Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Collection and Preparation of Saliva Samples
2.3. Biochemical Analysis of Saliva Samples
2.4. Statistical Data Processing
3. Results
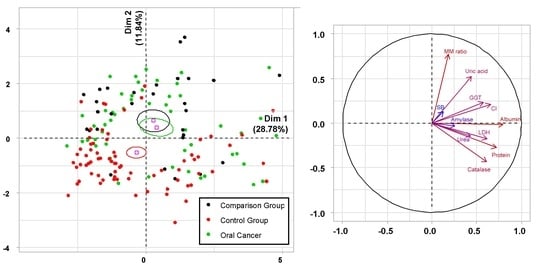
3.1. Biochemical Saliva Markers in the Diagnosis of OSCC
3.2. Prognostic Value of Biochemical Saliva Markers in OSCC
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Panta, P.; Wong, D.T.W. Salivary Biomarkers in Oral Cancer. Oral Cancer Detect. 2019, 265–295. [Google Scholar] [CrossRef]
- Blatt, S.; Krüger, M.; Ziebart, T.; Sagheb, K.; Schiegnitz, E.; Goetze, E.; Al-Nawas, B.; Pabst, A.M. Biomarkers in diagnosis and therapy of oral squamous cell carcinoma: A review of the literature. J. Cranio-Maxillofac. Surg. 2017, 45, 722–730. [Google Scholar] [CrossRef]
- Bouza, M.; Gonzalez-Soto, J.; Pereiro, R.; De Vicente, J.C.; Sanz-Medel, A. Exhaled breath and oral cavity VOCs as potential biomarkers in oral cancer patients. J. Breath Res. 2017, 11, 016015. [Google Scholar] [CrossRef] [PubMed]
- Blatt, S.; Ziebart, T.; Krüger, M.; Pabst, A.; Information, P.E.K.F.C. Diagnosing oral squamous cell carcinoma: How much imaging do we really need? A review of the current literature. J. Cranio-Maxillofac. Surg. 2016, 44, 538–549. [Google Scholar] [CrossRef]
- Chi, A.C.; Day, T.A.; Neville, B.W. Oral cavity and oropharyngeal squamous cell carcinoma-an update. CA A Cancer J. Clin. 2015, 65, 401–421. [Google Scholar] [CrossRef]
- Kaprin, A.D.; Starinskiy, V.V.; Petrova, G.V. The Status of Cancer Care for the Population of Russia in 2018; Moscow MNIOI im. Herzen of the Ministry Of Health Of Russia: Gertsena, Russia, 2019. (In Russian) [Google Scholar]
- Chatterjee, A.; Laskar, S.G.; Chaukar, D. Management of early oral cavity squamous cancers. Oral Oncol. 2020, 104, 104627. [Google Scholar] [CrossRef]
- Osipyan, Y.O.; Mudunov, A.M. Computed tomography and magnetic resonance imaging in assessment of the local advancement of oral and oropharyngeal cancer as the key factor of treatment selection (literature review). Head Neck Tumors (Hnt) 2017, 7, 53–62. [Google Scholar] [CrossRef][Green Version]
- Wolff, K.-D.; Follmann, M.; Nast, A. The Diagnosis and Treatment of Oral Cavity Cancer. Dtsch. Aerzteblatt Online 2012, 109, 829–835. [Google Scholar] [CrossRef]
- Rai, V.; Mukherjee, R.; Ghosh, A.K.; Routray, A.; Chakraborty, C. “Omics” in oral cancer: New approaches for biomarker discovery. Arch. Oral Biol. 2018, 87, 15–34. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Shahid, N.; Naqvi, S.M.A.; Saleem, M.; Siddiqui, A.J.; Ali, A. Metabolite Profiling of Preneoplastic and Neoplastic Lesions of Oral Cavity Tissue Samples Revealed a Biomarker Pattern. Sci. Rep. 2016, 6, 38985. [Google Scholar] [CrossRef]
