PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Definitions
2.2. LSM and Controlled Attenuation Parameter Measurement
2.3. Genotyping for SNPs
2.4. Statistical Analyses
2.5. Ethics
3. Results
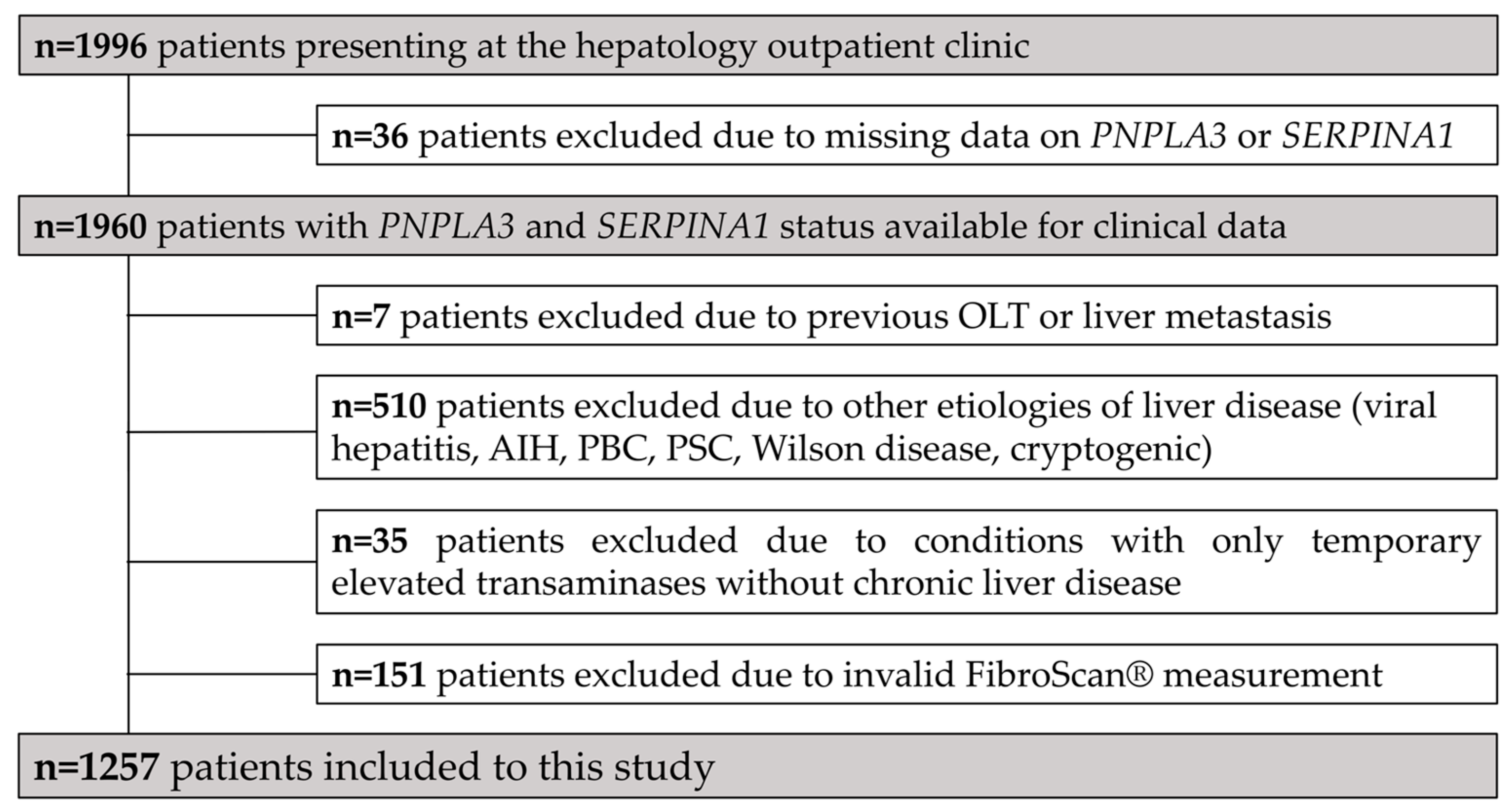
3.1. Study Population and Patient Characteristics
3.2. Prevalence of Risk Alleles
3.3. Differences between PNPLA3 and SERPINA1 Genotype Variants
3.4. Association with Liver Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sookoian, S.; Pirola, C.J. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 2011, 53, 1883–1894. [Google Scholar] [CrossRef]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Buch, S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Salameh, H.; Raff, E.; Erwin, A.; Seth, D.; Nischalke, H.D.; Falleti, E.; Burza, M.A.; Leathert, J.; Romeo, S.; Molinaro, A.; et al. PNPLA3 Gene Polymorphism Is Associated With Predisposition to and Severity of Alcoholic Liver Disease. Am. J. Gastroenterol. 2015, 110, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Stickel, F.; Buch, S.; Lau, K.; Meyer zu Schwabedissen, H.; Berg, T.; Ridinger, M.; Rietschel, M.; Schafmayer, C.; Braun, F.; Hinrichsen, H.; et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology 2011, 53, 86–95. [Google Scholar] [CrossRef]
- Trépo, E.; Gustot, T.; Degré, D.; Lemmers, A.; Verset, L.; Demetter, P.; Ouziel, R.; Quertinmont, E.; Vercruysse, V.; Amininejad, L.; et al. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J. Hepatol. 2011, 55, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.R.; Way, M.J.; McQuillin, A.; Morgan, M.Y.; Thursz, M.R. Homozygosity for rs738409:G in PNPLA3 is associated with increased mortality following an episode of severe alcoholic hepatitis. J. Hepatol. 2017, 67, 120–127. [Google Scholar] [CrossRef]
- Stättermayer, A.F.; Scherzer, T.; Beinhardt, S.; Rutter, K.; Hofer, H.; Ferenci, P. Review article: Genetic factors that modify the outcome of viral hepatitis. Aliment Pharm. 2014, 39, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Romeo, S.; Valenti, L. Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis. Biomed. Res. Int. 2015, 2015, 460190. [Google Scholar] [CrossRef]
- Unalp-Arida, A.; Ruhl, C.E. Patatin-Like Phospholipase Domain-Containing Protein 3 I148M and Liver Fat and Fibrosis Scores Predict Liver Disease Mortality in the U.S. Population. Hepatology 2020, 71, 820–834. [Google Scholar] [CrossRef]
