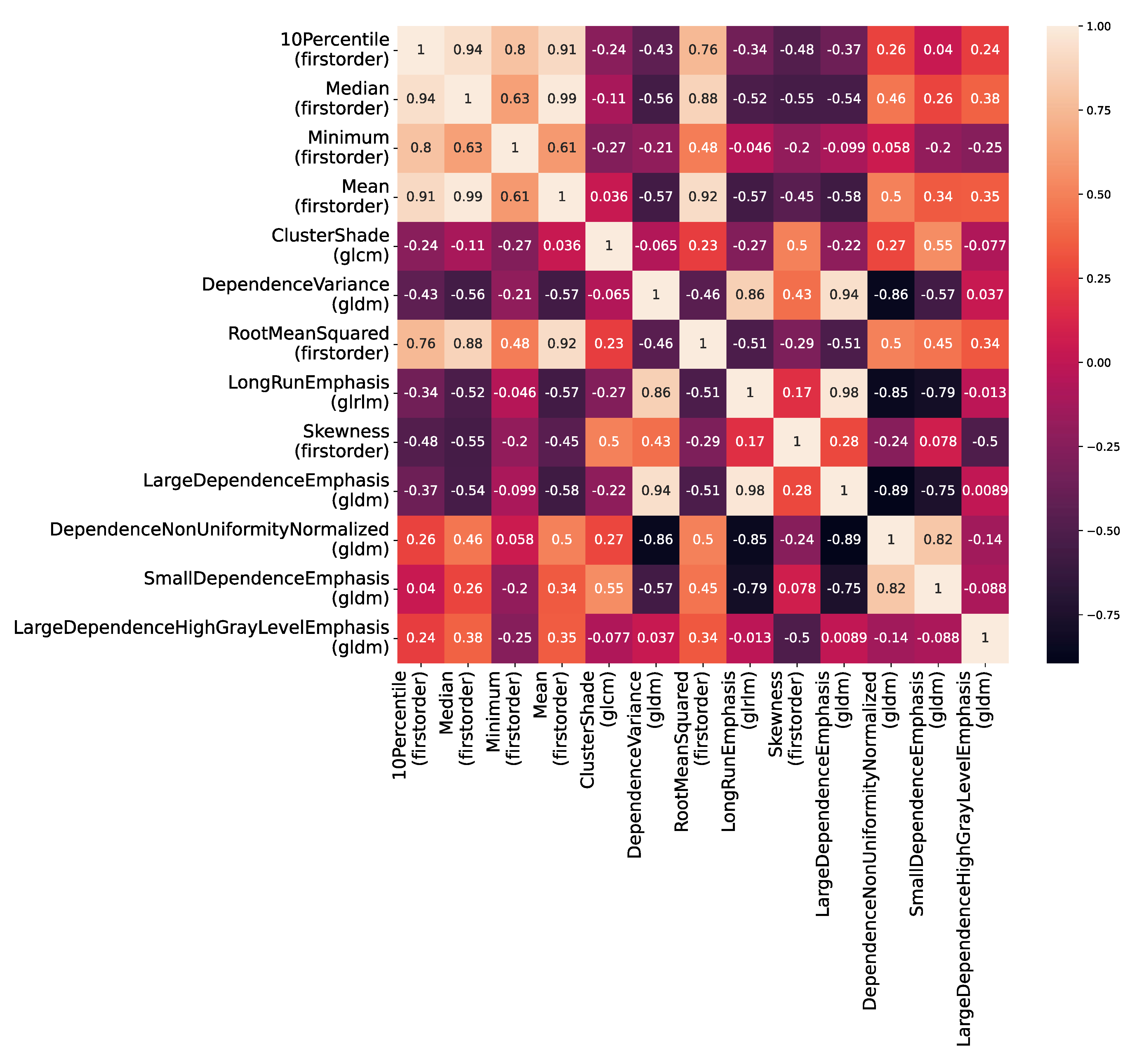
2.3. Radiomics Processing
3. Statistical Analysis
4. Results
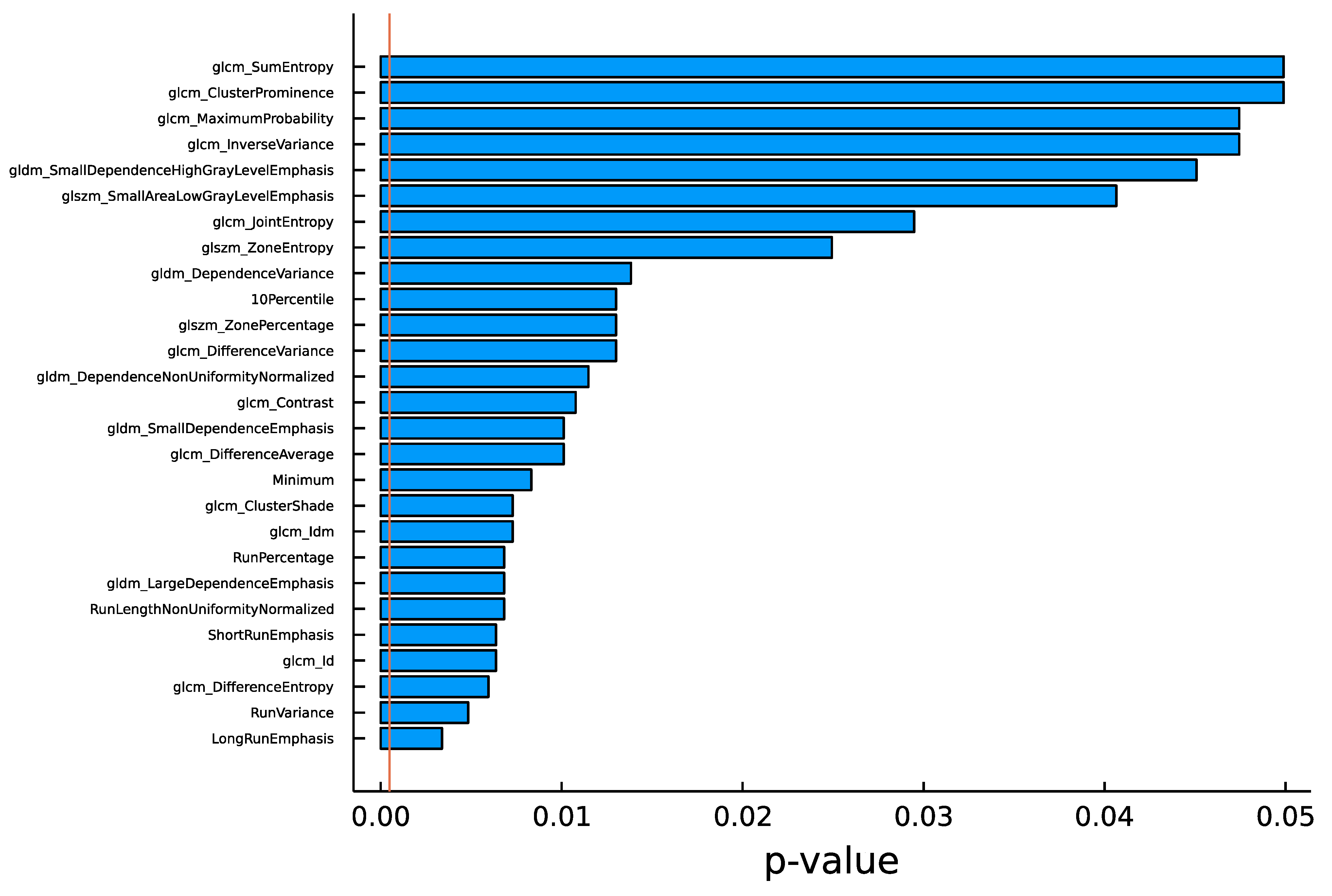
4.1. Univariate Analysis
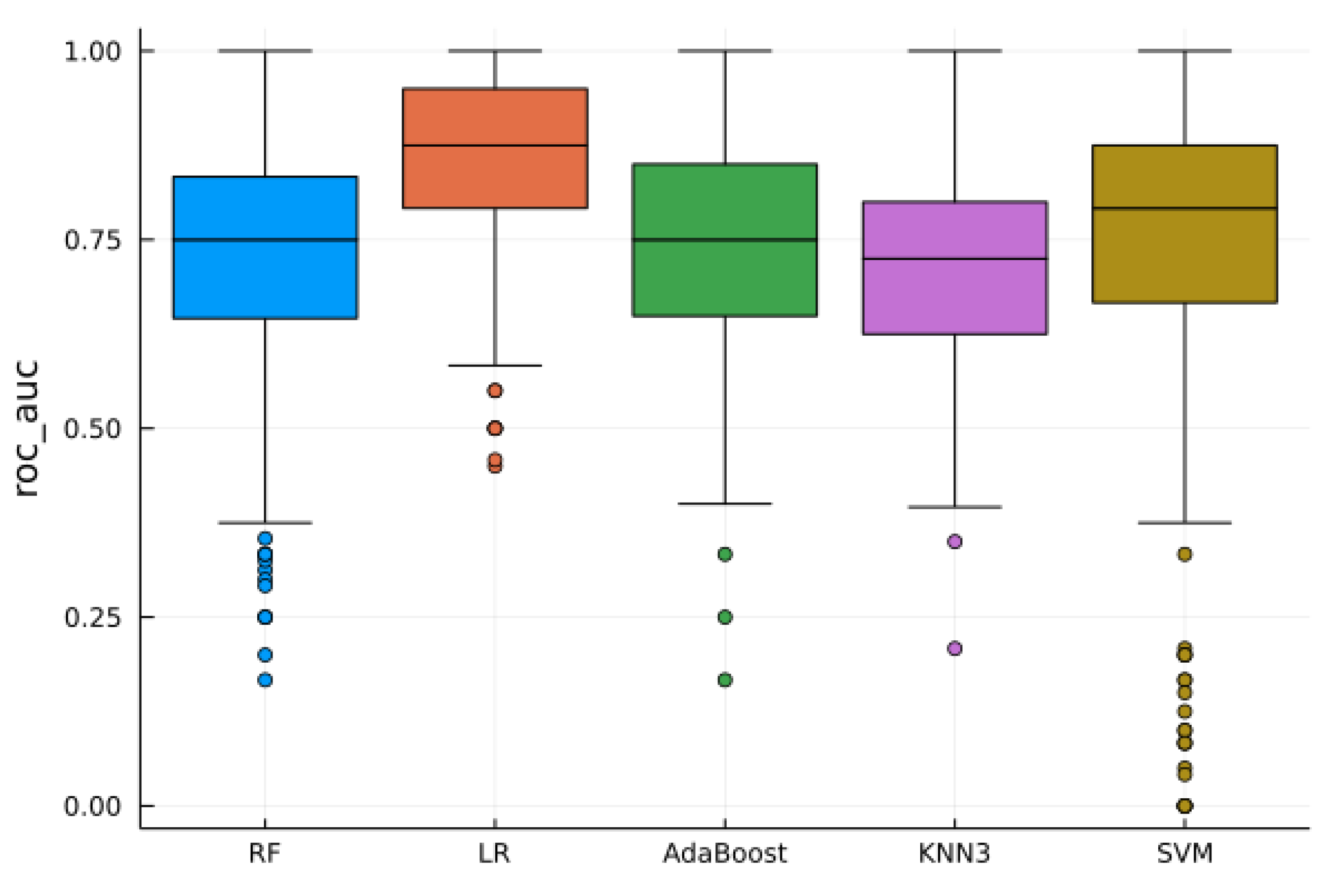
4.2. Supervised Analysis
4.3. Unsupervised Analysis
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Almassi, N.; Gill, B.C.; Rini, B.; Fareed, K. Management of the small renal mass. Transl. Androl. Urol. 2017, 6, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Gill, I.S.; Aron, M.; Gervais, D.A.; Jewett, M.A.S. Small Renal Mass. N. Engl. J. Med. 2010, 362, 624–634. [Google Scholar] [CrossRef]
- Nguyen, M.M.; Gill, I.S.; Ellison, L.M. The Evolving Presentation of Renal Carcinoma in the United States: Trends from the Surveillance, Epidemiology, and End Results Program. J. Urol. 2006, 176, 2397–2400. [Google Scholar] [CrossRef]
- Marconi, L. Systematic Review and Meta-analysis of Diagnostic Accuracy of Percutaneous Renal Tumour Biopsy. Eur. Urol. 2016, 14, 660–673. [Google Scholar] [CrossRef]
- Silverman, S.G.; Gan, Y.U.; Mortele, K.J.; Tuncali, K.; Cibas, E.S. Renal Masses in the Adult Patient: The Role of Percutaneous Biopsy. Radiology 2006, 240, 6–22. [Google Scholar] [CrossRef]
- Lebret, T.; Poulain, J.E.; Molinie, V.; Herve, J.M.; Denoux, Y.; Guth, A.; Scherrer, A.; Botto, H. Percutaneous Core Biopsy for Renal Masses: Indications, Accuracy and Results. J. Urol. 2007, 178, 1184–1188. [Google Scholar] [CrossRef]