- Chi, L.-M.; Hsiao, Y.-C.; Chien, K.-Y.; Chen, S.-F.; Chuang, Y.-N.; Lin, S.-Y.; Wang, W.-S.; Chang, I.Y.-F.; Yang, C.; Chu, L.J.; et al. Assessment of candidate biomarkers in paired saliva and plasma samples from oral cancer patients by targeted mass spectrometry. J. Proteom. 2019, 211, 103571. [Google Scholar] [CrossRef] [PubMed]
- Roblegg, E.; Coughran, A.; Sirjani, D. Saliva: An all-rounder of our body. Eur. J. Pharm. Biopharm. 2019, 142, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V. Possibilities of using saliva for the diagnosis of cancer. Russ. Clin. Lab. Diagn. 2019, 64, 333–336. [Google Scholar]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of Salivary Biomarkers in Oral Cancer Detection. Int. Rev. Cytol. 2018, 86, 23–70. [Google Scholar] [CrossRef]
- Amenábar, J.M.; Da Silva, B.M.; Punyadeera, C. Salivary protein biomarkers for head and neck cancer. Expert Rev. Mol. Diagn. 2020, 20, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, J.; Lu, J.; Xiong, H.; Shi, X.; Gong, L. Significance of CD44 expression in head and neck cancer: A systemic review and meta-analysis. BMC Cancer 2014, 14, 15. [Google Scholar] [CrossRef]
- Geng, X.-F.; Du, M.; Han, J.-X.; Zhang, M.; Tang, X.-F.; Xing, R.-D. Saliva CA125 and TPS Levels in Patients with Oral Squamous Cell Carcinoma. Int. J. Biol. Markers 2013, 28, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, K.; Ramya, R.; Nandhini, G.; Rajashree, P.; Kumar, A.R.; Anandan, S.N. Salivary and serum level of CYFRA 21-1 in oral precancer and oral squamous cell carcinoma. Oral Dis. 2013, 21, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Jou, Y.-J.; Lin, C.-D.; Lai, C.-H.; Tang, C.-H.; Huang, S.-H.; Tsai, M.-H.; Chen, S.-Y.; Kao, J.-Y.; Lin, C.-W. Salivary zinc finger protein 510 peptide as a novel biomarker for detection of oral squamous cell carcinoma in early stages. Clin. Chim. Acta 2011, 412, 1357–1365. [Google Scholar] [CrossRef]
- Weng, L.-P.; Wu, C.-C.; Hsu, B.-L.; Chi, L.-M.; Liang, Y.; Tseng, C.-P.; Hsieh, L.-L.; Yu, J.-S. Secretome-Based Identification of Mac-2 Binding Protein as a Potential Oral Cancer Marker Involved in Cell Growth and Motility. J. Proteome Res. 2008, 7, 3765–3775. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Q.; Chen, J.; Yi, P.; Xu, X.; Fan, Y.; Cui, B.; Yu, Y.; Li, X.; Du, Y.; et al. Salivary protease spectrum biomarkers of oral cancer. Int. J. Oral Sci. 2019, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Stott-Miller, M.; Houck, J.R.; Lohavanichbutr, P.; Méndez, E.; Upton, M.P.; Futran, N.D.; Schwartz, S.M.; Chen, C. Tumor and salivary matrix metalloproteinase levels are strong diagnostic markers of oral squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2628–2636. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-C.; Kim, M.-Y.; Kim, C.-H. C-reactive protein/albumin ratio as prognostic score in oral squamous cell carcinoma. J. Korean Assoc. Oral Maxillofac. Surg. 2016, 42, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-S.L.; Jordan, L.; Gorugantula, L.M.; Schneiderman, E.; Chen, H.-S.; Rees, T. Salivary Interleukin-6 and -8 in Patients With Oral Cancer and Patients With Chronic Oral Inflammatory Diseases. J. Periodontol. 2014, 85, 956–965. [Google Scholar] [CrossRef]