- Fischer, H.-P.; Ortiz-Pallardó, M.E.; Ko, Y.; Esch, C.; Zhou, H. Chronic liver disease in heterozygous α1-antitrypsin deficiency PiZ. J. Hepatol. 2000, 33, 883–892. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Salomaa, V.; Åberg, F. The Pi*MZ Allele in Alpha-1 Antitrypsin Increases Liver-Related Outcomes in a Population-Based Study. Gastroenterology 2020. [Google Scholar] [CrossRef]
- Strnad, P.; Buch, S.; Hamesch, K.; Fischer, J.; Rosendahl, J.; Schmelz, R.; Brueckner, S.; Brosch, M.; Heimes, C.V.; Woditsch, V.; et al. Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut 2019, 68, 1099–1107. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1388–1402. [CrossRef] [PubMed]
- Pons, M.; Rodríguez-Tajes, S.; Esteban, J.I.; Mariño, Z.; Vargas, V.; Lens, S.; Buti, M.; Augustin, S.; Forns, X.; Mínguez, B.; et al. Non-invasive prediction of liver-related events in patients with HCV-associated compensated advanced chronic liver disease after oral antivirals. J. Hepatol. 2020, 72, 472–480. [Google Scholar] [CrossRef]
- Semmler, G.; Wöran, K.; Scheiner, B.; Unger, L.W.; Paternostro, R.; Stift, J.; Schwabl, P.; Bucsics, T.; Bauer, D.; Simbrunner, B.; et al. Novel reliability criteria for controlled attenuation parameter assessments for non-invasive evaluation of hepatic steatosis. United Eur. Gastroenterol. J. 2020, 8, 321–331. [Google Scholar] [CrossRef]
- Boursier, J.; Zarski, J.P.; de Ledinghen, V.; Rousselet, M.C.; Sturm, N.; Lebail, B.; Fouchard-Hubert, I.; Gallois, Y.; Oberti, F.; Bertrais, S.; et al. Determination of reliability criteria for liver stiffness evaluation by transient elastography. Hepatology 2013, 57, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Kedenko, L.; Lamina, C.; Kedenko, I.; Kollerits, B.; Kiesslich, T.; Iglseder, B.; Kronenberg, F.; Paulweber, B. Genetic polymorphisms at SIRT1 and FOXO1 are associated with carotid atherosclerosis in the SAPHIR cohort. BMC Med. Genet. 2014, 15, 112. [Google Scholar] [CrossRef]
- Rodriguez, S.; Gaunt, T.R.; Day, I.N. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009, 169, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, B.; Mandorfer, M.; Viveiros, A.; Finkenstedt, A.; Ferenci, P.; Schneeberger, S.; Tilg, H.; Zoller, H. Heterozygosity for the alpha-1-antitrypsin Z allele in cirrhosis is associated with more advanced disease. Liver Transpl. 2018, 24, 744–751. [Google Scholar] [CrossRef]
- Krawczyk, M.; Liebe, R.; Lammert, F. Toward Genetic Prediction of Nonalcoholic Fatty Liver Disease Trajectories: PNPLA3 and Beyond. Gastroenterology 2020, 158, 1865–1880.e1861. [Google Scholar] [CrossRef]