- Rybicki, F.J.; Shu, K.M.; Cibas, E.S.; Fielding, J.R. Percutaneous Biopsy of Renal Masses: Sensitivity and Negative Predictive Value Stratified by Clinical Setting and Size of Masses. Am. J. Roentgenol. 2003, 7, 1281–1287. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.D.; Druskin, S.C.; Rowe, S.P.; Pierorazio, P.M.; Gorin, M.A.; Allaf, M.E. Surgical histopathology for suspected oncocytoma on renal mass biopsy: A systematic review and meta-analysis. BJU Int. 2017, 119, 661–666. [Google Scholar] [CrossRef]
- Cornelis, F.; Grenier, N. Multiparametric Magnetic Resonance Imaging of Solid Renal Tumors: A Practical Algorithm. Semin. Ultrasound CT MRI 2017, 38, 47–58. [Google Scholar] [CrossRef]
- Bosniak, M.A.; Megibow, A.J.; Hulnick, D.H.; Horii, S.; Raghavendra, B.N. CT diagnosis of renal angiomyolipoma: The importance of detecting small amounts of fat. Am. J. Roentgenol. 1988, 151, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Compérat, E.; Camparo, P.; Vieillefond, A. Classification anatomo pathologique des tumeurs du rein. J. Radiol. 2006, 87, 1015–1024. [Google Scholar] [CrossRef]
- Bhatt, N.R.; Davis, N.F.; Flynn, R.; McDermott, T.; Thornhill, J.A.; Manecksha, R.P. Dilemmas in diagnosis and natural history of renal oncocytoma and implications for management. Can. Urol. Assoc. J. 2015, 9, E709–E712. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Xi, I.L.; Zhao, Y.; Wang, R.; Chang, M.; Purkayastha, S.; Chang, K.; Huang, R.Y.; Silva, A.C.; Vallières, M.; Habibollahi, P. Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin. Cancer Res. 2020, 26, 1944–1952. [Google Scholar] [CrossRef]