- Elashoff, D.; Zhou, H.; Reiss, J.; Wang, J.; Xiao, H.; Henson, B.; Hu, S.; Arellano, M.; Sinha, U.; Le, A.; et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol. Biomark. Prev. 2012, 21, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Zahran, F.; Ghalwash, D.; Shaker, O.; Al-Johani, K.; Scully, C. Salivary microRNAs in oral cancer. Oral Dis. 2015, 21, 739–747. [Google Scholar] [CrossRef]
- Salazar, C.; Nagadia, R.; Pandit, P.; Cooper-White, J.J.; Banerjee, N.; Dimitrova, N.; Coman, W.B.; Punyadeera, C. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell. Oncol. 2014, 37, 331–338. [Google Scholar] [CrossRef]
- Qu, S.; Zhong, Y.; Shang, R.; Zhang, X.; Song, W.; Kjems, J.; Li, H. The emerging landscape of circular RNA in life processes. RNA Biol. 2016, 14, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Wong, Y.; Hsiao, H.; Wang, Y.; Chan, M.; Chang, K. Evaluation of saliva and plasma cytokine biomarkers in patients with oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2018, 47, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-F.; Huang, H.-D.; Fan, W.-L.; Jong, Y.-J.; Chen, M.-K.; Huang, C.-N.; Kuo, Y.-L.; Chung, W.-H.; Su, S.-C. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 2018, 77, 1–8. [Google Scholar] [CrossRef]
- Kaur, J.; Politis, C.; Jacobs, R. Salivary 8-hydroxy-2-deoxyguanosine, malondialdehyde, vitamin C, and vitamin E in oral pre-cancer and cancer: Diagnostic value and free radical mechanism of action. Clin. Oral Investig. 2015, 20, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Dziewulska, A.; Janiszewska-Olszowska, J.; Bachanek, T.; Grocholewicz, K. Salivary mineral composition in patients with oral cancer. Magnes. Res. 2013, 26, 120–124. [Google Scholar] [CrossRef]
- Shigeyama, H.; Wang, T.; Ichinose, M.; Ansai, T.; Lee, S.-W. Identification of volatile metabolites in human saliva from patients with oral squamous cell carcinoma via zeolite-based thin-film microextraction coupled with GC–MS. J. Chromatogr. B 2019, 1104, 49–58. [Google Scholar] [CrossRef]
- Kutukova, S.I.; Manikhas, G.M.; Yaremenko, A.I.; Chukhlovin, A.B.; Belyak, N.P.; Bozhor, S.S.; Dibirov, R.K.; Ermakova, T.S. Prognostic role of laboratory and immunohistochemical markers in the recurrence of oral mucosal squamous cell carcinoma. Head Neck Tumors 2014, 3, 47–50. [Google Scholar]
- Barak, V.; Meirovitz, A.; Leibovici, V.; Rachmut, J.; Peretz, T.; Eliashar, R.; Gross, M. The Diagnostic and Prognostic Value of Tumor Markers (CEA, SCC, CYFRA 21-1, TPS) in Head and Neck Cancer Patients. Anticancer. Res. 2015, 35, 5519–5524. [Google Scholar]