- Hassan, M.M.; Kaseb, A.; Etzel, C.J.; El-Serag, H.; Spitz, M.R.; Chang, P.; Hale, K.S.; Liu, M.; Rashid, A.; Shama, M.; et al. Genetic variation in the PNPLA3 gene and hepatocellular carcinoma in USA: Risk and prognosis prediction. Mol. Carcinog. 2013, 52 (Suppl. 1), E139–E147. [Google Scholar] [CrossRef]
- Trépo, E.; Romeo, S.; Zucman-Rossi, J.; Nahon, P. PNPLA3 gene in liver diseases. J. Hepatol. 2016, 65, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Jamialahmadi, O.; Pelusi, S.; Baselli, G.; Dongiovanni, P.; Zanoni, I.; Santoro, L.; Maier, S.; Liguori, A.; Meroni, M.; et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Shima, T.; Mizuno, M.; Mitsumoto, Y.; Umemura, A.; Kanbara, Y.; Tanaka, S.; Sumida, Y.; Yasui, K.; Takahashi, M.; et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS ONE 2018, 13, e0185490. [Google Scholar] [CrossRef] [PubMed]
- León-Mimila, P.; Vega-Badillo, J.; Gutiérrez-Vidal, R.; Villamil-Ramírez, H.; Villareal-Molina, T.; Larrieta-Carrasco, E.; López-Contreras, B.E.; Kauffer, L.R.; Maldonado-Pintado, D.G.; Méndez-Sánchez, N.; et al. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp. Mol. Pathol. 2015, 98, 178–183. [Google Scholar] [CrossRef]
- Mandorfer, M.; Bucsics, T.; Hutya, V.; Schmid-Scherzer, K.; Schaefer, B.; Zoller, H.; Ferlitsch, A.; Peck-Radosavljevic, M.; Trauner, M.; Ferenci, P.; et al. Liver disease in adults with α1-antitrypsin deficiency. United Eur. Gastroenterol. J. 2018, 6, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Wagenknecht, L.E.; Palmer, N.D.; Bowden, D.W.; Rotter, J.I.; Norris, J.M.; Ziegler, J.; Chen, Y.D.; Haffner, S.; Scherzinger, A.; Langefeld, C.D. Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: The Insulin Resistance Atherosclerosis Family Study. Liver Int. Off. J. Int. Assoc. Study Liver 2011, 31, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, P.; Romeo, S. The role of PNPLA3 in health and disease. Biochim. Et Biophys. Acta. Mol. Cell Biol. Lipids 2019, 1864, 900–906. [Google Scholar] [CrossRef]
- Teckman, J.H.; An, J.K.; Blomenkamp, K.; Schmidt, B.; Perlmutter, D. Mitochondrial autophagy and injury in the liver in alpha 1-antitrypsin deficiency. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G851–G862. [Google Scholar] [CrossRef]
- Strnad, P.; McElvaney, N.G.; Lomas, D.A. Alpha(1)-Antitrypsin Deficiency. N. Engl. J. Med. 2020, 382, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Hamesch, K.; Mandorfer, M.; Pereira, V.M.; Moeller, L.S.; Pons, M.; Dolman, G.E.; Reichert, M.C.; Schneider, C.V.; Woditsch, V.; Voss, J.; et al. Liver Fibrosis and Metabolic Alterations in Adults With alpha-1-antitrypsin Deficiency Caused by the