- Hoang, U.N.; Mojdeh Mirmomen, S.; Meirelles, O.; Yao, J.; Merino, M.; Metwalli, A.; Marston Linehan, W.; Malayeri, A.A. Assessment of multiphasic contrast-enhanced MR textures in differentiating small renal mass subtypes. Abdom. Radiol. 2018, 43, 3400–3409. [Google Scholar] [CrossRef]
- Coy, H.; Hsieh, K.; Wu, W.; Nagarajan, M.B.; Young, J.R.; Douek, M.L.; Brown, M.S.; Scalzo, F.; Raman, S.S. Deep learning and radiomics: The utility of Google TensorFlowTM Inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT. Abdom. Radiol. 2019, 44, 2009–2020. [Google Scholar] [CrossRef]
- Kocak, B.; Yardimci, A.H.; Bektas, C.T.; Turkcanoglu, M.H.; Erdim, C.; Yucetas, U.; Koca, S.B.; Kilickesmez, O. Textural differences between renal cell carcinoma subtypes: Machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur. J. Radiol. 2018, 107, 149–157. [Google Scholar] [CrossRef]
- Bektas, C.T.; Kocak, B.; Yardimci, A.H.; Turkcanoglu, M.H.; Yucetas, U.; Koca, S.B.; Erdim, C.; Kilickesmez, O. Clear Cell Renal Cell Carcinoma: Machine Learning-Based Quantitative Computed Tomography Texture Analysis for Prediction of Fuhrman Nuclear Grade. Eur. Radiol. 2019, 29, 1153–1163. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Kaya, O.K.; Kilickesmez, O. Machine learning-based unenhanced CT texture analysis for predicting BAP1 mutation status of clear cell renal cell carcinomas. Acta Radiol. 2019, 61, 856–864. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Sun, X.-Y.; Feng, Q.-X.; Xu, X.; Zhang, J.; Zhu, F.-P.; Yang, Y.-H.; Zhang, Y.-D. Radiologic-Radiomic Machine Learning Models for Differentiation of Benign and Malignant Solid Renal Masses: Comparison with Expert-Level Radiologists. Am. J. Roentgenol. 