- Bel’Skaya, L.V.; Sarf, E.A.; Kosenok, V.K.; Gundyrev, I.A. Biochemical Markers of Saliva in Lung Cancer: Diagnostic and Prognostic Perspectives. Diagnostics 2020, 10, 186. [Google Scholar] [CrossRef]
- Bel’Skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Correlation interrelations between the composition of saliva and blood plasmain norm. Klin. Lab. Diagn. 2018, 63, 477–482. [Google Scholar]
- Bel’Skaya, L.V.; Kosenok, V.K.; Sarf, E.A. Chronophysiological features of the normal mineral composition of human saliva. Arch. Oral Biol. 2017, 82, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Ogawa, T.; Washio, J.; Takahashi, T.; Echigo, S.; Takahashi, N. Glucose and glutamine metabolism in oral squamous cell carcinoma: Insight from a quantitative metabolomic approach. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 118, 218–225. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M.; Ota, S.; Hiwatari, K.; Enomoto, A.; Soga, T.; et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016, 6, 31520. [Google Scholar] [CrossRef] [PubMed]
- Reznick, A.Z.; Hershkovich, O.; Nagler, R.M. Saliva—A pivotal player in the pathogenesis of oropharyngeal cancer. Br. J. Cancer 2004, 91, 111–118. [Google Scholar] [CrossRef][Green Version]
- Choudhari, S.K.; Chaudhary, M.; Gadbail, A.R.; Sharma, A.; Tekade, S. Oxidative and antioxidative mechanisms in oral cancer and precancer: A review. Oral Oncol. 2014, 50, 10–18. [Google Scholar] [CrossRef]
- Khoubnasabjafari, M.; Ansarin, K.; Jouyban, A. Salivary malondialdehyde as an oxidative stress biomarker in oral and systemic diseases. J. Dent. Res. Dent. Clin. Dent. Prospect. 2016, 10, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Chole, R.; Patil, R.; Basak, A.; Palandurkar, K.; Bhowate, R.R. Estimation of serum malondialdehyde in oral cancer and precancer and its association with healthy individuals, gender, alcohol, and tobacco abuse. J. Cancer Res. Ther. 2010, 6, 487. [Google Scholar] [CrossRef]
- Peluso, I.; Raguzzini, A. Salivary and Urinary Total Antioxidant Capacity as Biomarkers of Oxidative Stress in Humans. Pathol. Res. Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary Proteomics for Oral Cancer Biomarker Discovery. Clin. Cancer Res. 2008, 14, 6246–6252. [Google Scholar] [CrossRef]
- Patel, S.; Metgud, R. Estimation of salivary lactate dehydrogenase in oral leukoplakia and oral squamous cell carcinoma: A biochemical study. J. Cancer Res. Ther. 2015, 11, 119. [Google Scholar] [CrossRef]
- Mafessoni, T.P.; Mazur, C.E.; Amenábar, J.M.; Mazur, C.E. Salivary lactate dehydrogenase (LDH) as a tool for early diagnosis of oral cancer in individuals with Fanconi anemia. Med. Hypotheses 2018, 119, 29–31. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Panda, M. Recent trends of saliva omics biomarkers for the diagnosis and treatment of oral cancer. J. Oral Biosci. 2019, 61, 84–94. [Google Scholar] [CrossRef]