Pi*ZZ Mutation. Gastroenterology 2019, 157, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.V.; Hamesch, K.; Gross, A.; Mandorfer, M.; Moeller, L.S.; Pereira, V.; Pons, M.; Kuca, P.; Reichert, M.C.; Benini, F.; et al. Liver Phenotypes of European Adults Heterozygous or Homozygous for Pi∗Z Variant of AAT (Pi∗MZ vs. Pi∗ZZ genotype) and Noncarriers. Gastroenterology 2020, 159, 534–548. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Overall n = 1257 (100%) | |||
|---|---|---|---|---|
| Clinical Data | Laboratory Measurements | |||
| Age, years | 52.7 ± 15.2 | Bilirubin, mg/dL | 0.6 (0.4–0.9) | |
| Male sex | 759 (60.4%) | Albumin, g/dL | 45.1 ± 4 | |
| BMI, kg/m2 | 27.2 ± 5 | INR | 1.2 (1.2–1.2) | |
| LSM, kPa | 5.9 (4.5–9.2) | Creatinine, mg/dL | 0.9 (0.8–1) | |
| LSM ≥ 10 kPa | 286 (22.8%) | Na, mmol/L | 139.7 ± 2.5 | |
| LSM ≥ 15 kPa | 176 (14%) | Platelets, ×109/L | 237 (195–283) | |
| LSM ≥ 20 kPa | 144 (11.5%) | Bilirubin, mg/dL | 0.6 (0.4–0.9) | |
| CAP, dB | 277 ± 68 | AST (U/L) | 33 (26–47) | |
| ACLD | 309 (24.6%) | ALT(U/L) | 40 (27–64) | |
| CSPH | 185 (14.7%) | GGT (U/L) | 70 (32–157) | |
| dACLD | 76 (6%) | White-cell count, ×109/L | 6.3 (5.3–7.7) | |
| HCC | 18 (1.4%) | CRP, mg/dL | 0.2 (0.1–0.5) | |
| Child–Pugh Score | 5 ± 1 | |||
| MELD Score | 9.5 ± 2.3 | |||
| Fib-4 Score | 1.14 (0.76–1.80) | |||
| Etiology of liver disease | ||||
| NAFLD | 1048 (83.4%) | |||
| ALD | 209 (16.6%) | |||
| Genotypes | χ2 | p | q | |
| PNPLA3 C/C | 623 (49.6%) | 7.1308 | 0.6925 | 0.3075 |
| PNPLA3 G/C | 495 (39.4%) | |||
| PNPLA3 G/G | 139 (11.1%) | |||
| SERPINA1 M/M | 1147 (91.2%) | 0.0074 | 0.9730 | 0.0270 |
| SERPINA1 M/S | 43 (3.4%) | |||
| SERPINA1 M/Z | 66 (5.3%) | |||
| SERPINA1 Z/Z | 1 (0.1%) | |||
| A | LSM, kPa | ACLD | CSPH |
| Age, year | 0.114 (0.072–0.157), p < 0.001 | 1.055 (1.042–1.068), p < 0.001 | 1.042 (1.028–1.056), p < 0.001 |
| BMI, kg/m2 | 0.327 (0.199–0.454), p < 0.001 | 1.124 (1.089–1.160), p < 0.001 | 1.053 (1.017–1.091), p = 0.004 |
| Alcohol abuse | 13.224 (11.580–14.867), p < 0.001 | 7.718 (5.457–10.915), p < 0.001 | 7.280 (5.082–10.428), p < 0.001 |
| PNPLA3 G-allele | 2.707 (1.435–3.979), p < 0.001 | 1.971 (1.448–2.681), p < 0.001 | 1.685 (1.180–2.406), p = 0.004 |
| B | LSM, kPa | ACLD | CSPH |
| Age, year | 0.111 (0.068–0.154), p < 0.001 | 1.053 (1.041–1.065), p < 0.001 | 1.041 (1.027–1.055), p < 0.001 |
| BMI, kg/m2 | 0.348 (0.220–0.476), p < 0.001 | 1.127 (1.092–1.163), p < 0.001 | 1.056 (1.020–1.094), p = 0.002 |
| Alcohol abuse | 13.462 (11.809–15.116), p < 0.001 | 7.851 (5.563–11.081), p < 0.001 | 7.598 (5.297–10.900), p < 0.001 |