2020, 214, W44–W54. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, F.; Tricaud, E.; Lasserre, A.S.; Petitpierre, F.; Bernhard, J.C.; Le Bras, Y.; Yacoub, M.; Bouzgarrou, M.; Ravaud, A.; Grenier, N. Routinely performed multiparametric magnetic resonance imaging helps to differentiate common subtypes of renal tumours. Eur. Radiol. 2014, 24, 1068–1080. [Google Scholar] [CrossRef]
- Cornelis, F.; Lasserre, A.-S.; Tourdias, T.; Deminière, C.; Ferrière, J.-M.; Le Bras, Y.; Grenier, N. Combined Late Gadolinium-Enhanced and Double-Echo Chemical-Shift MRI Help to Differentiate Renal Oncocytomas with High Central T2 Signal Intensity from Renal Cell Carcinomas. Am. J. Roentgenol. 2013, 200, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Canvasser, N.E.; Kay, F.U.; Xi, Y.; Pinho, D.F.; Costa, D.; de Leon, A.D.; Khatri, G.; Leyendecker, J.R.; Yokoo, T.; Lay, A.; et al. Diagnostic Accuracy of Multiparametric Magnetic Resonance Imaging to Identify Clear Cell Renal Cell Carcinoma in cT1a Renal Masses. J. Urol. 2017, 198, 780–786. [Google Scholar] [CrossRef]
- Garnier, C.; Ferrer, L.; Vargas, J.; Gallinato, O.; Jambon, E.; Le Bras, Y.; Bernhard, J.-C.; Colin, T.; Grenier, N.; Marcelin, C. A CT-Based Clinical, Radiological and Radiomic Machine Learning Model for Predicting Malignancy of Solid Renal Tumors (UroCCR-75). Diagnostics 2023, 13, 2548. [Google Scholar] [CrossRef] [PubMed]
- Tufano, A.; Antonelli, L.; Di Pierro, G.B.; Flammia, R.S.; Minelli, R.; Anceschi, U.; Leonardo, C.; Franco, G.; Drudi, F.M.; Cantisani, V. Diagnostic Performance of Contrast-Enhanced Ultrasound in the Evaluation of Small Renal Masses: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 2310. [Google Scholar] [CrossRef]