- Jean, J.-C.; Liu, Y.; Joyce-Brady, M. The importance of gamma-glutamyl transferase in lung glutathione homeostasis and antioxidant defense. BioFactors 2003, 17, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Maellaro, E.; Dominici, S.; Del Bello, B.; Valentini, M.A.; Pieri, L.; Perego, P.; Supino, R.; Zunino, F.; Lorenzini, E.; Paolicchi, A.; et al. Membrane gamma-glutamyl transpeptidase activity of melanoma cells: Effects on cellular H2O2 production, cell surface protein thiol oxidation and NF-kappa B activation status. J. Cell Sci. 2000, 113, 2671–2678. [Google Scholar]
- Radhika, T.; Jeddy, N.; Nithya, S.; Muthumeenakshi, R.M. Salivary biomarkers in oral squamous cell carcinoma—An insight. J. Oral Biol. Craniofac. Res. 2016, 6, S51–S54. [Google Scholar] [CrossRef]
- Błoniarz, J.; Rahnama, M.; Zareba, S. Influence of carcinogenesis in the oral cavity on the level of some bioelements in the saliva. Rocz. Państwowego Zakładu Hig. 2003, 54, 295–300. [Google Scholar]
- Fuchs, P.N.; Rogić, D.; Vidović-Juras, D.; Susić, M.; Milenović, A.; Brailo, V.; Boras, V.V. Salivary analytes in patients with oral squamous cell carcinoma. Coll. Antropol. 2011, 35, 359–362. [Google Scholar]
- Peretti, M.; Angelini, M.; Savalli, N.; Florio, T.; Yuspa, S.H.; Mazzanti, M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim. Et Biophys. Acta (BBA) Biomembr. 2015, 1848, 2523–2531. [Google Scholar] [CrossRef]
- Miyazaki, H.; Shiozaki, A.; Niisato, N.; Ohsawa, R.; Itoi, H.; Ueda, Y.; Otsuji, E.; Yamagishi, H.; Iwasaki, Y.; Nakano, T.; et al. Chloride ions control the G1/S cell-cycle checkpoint by regulating the expression of p21 through a p53-independent pathway in human gastric cancer cells. Biochem. Biophys. Res. Commun. 2008, 366, 506–512. [Google Scholar] [CrossRef]
- Mazumder, S.; Datta, S.; Ray, J.G.; Chaudhuri, K.; Chatterjee, R. Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 2019, 58, 137–145. [Google Scholar] [CrossRef]





| Indicators | Control Group, n = 114 (1) | Comparison Group, n = 50 (2) | Oral Cancer, n = 68 (3) | Kruskal–Wallis Test (H, p) |
|---|---|---|---|---|
| Flow rate, mL/min | 0.87 (0.72; 0.99) | 0.81 (0.74; 0.94) | 0.78 (0.64; 0.97) | 1.079, 0.5836 |
| Electrolytes | ||||
| Calcium, mmol/L | 1.26 (1.03; 1.61) | 1.38 (1.10; 1.69) | 1.39 (1.14; 1.85) | 4.599, 0.1003 |
| - | - | p1–3 = 0.0426 | ||
| Phosphorus, mmol/L | 4.36 (3.39; 5.58) | 4.70 (3.62; 6.38) | 4.29 (3.03; 6.19) | 1.350, 0.5091 |
| Ca/P | 0.290 (0.226; 0.399) | 0.267 (0.199; 0.474) | 0.316 (0.241; 0.601) | 2.762, 0.2514 |
| Sodium, mmol/L | 7.5 (4.9; 11.2) | 7.6 (5.3; 10.9) | 9.2 (6.3; 13.9) | 4.222, 0.1211 |
| - | - | p1–3 = 0.0403 | ||
| Potassium, mmol/L | 10.5 (8.6; 13.3) | 12.0 (8.9; 15.6) | 11.0 (8.3; 16.1) | 2.291, 0.3181 |