| SERPINA1 Z-allele | 2.581 (−0.244–5.406), p = 0.073 | 1.748 (0.925–3.307), p = 0.086 | 2.122 (1.067–4.218), p = 0.032 |
| C | LSM, kPa | ACLD | CSPH |
| Age, year | 0.113 (0.070–0.156), p < 0.001 | 1.055 (1.042–1.067), p < 0.001 | 1.042 (1.028–1.056), p < 0.001 |
| BMI, kg/m2 | 0.332 (0.205–0.459), p < 0.001 | 1.125 (1.090–1.161), p < 0.001 | 1.054 (1.018–1.092), p = 0.003 |
| Alcohol abuse | 13.302 (11.658–14.947), p < 0.001 | 7.896 (5.573–11.188), p < 0.001 | 7.553 (5.252–10.862), p < 0.001 |
| PNPLA3 G-allele | 2.715 (1.444–3.986), p < 0.001 | 1.989 (1.461–2.709), p < 0.001 | 1.707 (1.194–2.441), p = 0.003 |
| SERPINA1 Z-allele | 2.624 (−0.182–5.430), p = 0.067 | 1.832 (0.963–3.483), p = 0.065 | 2.196 (1.103–4.371), p = 0.025 |
| D | ACLD | CSPH | |
| Age, year | 1.054 (1.042–1.067), p < 0.001 | 1.042 (1.028–1.056), p < 0.001 | |
| BMI, kg/m2 | 1.125 (1.091–1.161), p < 0.001 | 1.054 (1.018–1.092), p = 0.003 | |
| Alcohol abuse | 7.918 (5.589–11.216), p < 0.001 | 7.559 (5.258–10.866), p < 0.001 | |
| No risk allele | Reference | Reference | |
| One risk allele | 1.955 (1.427–2.678), p < 0.001 | 1.675 (1.161–2.415), p = 0.006 | |
| Two risk alleles | 3.892 (1.561–9.706), p = 0.004 | 4.282 (1.667–10.996), p = 0.003 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semmler, G.; Balcar, L.; Oberkofler, H.; Zandanell, S.; Strasser, M.; Niederseer, D.; Feldman, A.; Stickel, F.; Strnad, P.; Datz, C.; et al. PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center. J. Pers. Med. 2021, 11, 165. https://doi.org/10.3390/jpm11030165
Semmler G, Balcar L, Oberkofler H, Zandanell S, Strasser M, Niederseer D, Feldman A, Stickel F, Strnad P, Datz C, et al. PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center. Journal of Personalized Medicine. 2021; 11(3):165. https://doi.org/10.3390/jpm11030165
Chicago/Turabian StyleSemmler, Georg, Lorenz Balcar, Hannes Oberkofler, Stephan Zandanell, Michael Strasser, David Niederseer, Alexandra Feldman, Felix Stickel, Pavel Strnad, Christian Datz, and et al. 2021. "PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center" Journal of Personalized Medicine 11, no. 3: 165. https://doi.org/10.3390/jpm11030165
APA StyleSemmler, G., Balcar, L., Oberkofler, H., Zandanell, S., Strasser, M., Niederseer, D., Feldman, A., Stickel, F., Strnad, P., Datz, C., Paulweber, B., & Aigner, E. (2021). PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center. Journal of Personalized Medicine, 11(3), 165. https://doi.org/10.3390/jpm11030165