- Tufano, A.; Leonardo, C.; Di Bella, C.; Lucarelli, G.; Dolcetti, V.; Dipinto, P.; Proietti, F.; Flammia, R.S.; Anceschi, U.; Perdonà, S.; et al. Qualitative Assessment of Contrast-Enhanced Ultrasound in Differentiating Clear Cell Renal Cell Carcinoma and Oncocytoma. J. Clin. Med. 2023, 12, 3070. [Google Scholar] [CrossRef]
- Gulati, S.; Martinez, P.; Joshi, T.; Birkbak, N.J.; Santos, C.R.; Rowan, A.J.; Pickering, L.; Gore, M.; Larkin, J.; Szallasi, Z.; et al. Systematic Evaluation of the Prognostic Impact and Intratumour Heterogeneity of Clear Cell Renal Cell Carcinoma Biomarkers. Eur. Urol. 2014, 66, 936–948. [Google Scholar] [CrossRef]
- Alessandrino, F.; Shinagare, A.B.; Bossé, D.; Choueiri, T.K.; Krajewski, K.M. Radiogenomics in renal cell carcinoma. Abdom. Imaging 2019, 44, 1990–1998. [Google Scholar] [CrossRef]
- Karlo, C.A.; Di Paolo, P.L.; Chaim, J.; Hakimi, A.A.; Ostrovnaya, I.; Russo, P.; Hricak, H.; Motzer, R.; Hsieh, J.J.; Akin, O.; et al. Radiogenomics of Clear Cell Renal Cell Carcinoma: Associations between CT Imaging Features and Mutations. Radiology 2014, 270, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Bratu, O.; Mischianu, D.; Marcu, D.; Spinu, D.; Iorga, L.; Cherciu, A.; Balescu, I.; Bacalbasa, N.; Diaconu, C.; Savu, C.; et al. Renal tumor biomarkers (Review). Exp. Ther. Med. 2021, 22, 1297. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Steinmann, A.; Ding, Y.; Lee, H.; Owens, C.; Wang, J.; Yang, J.; Followill, D.; Ger, R.; MacKin, D.; et al. Radiomics feature robustness as measured using an MRI phantom. Sci. Rep. 2021, 11, 3973. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cao, K.; Jin, S.; Zhu, X.; Ding, J.; Peng, W. Differentiation of renal cell carcinoma subtypes through MRI-based radiomics analysis. Eur. Radiol. 2020, 30, 5738–5747. [Google Scholar] [CrossRef]



| SIEMENS Avanto (1.5T) | Philips Medical Systems Achieva (1.5T) | GE Medical Systems Discovery MR750W (3T) | |
|---|---|---|---|
| Sequences | T2 | T2 | T2 |
| Slices | 32 | 55 | 30 |
| Thickness (mm) | 4 | 3 | 4 |
| Intersection gap (mm) | 0.4 | 0.3 | 0.5 |
| Flip angle (°) | 140 | 90 | 142 |
| FOV (mm) | 380 | 288 | 360 |
| Matrix (pixels) | 384 × 384 | 252 × 252 | 384 × 288 |
| Resolution (mm) | 1 × 1 | 1.14 × 1.14 | 0.9 × 1.25 |
| TR (ms) | 7152.9 | 2874 | 12,000 |
| TE (ms) | 110 | 100 | 160 |
| Duration (s) | 170 | 144 | 216 |
| Clear Cell Renal Cell Carcinoma (Mean, Range) | Oncocytoma (Mean, Range) | ||
|---|---|---|---|
| Age (years) | 59.1 (19–83) | 67.1 (43–88) | |
| Sex (n, %) | Male | 20/28 (71.4%) | 9/20 (45%) |
| Female | 8/28 (28.6%) | 11/20 (55%) | |
| Localization (n,%) | Right kidney | 18/28 (64.3%) | 11/20 (55%) |
| Left kidney | 10/28 (35.7%) | 9/20 (45%) | |
| Topography (n,%) | Superior pole | 14/28 (50%) | 3/20 (15%) |
| Equatorial pole | 8/28 (29%) | 13/20 (65%) | |
| Inferior pole | 6/28 (21%) | 4/20 (20%) | |
| Average size (mm) | 28.8 (15–40) | 26.3 (15–40) | |
| Histology (%) | Biopsy | 10/28 (35.7%) | 6/20 (30%) |