| Na/K | 0.68 (0.48; 1.08) | 0.67 (0.49; 1.04) | 0.88 (0.59; 1.40) | 5.575, 0.0616 |
| - | - | p1–3 = 0.0220 | ||
| Chlorides, mmol/L | 23.7 (19.4; 29.6) | 26.1 (21.7; 33.9) | 30.1 (21.9; 39.5) | 12.38, 0.0020 * |
| - | p1–2 = 0.0325 | p1–3 = 0.0009 | ||
| Protein Metabolism | ||||
| Protein, g/L | 1.11 (0.78; 1.53) | 0.75 (0.49; 1.23) | 0.84 (0.47; 1.59) | 8.581, 0.0137 * |
| - | p1–2 = 0.0051 | - | ||
| Albumin, g/L | 0.24 (0.17; 0.36) | 0.34 (0.19; 0.48) | 0.36 (0.19; 0.67) | 11.53, 0.0031 * |
| - | p1–2 = 0.0108 | p1–3 = 0.0031 | ||
| Urea, mmol/L | 6.76 (4.35; 8.78) | 8.67 (5.73; 13.48) | 8.66 (4.62; 11.29) | 7.823, 0.0200 * |
| - | p1–2 = 0.0072 | - | ||
| Uric acid, nmol/mL | 76.8 (25.2; 161.2) | 124.5 (67.3; 193.6) | 93.0 (43.2; 182.9) | 9.346, 0.0093 * |
| - | p1–2 = 0.0028 | - | ||
| Sialic acids, mmol/L | 0.189 (0.134; 0.311) | 0.186 (0.131; 0.250) | 0.220 (0.140; 0.336) | 1.147, 0.5634 |
| Enzymes | ||||
| ALP, U/L | 60.84 (39.11; 84.75) | 70.62 (47.81; 99.96) | 73.88 (49.98; 109.74) | 5.128, 0.0770 |
| - | - | p1–3 = 0.0439 | ||
| LDH, U/L | 1008.0 (607.9; 1702.0) | 1471.0 (1121.0; 2098.0) | 1441.0 (864.8; 2028.0) | 11.30, 0.0035 * |
| - | p1–2 = 0.0026 | p1–3 = 0.0153 | ||
| GGT, U/L | 18.6 (15.4; 23.0) | 22.4 (19.7; 24.7) | 23.2 (17.9; 27.0) | 21.45, 0.0000 * |
| - | p1–2 = 0.0002 | p1–3 = 0.0001 | ||
| α-amylase, U/L | 178.8 (74.3; 382.9) | 316.1 (127.2; 503.7) | 315.1 (196.3; 519.5) | 8.345, 0.0154 * |
| - | - | p1–3 = 0.0100 | ||
| Catalase, mcat/L | 4.52 (3.60; 5.90) | 3.48 (2.34; 5.13) | 3.00 (2.18; 4.56) | 27.79, 0.0000 * |
| - | p1–2 = 0.0005 | p1–3 = 0.0000 | ||
| SOD, c.u. | 61.8 (36.8; 115.8) | 63.2 (28.9; 131.6) | 68.4 (26.3; 123.7) | 0.0521, 0.9743 |
| Lipoperoxidation Products and Endogenous Intoxication Rates | ||||
| Diene Conjugates, c.u. | 3.90 (3.74; 4.02) | 3.94 (3.78; 4.11) | 3.93 (3.76; 4.16) | 3.259, 0.1960 |
| Triene Conjugates, c.u. | 0.885 (0.816; 1.042) | 0.911 (0.810; 1.080) | 0.923 (0.837; 1.047) | 0.5642, 0.7542 |
| Schiff Bases, c.u. | 0.534 (0.494; 0.570) | 0.545 (0.497; 0.653) | 0.558 (0.513; 0.715) | 12.95, 0.0015 * |
| - | - | p1–3 = 0.0004 | ||
| MDA, nmol/mL | 6.92 (6.15; 9.06) | 6.84 (5.73; 8.72) | 6.58 (5.56; 7.78) | 2.716, 0.2572 |
| MM 280/254 nm | 0.778 (0.694; 0.878) | 0.918 (0.818; 1.036) | 0.895 (0.795; 0.990) | 30.09, 0.0000 * |
| - | p1–2 = 0.0000 | p1–3 = 0.0000 | ||
| Indicator | St I, n = 10 | St II, n = 16 | St III, n = 20 | St IV, n = 22 |
|---|---|---|---|---|
| Flow rate, mL/min | 0.80 (0.67; 0.95) | 0.78 (0.62; 0.98) | 0.81 (0.70; 1.01) | 0.76 (0.62; 0.93) |
| Electrolytes | ||||
| Calcium, mmol/L | 1.23 (1.00; 1.45) | 1.52 (1.31; 2.24) | 1.55 (1.20; 1.86) | 1.27 (0.79; 1.94) |
| - | p = 0.0160 | p = 0.0492 | - | |
| Phosphorus, mmol/L | 4.40 (3.80; 7.13) | 4.43 (3.61; 6.82) | 5.19 (3.83; 8.64) | 3.56 (1.92; 5.50) |
| - | - | p = 0.0394 | p = 0.0255 | |
| Ca/P | 0.238 (0.203; 0.276) | 0.375 (0.215; 0.550) | 0.275 (0.185; 0.568) | 0.441 (0.254; 0.861) |
| p = 0.0002 | p = 0.0001 | p = 0.0000 | p = 0.0191 | |
| Sodium, mmol/L | 14.9 (4.5; 16.1) | 8.6 (6.5; 11.9) | 7.1 (5.4; 10.9) | 10.0 (6.0; 14.0) |
| Potassium, mmol/L | 14.4 (10.5; 18.6) | 14.1 (8.5; 17.7) | 13.3 (9.0; 16.5) | 9.2 (4.0; 15.6) |
| p = 0.0376 | - | - | - | |
| Na/K | 0.810 (0.386; 1.198) | 0.668 (0.598; 0.917) | 0.733 (0.327; 0.840) | 1.282 (0.880; 1.984) |
| p = 0.0005 | p = 0.0000 | p = 0.0000 | p = 0.0000 | |
| Chlorides, mmol/L | 27.3 (16.2; 38.7) | 31.7 (27.3; 40.0) | 35.1 (25.8; 45.5) | 24.5 (20.7; 32.9) |
| - | p = 0.0004 | p = 0.0007 | - | |
| Protein Metabolism | ||||
| Protein, g/L | 0.68 (0.55; 1.32) | 0.87 (0.53; 1.63) | 0.94 (0.44; 1.71) | 0.84 (0.45; 1.31) |
| Albumin, g/L | 0.39 (0.10; 0.57) | 0.39 (0.22; 0.53) | 0.48 (0.25; 0.91) | 0.35 (0.15; 0.57) |
| - | p = 0.0353 | p = 0.0022 | - | |
| Urea, mmol/L | 8.71 (5.09; 13.39) | 9.21 (4.98; 13.94) | 7.90 (5.08; 10.30) | 8.02 (3.02; 10.22) |
| Uric acid, nmol/mL | 106.85 (41.83; 187.51) | 102.16 (56.23; 188.14) | 117.31 (35.31; 192.48) | 71.96 (40.00; 136.11) |
| Sialic acids, mmol/L | 0.244 (0.150; 0.375) | 0.317 (0.098; 0.336) | 0.269 (0.174; 0.391) | 0.165 (0.119; 0.229) |
| Enzymes | ||||
| ALP, U/L | 94.53 (33.68; 123.86) | 78.23 (64.10; 139.07) | 59.76 (47.81; 98.87) | 72.80 (54.33; 110.82) |
| LDH, U/L | 1094.8 (586.5; 1546.0) | 1903.5 (1226.5; 2316.5) | 1756.0 (938.7; 2179.0) | 1295.5 (702.9; 1928.0) |
| - | p = 0.0151 | p = 0.0248 | - | |
| GGT, U/L | 23.7 (22.9; 28.8) | 24.9 (17.3; 27.4) | 24.6 (17.1; 28.9) | 19.8 (17.9; 23.9) |
| p = 0.0105 | p = 0.0256 | p = 0.0052 | - | |
| α-amylase, U/L | 272.1 (183.2; 545.9) | 463.8 (279.2; 756.0) | 422.1 (265.7; 1073.0) | 261.6 (106.1; 399.2) |
| - | p = 0.0096 | p = 0.0164 | - | |
| Catalase, mcat/L | 2.35 (1.44; 2.69) | 3.00 (2.50; 4.73) | 3.45 (1.94; 4.61) | 3.03 (1.85; 6.39) |
| p = 0.0001 | p = 0.0040 | p = 0.0045 | p = 0.0178 | |
| SOD, c.u. | 55.3 (23.7; 182.9) | 71.1 (28.9; 150.0) | 65.8 (25.0; 128.9) | 67.1 (30.3; 88.2) |
| Lipoperoxidation Products and Endogenous Intoxication Rates | ||||
| Diene Conjugates, c.u. | 3.88 (3.53; 4.16) | 3.87 (3.64; 4.11) | 4.09 (3.85; 4.26) | 3.86 (3.75; 4.04) |
| - | - | p = 0.0091 | - | |
| Triene Conjugates, c.u. | 0.919 (0.760; 1.081) | 0.886 (0.837; 0.960) | 0.923 (0.825; 1.015) | 1.011 (0.882; 1.181) |
| - | - | - | p = 0.0093 | |
| Schiff Bases, c.u. | 0.556 (0.514; 0.621) | 0.546 (0.517; 0.680) | 0.554 (0.512; 0.691) | 0.699 (0.536; 0.907) |
| - | - | - | p = 0.0000 | |
| MDA, nmol/mL | 5.98 (5.21; 6.32) | 5.38 (5.13; 7.61) | 7.09 (6.15; 8.03) | 7.18 (6.41; 8.97) |
| p = 0.0175 | p = 0.0391 | - | - | |
| MM 280/254 nm | 0.930 (0.576; 1.045) | 0.839 (0.730; 0.957) | 0.937 (0.784; 1.039) | 0.901 (0.818; 0.924) |