| Tumorectomy | 15/28 (53.5%) | 6/20 (30%) | |
| Partial nephrectomy | 3/28 (10.8%) | 8/20 (40%) |
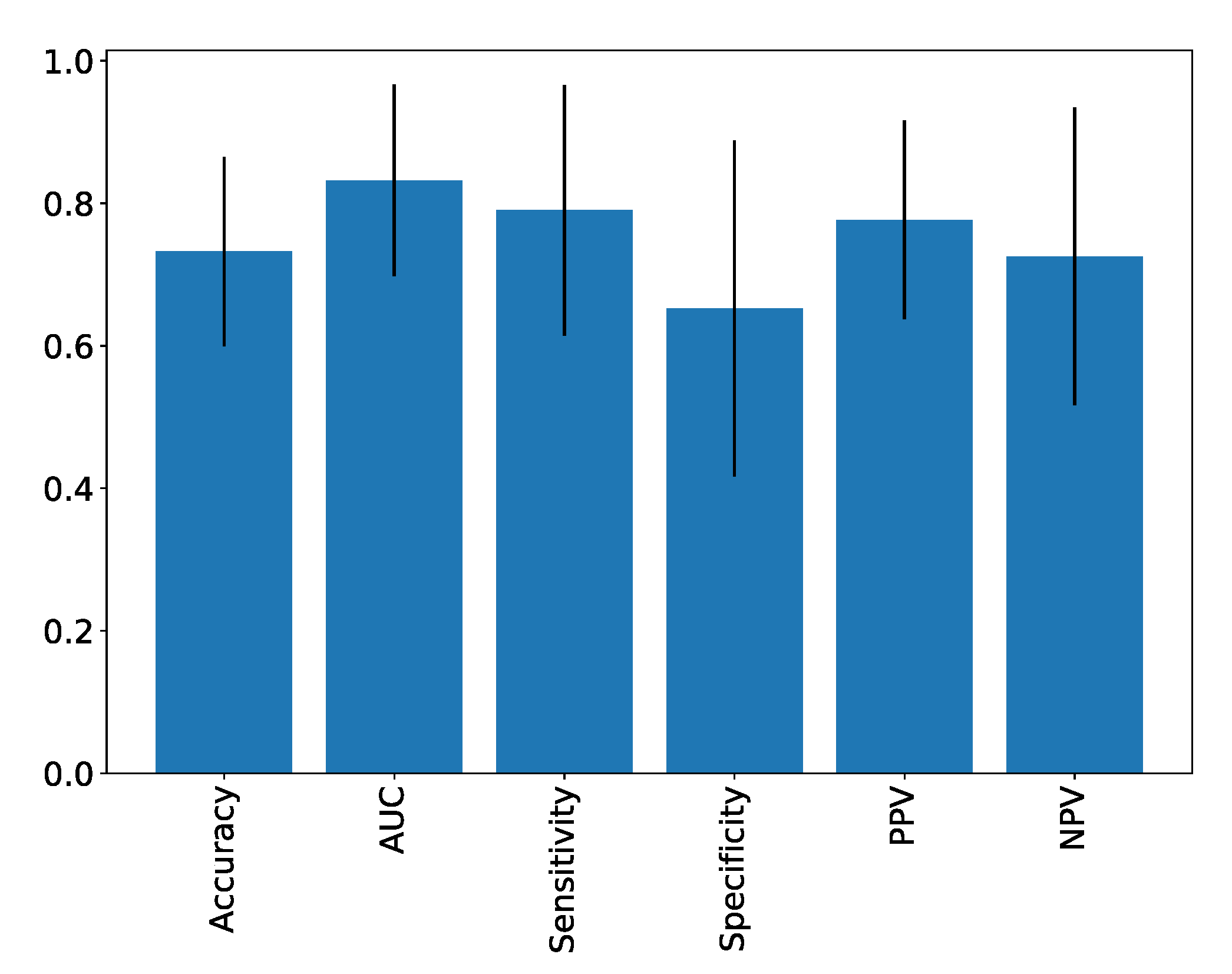
| Accuracy | AUC | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|
| Scores | 0.73 ± 0.13 | 0.83 ± 0.13 | 0.79 ± 0.18 | 0.65 ± 0.24 | 0.78 ± 0.14 | 0.73 ± 0.21 |
| Accuracy | AUC | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|
| Scores | 0.77 ± 0.12 | 0.90 ± 0.10 | 0.83 ± 0.16 | 0.69 ± 0.22 | 0.81 ± 0.12 | 0.78 ± 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toffoli, T.; Saut, O.; Etchegaray, C.; Jambon, E.; Le Bras, Y.; Grenier, N.; Marcelin, C. Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy. J. Pers. Med. 2023, 13, 1444. https://doi.org/10.3390/jpm13101444
Toffoli T, Saut O, Etchegaray C, Jambon E, Le Bras Y, Grenier N, Marcelin C. Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy. Journal of Personalized Medicine. 2023; 13(10):1444. https://doi.org/10.3390/jpm13101444
Chicago/Turabian StyleToffoli, Thibault, Olivier Saut, Christele Etchegaray, Eva Jambon, Yann Le Bras, Nicolas Grenier, and Clément Marcelin. 2023. "Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy" Journal of Personalized Medicine 13, no. 10: 1444. https://doi.org/10.3390/jpm13101444
APA StyleToffoli, T., Saut, O., Etchegaray, C., Jambon, E., Le Bras, Y., Grenier, N., & Marcelin, C. (2023). Differentiation of Small Clear Renal Cell Carcinoma and Oncocytoma through Magnetic Resonance Imaging-Based Radiomics Analysis: Toward the End of Percutaneous Biopsy. Journal of Personalized Medicine, 13(10), 1444. https://doi.org/10.3390/jpm13101444