| - | - | p = 0.0018 | p = 0.0022 | |
| Indicators | Kruskal–Wallis Test (H, p) | |
|---|---|---|
| Control Group, n = 114 | Comparison Group, n = 50 | |
| Electrolytes | ||
| Calcium, mmol/L | 8.728, 0.0683 | 4.776, 0.3111 |
| Phosphorus, mmol/L | 10.72, 0.0299 * | 10.41, 0.0341 * |
| Ca/P | 7.739, 0.1016 | 6.252, 0.1811 |
| Sodium, mmol/L | 5.818, 0.2140 | 2.588, 0.6289 |
| Potassium, mmol/L | 8.750, 0.0677 | 4.840, 0.3041 |
| Na/K | 13.19, 0.0104 * | 12.58, 0.0135 * |
| Chlorides, mmol/L | 20.41, 0.0004 * | 8.900, 0.0637 |
| Protein Metabolism | ||
| Protein, g/L | 3.459, 0.4842 | 1.726, 0.7859 |
| Albumin, g/L | 12.01, 0.0173 * | 3.487, 0.4799 |
| Urea, mmol/L | 4.783, 0.3016 | 1.164, 0.8063 |
| Uric acid, nmol/mL | 2.906, 0.5738 | 1.972, 0.7410 |
| Sialic acids, mmol/L | 6.715, 0.1518 | 7.317, 0.1201 |
| Enzymes | ||
| ALP, U/L | 5.660, 0.2260 | 1.582, 0.8121 |
| LDH, U/L | 10.19, 0.0374 * | 4.283, 0.3690 |
| GGT, U/L | 16.42, 0.0025 * | 6.669, 0.1545 |
| α-amylase, U/L | 12.41, 0.0145 * | 6.313, 0.1770 |
| Catalase, mcat/L | 28.05, 0.0000 * | 4.612, 0.3294 |
| SOD, c.u. | 0.2268, 0.9940 | 0.2448, 0.9931 |
| Lipoperoxidation Products and Endogenous Intoxication Rates | ||
| Diene Conjugates, c.u. | 7.406, 0.1159 | 6.668, 0.1545 |
| Triene Conjugates, c.u. | 8.209, 0.0842 | 7.210, 0.1252 |
| Schiff Bases, c.u. | 20.59, 0.0004 * | 6.190, 0.1854 |
| MDA, nmol/mL | 9.623, 0.0473 * | 7.446, 0.1141 |
| MM 280/254 nm | 17.65, 0.0014 * | 3.365, 0.4987 |
| Indicators | Category | HR (95% CI) | p-Value | OS, Months |
|---|---|---|---|---|
| MDA, nmol/mL | ˂7.34, n = 42 | 1 | 0.17155 | 25.4 |
| >7.34, n = 26 | 0.45 (0.15–1.37) | 36.8 | ||
| Na/K, c.u. | ˂1.09, n = 39 | 1 | 0.06187 | 44.0 |
| >1.09, n = 29 | 1.49 (0.51–4.36) | 15.5 | ||
| MDA + Na/K | Favorable prognosis, n = 16 | 1 | 0.01876 | 49.3 |
| Other combinations, n = 35 | 1.40 (0.33–5.86) | 38.1 | ||
| Unfavorable prognosis, n = 17 | 7.88 (1.10–54.62) * | 9.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bel’skaya, L.V.; Sarf, E.A.; Solomatin, D.V.; Kosenok, V.K. Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma. Diagnostics 2020, 10, 818. https://doi.org/10.3390/diagnostics10100818
Bel’skaya LV, Sarf EA, Solomatin DV, Kosenok VK. Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma. Diagnostics. 2020; 10(10):818. https://doi.org/10.3390/diagnostics10100818
Chicago/Turabian StyleBel’skaya, Lyudmila V., Elena A. Sarf, Denis V. Solomatin, and Victor K. Kosenok. 2020. "Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma" Diagnostics 10, no. 10: 818. https://doi.org/10.3390/diagnostics10100818
APA StyleBel’skaya, L. V., Sarf, E. A., Solomatin, D. V., & Kosenok, V. K. (2020). Diagnostic and Prognostic Value of Salivary Biochemical Markers in Oral Squamous Cell Carcinoma. Diagnostics, 10(10), 818. https://doi.org/10.3390/diagnostics10100818